Abstract
Background
A heat shock response model of Escherichia coli developed by Srivastava, Peterson, and Bentley (2001) has multiscale nature due to its species numbers and reaction rate constants varying over wide ranges. Applying the method of separation of time-scales and model reduction for stochastic reaction networks extended by Kang and Kurtz (2012), we approximate the chemical network in the heat shock response model.
Results
Scaling the species numbers and the rate constants by powers of the scaling parameter, we embed the model into a one-parameter family of models, each of which is a continuous-time Markov chain. Choosing an appropriate set of scaling exponents for the species numbers and for the rate constants satisfying balance conditions, the behavior of the full network in the time scales of interest is approximated by limiting models in three time scales. Due to the subset of species whose numbers are either approximated as constants or are averaged in terms of other species numbers, the limiting models are located on lower dimensional spaces than the full model and have a simpler structure than the full model does.
Conclusions
The goal of this paper is to illustrate how to apply the multiscale approximation method to the biological model with significant complexity. We applied the method to the heat shock response model involving 9 species and 18 reactions and derived simplified models in three time scales which capture the dynamics of the full model. Convergence of the scaled species numbers to their limit is obtained and errors between the scaled species numbers and their limit are estimated using the central limit theorem.
Keywords: Multiscale, Markov chains, Chemical reaction, Reaction networks, Heat shock
Background
Stochasticity may play an important role in biochemical systems. For example, stochasticity may be beneficial to give variability in gene expression, to produce population heterogeneity, and to adjust or respond to fluctuations in environment [1]. We are interested in local dynamics of biochemical networks involving some species with a small number of molecules so that the system is assumed to be well-mixed and relative fluctuations of small species numbers may play a role in the system dynamics.
The conventional stochastic model for the well-stirred biochemical network is based on the chemical master equation. The chemical master equation governs the evolution of the probability density of species numbers and is expressed as the balanced equation between influx and outflux of the probability density. When the biochemical network involves many species or bimolecular reactions, it is rarely possible to obtain an exact solution of the master equation in a closed form. Instead of searching for the solution of the master equation, stochastic simulation algorithms are used to obtain the temporal evolution of the species numbers. For example, Gillespie’s Stochastic Simulation Algorithm (SSA, or the direct method) is well known [2,3] and provides a realization of the exact trajectory of the sample path for the species numbers. As the biochemical network has more species and reactions, SSA becomes computationally expensive and more efficient algorithms were suggested by many authors [4-6]. The detailed review of stochastic simulation methods, stochastic approximations, and hybrid simulation methods is given in [7]. For models with well-separated time scales, numerous authors suggested stochastic simulation algorithms for biochemical reaction networks by assuming that “fast” subnetworks have reached a “partial equilibrium” [6] or a “quasi-steady state” [4]. Using these assumptions, the approximate stochastic simulation algorithms involve a reduced number of species or reactions.
On the other hand, Ball et al. [8] described the state of the biochemical reaction network in the well-stirred system directly using stochastic equations for species numbers, and suggested an approximation of the reaction network via limiting models derived using different scalings for the species numbers and for the reaction rate constants. Kang and Kurtz [9] extended this multiscale approximation method and gave a systematic way to obtain limiting models in the time scales of interest. Conditions are given to help identify appropriate values for a set of scaling exponents which determine the time scale of each species and reaction. Using this method, nonstationary behavior of biochemical systems can be analyzed. Moreover, application of the method is flexible in the sense that the method does not require the exact parameter values but gives approximations valid for a range of parameter values. More recently, Crude et al. [10] also proposed a reduction method to derive simplified models with preserving stochastic properties and with key parameters using averaging and hybrid simplification.
The multiscale approximation method in [9] requires consideration of magnitude of both species numbers and rate constants of the reactions involving the corresponding species. When a moderately fast reaction involves two species, one with a small number of molecules and the other with a large number of molecules, the effects of this reaction on these species are different. Net molecule changes of species with large numbers due to the reaction is less noticeable than those of species with small numbers. Therefore, though the same reaction governs these species, their time scales may be different from each other. Letting N0 be a fixed constant and choosing a large value for N0, for example N0=100, we express magnitudes of species numbers and reaction rate constants in terms of powers of N0 with different scaling exponents. For instance, 1 to 10molecules are expressed as to , 500 to 800molecules are rewritten as 5×N0 to 8×N0molecules, and 0.0002 sec becomes . Assuming N0 is large, we replace N0by a large parameter N and stochastic equations for species numbers are expressed in terms of N. Then, N is an analogue of 1/ε where ε is a small parameter in perturbation theory.
A specific time scale of interest is expressed in terms of a power of N, and its exponent contributes to reaction rates due to change of variables in time. For each species (or linear combination of species), we compare a power of N for the species number and those for reaction rates involving this species. Consider a case when the power for the species number is larger than those for the rates of all reactions where the species is involved. Then net molecule changes due to the reactions are not large enough to be noticeable in this time scale, and the species number is approximated as constant. Next, consider a case when the power for the species number is smaller than those for some reaction rates involving the species. In this case, the species number fluctuates very rapidly due to the fast reactions in this time scale, and the averaged behavior of the species number can be described in terms of other species numbers. The method of averaging is similar to approximation of one variable in terms of others using a quasi-steady state assumption. Last, when the power for the species number is equal to those for the rates of reactions where the species is involved, the scaled species number is approximated by a nondegenerate limit describing nonstationary behavior of the species number in the specific time scale of interest. The limit could be described in various kinds of variables: a continuous time Markov chain, a deterministic model given by a system of ordinary differential equations, or a hybrid model with both discrete and continuous variables. Since some of the scaled species numbers are approximated as constants or the averaged behavior of some species numbers is expressed in terms of other variables, dimension of species in the approximation of the biochemical network is reduced.
In the multiscale approximation method, scaling exponents for species numbers and for reaction rate constants are not uniquely determined, since the choice of values for the exponents is flexible. For example, 0.005 sec can be expressed as or when N0=100. The goal in this method is to find an appropriate set of scaling exponents to obtain a nondegenerate limit of the scaled species numbers. Orders of magnitude of species numbers in the propensities affect reaction rates, and reaction rates contribute to determining rates of net molecule changes of the species involved in the reactions. Since species numbers and reaction rates interact, it is not easy to determine scaling exponents for all species numbers and reaction rate constants so that the limits of the scaled species numbers become balanced.
Kang and Kurtz [9] introduced balance conditions for the scaling exponents, which help to determine values for a set of exponents. The key idea in these conditions is that for each species (or linear combination of species) the maximum of scaling exponents in the rates of the reactions where this species is produced should be the same as that in the rates of the reactions where this species is consumed, i.e. maximal production and consumption rates of the species should be balanced in the order of magnitude. In case the maximums of scaling exponents for productions and consumptions are not balanced for some species, an increase or decrease of the scaled species number can be described by its limit during a certain time period. However after this time period, the scaled species number will either become zero or blow up to infinity. Therefore, if some of the scaled species numbers are not balanced due to a difference between orders of magnitude of production and consumption rates, the chosen scaling is valid up to a certain time scale. After this time scale, we need to choose different values for scaling exponents. In each time scale of interest we derive a limiting model including a subset of species and reactions, which is used to approximate the state of the full reaction network. The multiscale approximation method is applicable in case some of reaction rates are not known accurately, since the chosen scaling is applicable in some ranges of the parameters. Therefore, based on the behavior of the limiting models, we may be able to estimate behavior for a range of parameter values without performing a huge number of stochastic simulations.
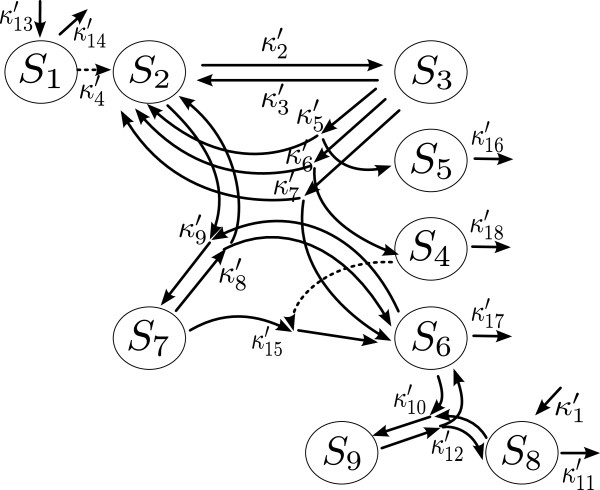
The paper [9] included several simple examples of biochemical networks involving two to four species, and derived limiting models in each time scale of interest. To apply this method, more scaling exponents must be determined as the biochemical network involves more species or reactions. Therefore, it is challenging to apply the method to complex biochemical systems and to determine appropriate values for scaling exponents so that the corresponding limiting models preserve important dynamical features of the full system. One of the goals of this paper is to illustrate how to apply this method to an example with significant complexity. In this paper, using a significantly complicated biochemical network, we derive limiting models, show convergence of the scaled species numbers to their limit, and estimate the error analytically between the scaled species numbers and their limit. We analyze a heat shock response model of Escherichia coli (E. coli) developed by Srivastava, Peterson, and Bentley in [11]. The model involves 9 species and 18 reactions with significant complexity as shown in Figure 1, and it has various time scales due to wide ranges of species numbers and reaction rate constants. Because of various scales involved, this model has been used as an example to show accuracy of the stochastic simulation algorithms which are developed to increase computational efficiency using the multiscale nature of the chemical reaction network [12,13]. Another version of a heat shock response model of E. coli is studied in [6] using an accelerated SSA that also exploits the multiscale nature of the system.
Figure 1.
A chemical reaction network in the heat shock response model of E. coli. A dotted line represents the effect of the species acting as catalysts. ’s represent stochastic reaction rate constants.
Applying the multiscale approximation method to the heat shock response model of E. coli, we derive limiting models in three time scales of our interests, which approximate the full network given in Figure 1. Denote ∅ as species we are not interested in. Let Si represent the ith species and S23be addition of species S2and S3. A→B denotes a reaction where one molecule of species A is converted to one molecule of species B. In the early stage of time period of order 1 sec, we obtain the following reduced network:
The reduced network in the early stage has very simple structure without any bimolecular reactions, and all reactions involved are either production from a source or conversion. Moreover, the reduced network is well separated into two due to independence of S8from S2and S3.
In the medium stage of time period of order 100 sec, the full network is reduced to
where a species over the arrow accelerates or inhibits the corresponding reaction. The reaction does not change this species number, but the propensity of the corresponding reaction is a function of this species number. In this time scale, conversion between S2 and S3 occurs very frequently and S2and S3play a role as a single “virtual” species rather than separate species. The species numbers of S23 and S8are described as two independent birth processes and the species number of S7 is governed by conversion. In this time scale, the species number of S8is normalized and treated as a continuous variable. The interesting thing is that the behavior of the species S8 which rapidly increases in time is well approximated in both first and second time scales.
In the late stage of time period of order 10,000 sec, we get a reduced network with more species involved than those in the previous time scales. However, the reduced network is still much simpler than the full network in Figure 1. At this time scale, we get
As we see in Figure 1, the full network involves reactions with more than two reactants or products. However, all reactions in the reduced network at the times of order 10,000 sec consist of either production or degradation of each species, though most of the species (6 species out of 9) are involved in the reduced model. As in the medium stage of time period, S2and S3play a role as a single species. In the early and medium stages of time period propensities are in a form following the law of mass action, while in the late stage of time period the propensity for degradation of S23 is a nonlinear function of the species numbers similar to the reaction rate appearing in the Michaelis-Menten approximation for an enzyme reaction. The nonlinear function involves the species numbers of S23, S8, and S9, which come from averaging of the species numbers of S2and S6which fluctuate rapidly in the third time scale. Similarly, the propensity of catalytic degradation of S8 is not proportional to the number of molecules of S8.
In the late stage of time period of order 10,000 sec, we study the error between the scaled species numbers and their limit analytically using the central limit theorem derived in [14] and show that the error is of order 10−1.
Methods
In the next several sections, we apply the multiscale approximation to the heat shock response model of E. coli and derive the limiting models. The multiscale approximation method is described in terms of the following steps so that the method can be applied to the general cases.
1. Write a chemical reaction network involving s0species and r0 reactions in the form of
where νik and are nonnegative integers. Rearrange the reactions so that the reaction rate constants are decreasing monotonically as k gets large.
2. Derive a system of stochastic equations for species numbers.
(a) Letting Xi(t) be the number of molecules of species Siat time t, the corresponding stochastic equation is
where counts the number of times that the kth reaction occurs up to time t.
(b) λk(x) is determined by a stochastic version of mass action kinetics, and is expressed as a product of the rate constant and the numbers of molecules of reactants. If the kth reaction is second-order () with different types of reactants, . When the reactants are two molecules of the same species, .
3. Derive a system of stochastic equations for the normalized species numbers after a time change, ZN,γ(t).
(a) In the equation for Xi(t) obtained in Step 2 (a), replace Xiby and divide reaction terms by Nαi. In the kth reaction term, put Nγ + ρk in the propensity and replace λk(X) by . Then, we have
(b) In the equation in Step 3 (a), .
(c) In the most reactions, is obtained by replacing by κkin λk. In case the kth reaction is second-order with reactants of the same species, is replaced by .
4. Write a set of species balance equations and their time-scale constraints.
(a) Define and as subsets of reactions where the species number of Siincreases or decreases every time the reaction occurs. Comparing ρk’s for and those for , set the balance equations as
(b) Time-scale constraints are given as
5. Find a minimum set of linear combinations of species whose maximum of collective production (or consumption) rates may be different from that of one of any species. We construct a minimum set of linear combinations of species by selecting a linear combination of species if any reaction term involving the species consisting of the linear combination is canceled in the equation for the linear combination of species.
6. For each selected linear combination of species, write a collective species balance equation and its time-scale constraint. They are obtained similarly to the ones in Step 4 using subsets of reactions where the number of molecules of linear combinations of species either increases or decreases instead of using and .
7. Select a large value for N0and choose an appropriate set of αi’s and βk’s so that
(a) the species number Xiand the reaction rate constant are approximately of orders and ;
(b) the normalized species number and the scaled reaction rate constant κkare of order 1;
(c) most of the balance equations obtained in Steps 4 and 6 are satisfied;
(d) βk’s are monotone decreasing among each class of reactions which have the same number of molecules of reactants.
8. Plugging the chosen values for αi’s and βk’s in the time-scale constraints obtained in Steps 4 and 6, compute an upper bound (denoted as γ0) for a time-scale exponent. Then, the chosen set of exponents αi’s and βk’s can be used for γsatisfying γ≤γ0. For γ>γ0, select another set of exponents αi’s and βk’s using Steps 7 and 8.
9. Using each set of values for αi’s and βk’s, identify a natural time scale exponent of each species (denoted as γi for species Si) so that γi satisfies
We collect γi’s with the same values, whose species are in the same time scales in the approximation.
10. Modify αi’s and βk’s so that the conditions in Step 7 are satisfied and that γi’s are divided into appropriate number of values, which gives the number of time scales, Nγ=Nγi, we are interested in.
11. For each chosen γ, derive a limiting equation for each species Siwith γi=γ. Using the stochastic equation obtained in Step 3 (a), we let N go to infinity.
(a) For , the kth reaction term converges to zero if αi>γ + ρk.
(b) If αi=γ + ρk, the kth reaction term appears as a limit in the limiting equation. The limit of the kth reaction term is discrete if αi=0, while it is a continuous variable with the limit of its propensity if αi>0.
(c) There is no k satisfying αi<γ + ρkin the equation for species Siwith γ=γidue to the definition of γigiven in Step 9.
12. In the limiting equation for each species Siwith γi=γ, we approximate propensities in the reaction terms. Suppose that the normalized species number for Sjappears in the propensities.
(a) If γj>γ, the limit of the normalized species number for Sjis its initial value.
(b) If γj=γ, the limit of the normalized species number for Sjappears as a variable in the propensities in the limiting equation.
(c) If γj<γ, the limit of the normalized species number for Sjis expressed as a function of the limits of the normalized species numbers for Siwith γi=γ. The function for Sjis obtained by dividing the equation for Sjby and letting N go to infinity.
13. If a limiting model is not closed, consider limiting equations for some linear combinations of species selected in Step 5 whose natural time scale exponents are equal to the chosen γ.
The method for multiscale approximation described above can be applied to general chemical reaction networks containing different scales in species numbers and reaction rate constants. We can apply the method in case the rates of chemical reactions are determined by law of mass action and when there is no species whose number is either zero or infinity at all times. As given in [9], in the reaction network involving ∅→S1, ∅→S2, ∅→S3, S1 + S2→∅, and S1 + S3→∅, convergence of the limit for the scaled species numbers may not be guaranteed at some time scales. Suppose that production rate of S1 is larger than that of S2but with the same order of magnitude, and that production rate of S3 is much smaller than those of S1and S2. Then, X1(t) may blow up to infinity and X2(t) may go to zero at some time scales. In this case, the method is not applicable.
Results and discussion
Model description
We analyze a heat shock response model of E. coli developed by Srivastava, Peterson, and Bentley [11]. The heat shock response model gives a simplified mechanism occurring in the E. coli to respond to high temperature. Heat causes unfolding, misfolding, or aggregation of proteins, and cells overcome the heat stress by producing heat shock proteins, which refold or degrade denatured proteins. In E. coli, σ32factors play an important role in recovery from the stress under the high temperature. σ32factors catalyze production of the heat shock proteins such as chaperon proteins and other proteases. In this model, J denotes a chaperon complex, FtsH represents a σ32-regulated stress protein, and GroEL is a σ32-mediated stress response protein.
σ32 factors are in three different forms, free σ32protein, σ32 combined with RNA polymerase (Eσ32), and σ32 combined with a chaperon complex (σ32-J). Under the normal situation without stress, most of the σ32 factors combine with chaperon complexes and form σ32-J. A chaperon complex J keeps σ32factors in an inactive form, and σ32factors can directly respond to the stress by changing into different forms. When there exist σ32factors combined with chaperon complexes, FtsH catalyzes degradation of σ32 factors. Thus, if enough σ32-regulated stress proteins are produced, σ32factors are degraded.
Not only σ32factors, but recombinant proteins also require chaperon complexes to form a complex so that denatured protein can be fixed. Therefore, σ32factors and recombinant proteins compete to bind chaperon complexes, and different levels of binding affinity of recombinant proteins to chaperon complexes change the evolution of the system state. In the model, we assume that σ32 factors and recombinant proteins have the same affinity to bind to chaperon complexes. The system is sensitive to the amount and forms of σ32 factors: a small decrease of σ32factors causes a large reduction of production of chaperon complexes and σ32-regulated stress proteins, and the ratio of three different forms of σ32factors determines system dynamics in the stress response [11]. The total initial number of molecules of σ32 factors in each cell is small [11] (also see initial values for S2, S3, and S7 which are 1, 1, and 7 in Table 1), and the stochastic model is appropriate to be considered.
Table 1.
Species in the heat shock response model of E. coli and their initial values
|
X1 |
= |
# of S1 |
σ32 mRNA |
X1(0) |
= |
10 |
|
X2 |
= |
# of S2 |
σ32 protein |
X2(0) |
= |
1 |
|
X3 |
= |
# of S3 |
Eσ32 |
X3(0) |
= |
1 |
|
X4 |
= |
# of S4 |
FtsH |
X4(0) |
= |
93 |
|
X5 |
= |
# of S5 |
GroEL |
X5(0) |
= |
172 |
|
X6 |
= |
# of S6 |
J |
X6(0) |
= |
54 |
|
X7 |
= |
# of S7 |
σ32-J |
X7(0) |
= |
7 |
|
X8 |
= |
# of S8 |
Recombinant protein |
X8(0) |
= |
50 |
| X9 | = | # of S9 | J-Recombinant protein | X9(0) | = | 0 |
The model involves 9 species and 18 reactions. Denote s0 as the number of species and r0 as the number of reactions. Let X(t) be a state vector whose ith component represents the number of molecules of species Si at time t for i=1,⋯,s0. Define a random process which counts the number of times that the kth reaction occurs by time t as
where λk(X) is the propensity of the kth reaction and the Yk’s are independent unit Poisson processes. Therefore, is a nonnegative integer-valued random process increasing by 1. As λk(·) gets large, the moment when increases becomes more frequent. Let νik() be the number of molecules of Si that are consumed (produced) in the kth reaction. Define νk() as an s0-dimensional vector whose ith component is νik(). Then, X(t) is given as
| (1) |
That is, species numbers at time t are expressed in terms of their initial values and sum of the number of times that each reaction occurs multiplied by net molecule changes in the corresponding reaction. In our model, the system of equations are derived using a set of reactions in Table 2 as:
Table 2.
Reactions in the heat shock response model of E. coli
| Reaction | Transition | |
|---|---|---|
| R1 |
|
Recombinant protein synthesis |
| R2 |
S2→S3 |
Holoenzyme association |
| R3 |
S3→S2 |
Holoenzyme disassociation |
| R4 |
|
σ32 translation |
| R5 |
|
GroEL synthesis |
| R6 |
|
FtsH synthesis |
| R7 |
|
J-production |
| R8 |
S7→S2 + S6 |
σ32-J-disassociation |
| R9 |
S2 + S6→S7 |
σ32-J-association |
| R10 |
S6 + S8→S9 |
Recombinant protein-J association |
| R11 |
S8→∅ |
Recombinant protein degradation |
| R12 |
S9→S6 + S8 |
Recombinant protein-J disassociation |
| R13 |
|
σ32 transcription |
| R14 |
S1→∅ |
σ32 mRNA decay |
| R15 |
|
σ32 degradation |
| R16 |
|
GroEL degradation |
| R17 |
|
J-disassociation |
| R18 | FtsH degradation |
In Reaction 5, 6, and 7, we assume that the number of molecules of each gene is 1 and that these reactions are effectively unimolecular. Similarly, Reactions 1 and 13 are treated as production from a source.
| (2) |
represents the stochastic reaction rate constant for the kth reaction, and their values from [11] are given in Table 3.
Table 3.
Stochastic reaction rate constants in the heat shock response model of E. coli
| Rates | Rates | ||
|---|---|---|---|
|
|
4.00×100 |
|
3.62×10−4 |
|
|
7.00×10−1 |
|
9.99×10−5 |
|
|
1.30×10−1 |
|
4.40×10−5 |
|
|
7.00×10−3 |
|
1.40×10−5 |
|
|
6.30×10−3 |
|
1.40×10−6 |
|
|
4.88×10−3 |
|
1.42×10−6 |
|
|
4.88×10−3 |
|
1.80×10−8 |
|
|
4.40×10−4 |
|
6.40×10−10 |
| 3.62×10−4 | 7.40×10−11 |
We convert deterministic rate constants in [11] using the volume of E. coli which is assumed to be 1.5×10−15L.
We derive the limiting models in three time scales, which approximate a full network in a certain time period involving a subset of species and reactions. In what follows, is a limit of the scaled species number of Si at some time scales depending on γ, and as γ gets larger the times are in the later stage. Note that the exponent γin does not imply (Zi)γ but it shows dependence of on γ. Let κk be a scaled reaction rate constant for the kth reaction. In the first time scale (when the times are in the early stage), the subnetwork governed by
| (3) |
approximates the network when the times are of order 1 sec. Denote as the limit of the addition of the scaled species numbers for S2and S3. In the second time scale (when the times are in the medium stage), the subnetwork governed by
| (4) |
approximates the network at the times of order 100 sec. In the third time scale, set the limit of the averaged scaled species numbers of fast-fluctuating species S2, S3, and S6 as
When the times are in a late stage, the subnetwork governed by
| (5) |
approximates the network at the times of order 10,000 sec. Detailed derivation is given in the later sections. Note that it is possible to identify different numbers of time scales depending on the scaling of the species numbers and reaction rate constants. In the heat shock response model of E. coli, it is possible to obtain approximate models with two or four time scales. However, if the number of time scales are too many, the limiting model in each time scale may involve one species and a few number of reactions and the model in this case may not be interesting to consider.
Derivation of the scaled models
The stochastic equations given in Equations (2) describe temporal evolution of the species numbers. For example, the equations for species S2and S3 are
| (6a) |
| (6b) |
In Equation (6), species numbers of S2and S3 are determined by the times when reactions occur and by the number of times that reactions happen. On the other hand, reaction time and frequency are determined by propensities which are some functions of species numbers. Therefore, reaction rates and species numbers interact one another. Reaction rates vary from O(10−11) to O(1) as we see in Table 3, and species numbers in this model are from O(1) to O(104) as we see later in the simulation of the full network. We express each species number and rate constant in terms of powers of a common number with different weights on exponents. Define N0=100 as a fixed unitless constant used to express the magnitude of the species numbers and the reaction rate constants. Define αi for i=1,⋯,s0 and βk for k=1,⋯,r0 as the scaling exponent for species Si and for the reaction rate constant . We express the reaction rate constants in a form of where κk is of order 1 and is determined so that . For example, we have and we can choose β6=−1 so that the reaction rate is expressed as . Assuming that N0 is large, we replace N0 by N and express the process as to show dependence of the species numbers on N. Note that {XN(t)} is a family of processes depending on N and when N=N0. Then, the equation for is given as
where is defined later so that when N=N0. Since the numbers of molecules of species are in different orders of magnitude, we scale the number of molecules of the ith species by Nαi and set a normalized species number as
The ith species number may have different orders of magnitude at different times so αi may have different values for different time scales. Now, we set the initial values as
| (7) |
so that and .
Next, we scale the propensities of reactions by replacing by and replacing by Nβkκk. For example, consider the 9th reaction term in (6a). Replacing by Nβ9κ9, by , and by , the 9th reaction term becomes
| (8) |
For simplicity, set ρ9=β9 + α2 + α6 and define a scaling exponent in the propensity of the kth reaction term as
where and . Here, νik gives the number of molecules of species Siconsumed in the kth reaction. Then, (8) is rewritten as
Dividing (6a) by Nα2 and (6b) by Nα3 and scaling the propensities, we get
| (9a) |
| (9b) |
For each reaction, ρkis given in terms of αiand βk in the Additional file 1: Table S1.
We are interested in dynamics of species numbers and in various stages of time period. In the early stage of time period, normalized species numbers of S2 and S3 are very close to their scaled initial values, since these species numbers have not changed yet. In the medium stage of time period, the normalized species numbers of S2and S3 are asymptotically equal to non-constant limits. In the late stage of time period, the normalized species numbers of S2 and S3fluctuate very rapidly and their averaged behavior is captured in terms of some function of other species numbers.
We want to express the time scale of each species in terms of power of N. First, we express order of magnitude of a specific time period of interest as a power of N with a time scale exponent γ. Applying a time change by replacing t by Nγtin , we define a variable for the normalized species numbers after a time change as
| (10) |
Then, gives a normalized species number at the times of order Nγ. A natural time scale of Siis the time when has a nonzero finite limit which is not constant and of order 1.
Changing a time variable by replacing t by Nγt in (9a) and (9b), the normalized species numbers of S2and S3after a time change satisfy
| (11a) |
| (11b) |
where Nγin each propensity comes from the change of the time variable. Here, the initial values may depend on γ, since we can choose different values for αifor each γdue to changes in order of magnitude of species numbers in time. The stochastic equations after scaling and a time change for all species are given in the Additional file 1: Section 1.
Balance conditions
Our goal is to approximate dynamics of the full network in the heat shock response model of E. coli in specific times of interest in terms of simplified subnetworks preserving significant biological features. In each time period of interest, we obtain a nondegenerate limiting model which is not equal to zero and does not blow up to infinity. In this section, we introduce balance conditions which help us to choose appropriate values for the scaling exponents αi’s and βk’s so that the limit is nonzero finite. For each time period of interest of order where N0=100, we choose values for scaling exponents so that orders of magnitude of the species number for Si and the kth reaction rate constant are about and , respectively. That is,
It is natural to choose βk’s in monotone decreasing manner in k, since ’s are in monotone decreasing order as shown in Table 3. In Table 3, the production rates from a source are the rates per second. The unimolecular reaction rates are the rates per molecule per second while the bimolecular reaction rates are the rates per a pair of molecules per second. Since the reaction rates are expressed in different units, we separate rate constants into three classes based on the number of reactants and assume that monotonicity of βk’s holds in each class of reactions. In other words, we choose βk’s so that
Next, in order to make the normalized specie number balanced, it is required that the rates of production and consumption of Sishould be in the same order of magnitude. If the order of magnitude of production rate is larger than that of consumption, the normalized species number asymptotically goes to infinity. In the opposite case, the normalized species number asymptotically becomes zero. Therefore, for each species Si, we set the balance equation for αi’s and βk’s so that the maximal exponent in the propensities of the reactions producing Si is equal to that in the propensities of the reactions consuming Si. For example, to obtain a balance equation for species S2, we compare the scaling exponents in propensities of reactions involving S2using (11a), and set the maximal exponents of production and consumption of S2 equal. Similarly, using (11b), we set the maximal exponents in the production rates and the consumption rates of S3 equal. Then, the balance equations for species S2and S3 are
| (12a) |
| (12b) |
If the maximal orders of magnitudes of production and consumption rates for S2 are different from each other, the species number of S2should be large enough so that a difference between production and consumption of Si is not noticeable. In other words if αi’s and βk’s do not satisfy (12a), α2should be at least as large as the scaling exponents located in all reaction terms in (11a) to prevent the limit becoming zero or blowing up to infinity. Similarly, in case (12b) is not satisfied, α3 should be at least as large as the scaling exponents located in the reaction terms in (11b) to prevent the limit becoming zero or blowing up to infinity.
| (13) |
Solving (13) for γ, we get the following time-scale constraints:
| (14a) |
| (14b) |
Inequalities in (14) mean that if maximal production and consumption rates are not balanced either for S2 or S3, the chosen set of values for scaling exponents can be used to approximate the dynamics of the full network up to times of order Nu2 or Nu3. For times later than those of order Nu2or Nu3, we need to choose another set of values for scaling exponents based on the balance equations. We call the balance equation and the time-scale constraint for each species as the species balance condition. If either (12a) or (??) is satisfied, we say that the species balance condition for S2 is satisfied.
Even though species balance conditions for S2and S3 are satisfied, the limit of the normalized species numbers for S2or S3 may become degenerate. Consider addition of species S2and S3 as a single virtual species, and compare the collective rates of production and consumption of this species. Recall that S23 denotes addition of species S2and S3. Since production of one species is canceled by consumption of the other species, maximal production rate of S23 may be different from that of S2or S3. Suppose that the maximal collective rates of production or consumption of S23 are slower than the maximal production or consumption rates of S2and S3. Also, suppose that the maximal collective rates of production and consumption of the complex have different orders of magnitude. Then, a limit of the normalized species number of S23can be zero or infinity, even though the species balance conditions for S2 and S3 are satisfied. Therefore, we need an additional condition to obtain balance between collective production and consumption rates for S23. To obtain a balance equation for S23, we unnormalize (11a) and (11b) by multiplying Nα2 and Nα3, respectively. Adding the unnormalized equations for species S2and S3 and dividing it by , we get
| (15) |
Comparing the maximal scaling exponents of production and consumption of S23 in (15), a balance equation for S23is given as
| (16) |
In case (16) is not satisfied, the order of magnitude of the species number for S23 should be larger than those of collective production and consumption rates so that a difference between production and consumption is not noticeable. This gives
| (17) |
Solving (??) for γ, we get
| (18) |
Similarly to the time-scale constraint in the species balance condition, (18) implies that if maximal collective production and consumption rates for S23are not balanced, our choice of values for scaling exponents are valid up to times of order Nu23.
We call (16) and (18) the collective species balance condition for S23, that is, either (16) or (18) must hold. The species balance conditions for all species and the collective species balance conditions for all positive linear combinations of species should be satisfied to obtain a nondegenerate limit of (Condition 3.2 in [9]). Condition 3.2 can be reduced by Lemma 3.4-3.8 and Remark 3.9 in [9]. A key idea is to find a minimum subset of linear combinations of species so that production of one species is canceled by consumption of the other species when we combine the species. In that case, maximal collective production rate of the linear combination of the species may be different from that of each species. Therefore, species balance conditions may not imply the collective species balance condition for the linear combination of the species. For example, a collective species balance condition for addition of S2 and S3 should be satisfied, since reactions producing S2or S3 may not increase the species number of S23. In Table 4, we choose linear combinations of species whose collective species balance conditions may not be satisfied by the species balance conditions. For other linear combinations of species, their collective species balance conditions are derived from the ones in Table 4. Satisfying all balance conditions in Table 4 guarantees satisfying balance conditions for all positive linear combination of species, and these conditions help to identify scaling exponents which give a nondegenerate limit of the normalized species numbers in the heat shock response model of E. coli. In most cases satisfying balance conditions gives nondegenerate limiting models in the times of interest, but we can still find counter examples as given in the last paragraph in the section for methods.
Table 4.
Balance equations and time-scale constraints for each species and for each collective species chosen
| Balance equations | Time-scale constraints | |
|---|---|---|
|
S1 |
ρ13=ρ14 |
|
|
S2 |
|
|
|
S3 |
|
|
|
S4 |
ρ6=ρ18 |
|
|
S5 |
ρ5=ρ16 |
|
|
S6 |
|
|
|
S7 |
|
|
|
S8 |
|
|
|
S9 |
ρ10=ρ12 |
|
|
S2 + S3 + S7 |
ρ4=ρ15 |
|
|
S2 + S3 |
|
|
|
S2 + S7 |
|
|
|
S6 + S7 + S9 |
ρ7=ρ17 |
|
|
S6 + S7 |
|
|
|
S6 + S9 |
|
|
| S8 + S9 | ρ1=ρ11 |
In each case, either the balance equation or the time-scale constraint must hold.
Based on species and collective species balance equations in Table 4, we choose appropriate values for αi’s and βk’s so that most of the balance equations are satisfied. If some of the balance equations are not satisfied, corresponding time-scale constraints give a range of γ where the chosen αi’s and βk’s are valid. The time-scale constraint, γ≤γ0, implies that the set of scaling exponents αi’s and βk’s chosen is appropriate only up to time whose order of magnitude is equal to Nγ0. For the times larger than O(Nγ0), we need to choose a different set of values for the scaling exponents, αi’s. Assuming that reaction rate constants do not change in time and that the species numbers vary in time, we in general use one set of βk’s for all time scales and may use several sets of αi’s. A large change of the species numbers in time requires different αi’s in different time scales. For the heat shock model we identify three different time scales as we will see in the section of limiting models in three time scales, and α1, α2, α3, α8, and α9 may depend on the time scale. α4, α5, α6, and α7 are the same for all time scales.
Before we determine scaling exponents for S1, S2, and S3, we run one realization of stochastic simulation to find ranges of the species numbers in time. Using initial values for species S1, S2, and S3, X1(0)=10 and X2(0)=X3(0)=1 as given in Table 1 and using N0=100, we set , , and with α1=1 and α2=α3=0 in the early stage of time period. Plugging in αi’s and βk’s in the balance equations for S2, S3, and S23, equality holds in (12a) and (12b) but not in (16). Therefore, (18) gives
Then, the first set of scaling exponents with α1=1 and α2=α3=0 is valid only when γ≤0. Next, based on the fact that X2(t)≈O(10) and X3(t)≈O(10) in the medium stage of time period, we choose α2=α3=0 for γ>0. At this stage of time period, we set with α1=0. Then, (12a) and (12b) are satisfied but not (16). The condition (18) gives γ≤1, and the second set of scaling exponents with α1=α2=α3=0 is valid when γ≤1. Finally, we set α1=0 and α2=α3=1 for γ>1 based on the fact that the numbers of molecules of S2and S3 grow in time and are of order 100. Then, (12a), (12b), and (16) are all satisfied, and the third set of scaling exponents with α1=0 and α2=α3=1 can be used for γ>1.
The three sets of values for the scaling exponents chosen are given in the Additional file 1: Table S4. With chosen values for the scaling exponents, we check whether each balance equation is satisfied and give a time-scale constraint in the Additional file 1: Table S6 in case the balance equation is not satisfied. Different choices of αi’s and βk’s from the ones in the Additional file 1: Table S4 give different limiting models. As long as the chosen values for αi’s and βk’s satisfy balance conditions, the limiting model will describe nontrivial behavior of the species numbers which are nonzero and finite in the specific time of interest.
Limiting models in three time scales
In the heat shock response model of E. coli, we identify a time scale of interest using the chosen set of scaling exponents and derive a limiting model which approximates dynamics of the full chemical reaction network. Each limiting model involves a subset of species and reactions, and gives features of the full network during the time interval of interest.
To identify a time scale involving a limiting model with interesting dynamics (nondegenerate), we first need to determine a natural time scale of each species. Recall that a natural time scale of species Si is the time period of order Nγi when is of order 1. The natural time scale exponent γifor species Si is rigorously determined by
| (19) |
where Γi + denotes the collection of reactions where the species number of Si increases every time the reaction occurs. Similarly, Γi− is the subset of reactions where the species number of Sidecreases every time the reaction occurs. In (19), the left-side term is the maximal order of magnitude of rates of reactions involving Siand the right-side term is the order of magnitude of the species number for Si. If times are earlier than those of order Nγi(γ<γi), fluctuations of species number of Si due to the reactions involving Siare not noticeable compared to magnitude of the species number of Si. Then, the species number of Si is approximated as its initial value. In the times of order Nγi(γ=γi), changes of species number of Si due to the reactions and the species number of Si are similar in magnitude and behavior of the species number of Siis described by its nondegenerate limit. If times are later than those of order Nγi(γ>γi), the species number of Si fluctuates very rapidly due to the reactions involving Si compared to the magnitude of the species number of Si. Then, the averaged behavior of the species number of Siis approximated by some function of other species numbers. Note that γi depends on αi’s and βk’s, and the time scale of the ith species may change if we use several sets of αi’s.
All values of αi’s and ρk’s for three scalings which are used to derive limiting models are given in the Additional file 1: Table S4. The equations for normalized species numbers and the equation for which are used later in this section are given in the Additional file 1: Section 1 and Section 2, respectively. When we derive limiting models in three time scales, boundedness of the normalized species numbers is required. For first two time scales, we define stopping times so that the normalized species numbers are bounded up to those times. For the last time scale, we proved stochastic boundedness of some normalized species numbers in a finite time interval. For more details, see Additional file 1: Section 5.
Consider a model with the first set of scaling exponents including α1=1 and α2=α3=0. Note that the first set of scaling exponents is valid when γ≤0 based on the time scale constraints given in Table 4. Substituting α2=0 and ρk’s for the first scaling to the equation for given in (11a), we have
| (20) |
When γ=γ2, the maximal scaling exponent in the propensities of all reaction terms in (20) should be equal to the scaling exponent for the species number of S2. Therefore, γ2satisfies
| (21) |
and we get γ2=0. Similarly, we get γ3=γ8=0.
Next, we plug α1=1 and ρk’s for the first scaling in the equation for and get
| (22) |
By comparing the maximal scaling exponent in the propensities of all reaction terms in (22) and the scaling exponent for the species number of S1, γ1 satisfies
| (23) |
and we get γ1=2. Similarly, we get γi>0 for i=4,5,6,7,9. Among all natural time scale exponents of species, we choose the smallest one, γ=0, and set t∼O(N0)=O(1) as the first time scale we are interested in. Since γ1>0, as N→∞. Similarly, for i=4,5,6,7,9 as N→∞. To sum up, in this time scale with γ=0, the species numbers of Si’s for i=1,4,5,6,7,9 change more slowly than other species numbers, and the species numbers with slow time scales are approximated as constant.
To derive the limiting equation for S2, we set γ=0 in (20). Since the 2nd, 3rd, and 4th reaction terms have propensities with N0=1 and the species number of S2 is of order 1, these reaction terms converge to nonzero limits in the limiting equation. On the other hand, the propensities of the 5th, 6th, 7th, 8th and 9th reaction terms are of order N−1 or N−2 which are smaller than the species number for S2of order 1. Therefore, these reaction terms converge to zero as N→∞at least in the finite time interval. In the 2nd and 3rd reaction terms in (20), and as N→∞ since γ2=γ3=0. Then, using as N→∞, the limit of satisfies
Similarly, we get a limiting model with , , and for γ=0 as given in (3).
Next, consider a model with the second set of scaling exponents including α1=α2=α3=0. Note that the second set of scaling exponents is valid when γ≤1 based on the time scale constraints given in Table 4. To determine the natural time scale of S6, substitute α6=0 and ρk’s for the second scaling in the equation for , and we have
| (24) |
Comparing the exponents inside and outside of the reaction terms in (24), γ6 satisfies
| (25) |
and we get γ6=1. Similarly, we get γ7=γ8=1, γi<1 for i=2,3, and γi>1 for i=1,4,5,9. We already get the temporal behavior of species numbers of S2, S3, and S8 through the limiting model when γ=0. Thus, we set t∼O(N1) as the second time scale we are interested in, and derive a limiting model for S6, S7, and S8 when γ=1. Note that species S8 is involved in the limiting models for both γ=0 and γ=1, since we use different sets of scaling exponents in these models. For i=1,4,5,9 as N→∞, since γi>1. Thus, in the 12th and 15th reaction terms in (24), and as N→∞. Since the propensities of the 8th, 9th, and 17th reaction terms in (24) are of order Nγ−2=N−1 for γ=1 and the species number of S6 is of order 1, these reaction terms go to zero as N→∞. In the 10th and 15th reaction terms in (24), , , and are asymptotically O(1) and converge to , , and as N→∞ since γ6=γ7=γ8=1.
Now, consider the asymptotic behavior of the 7th reaction term in (24) when γ=1. Since γ3<1, fluctuates very much, and there exists no functional limit as N→∞. However, still converges, which gives the averaged behavior of the normalized species number of S3. To get the limit of , we plug the second set of scaling exponents in the equation for and obtain
| (26) |
The law of large numbers of Poisson processes gives an asymptotic limit of the scaled reaction terms as
| (27) |
where the Yk’s are unit Poisson processes and αi>0. For example, the 2nd reaction term in (26) divided by N is approximated as
Dividing (26) by N and using the law of large numbers for Poisson processes, we get
| (28) |
as N→∞.
We introduce an auxiliary variable to make the limiting model closed and define
Plugging α2=α3=0 and ρk’s in the second scaling in the equation for , we get
| (29) |
Since where γ23 denotes a natural time scale exponent of S23, we compare the scaling exponents of N in the reaction terms in (29) and the scaling exponent of N outside the reaction terms. Then γ23satisfies
and we get γ23=1. Since these reaction terms have Nγ−2=N−1 in their propensities when γ=1, which is smaller than the species number for S23 of order 1, these reaction terms converge to zero as N→∞. Using , the limit of satisfies
Adding and subtracting terms in (28) and dividing the equation by −(κ2 + κ3), we get
as N→∞, and this is used to obtain the limit of the 7th reaction term in (24). Letting N→∞, the limiting equation for is given as
| (30) |
In (30), note that since X9(0)=0 as given in Table 1. Limiting equations for and can be derived similarly, and a limiting model with , , , and for γ=1 is given in (4).
Last, consider a model with the third scaling exponents with α1=0 and α2=α3=1. To derive a limiting equation for , we plug ρk’s and α2=α3=1 for the third scaling in the equation for and get
| (31) |
In (31), the 8th reaction term is asymptotically zero, since the term is of order N−1. Using the law of large numbers for Poisson processes in (27), the 4th and the 9th terms in (31) are asymptotically equal to
| (32) |
Since γ1=2, as N→∞. On the other hand, since γ2,γ6<2, both and in (32) fluctuate rapidly and we must identify the averaged limit. is also averaged, since γ3<2. We actually show convergence of the fast-fluctuating species numbers of S2 and S3 to some limits in the Additional file 1: Section 5.1. For any ε>0 and for any t such that ,
| (33) |
| (34) |
uniformly as N→∞.
On the other hand, since γ6<2, converges to a limit which gives averaged behavior of the normalized species number of S6. Using the equation for , we get
| (35) |
Dividing (35) by N2, using the law of large numbers for Poisson processes in (27), and using the stochastic boundedness of the propensities of the 8th, 9th, 15th, and 17th reaction terms in the finite time interval shown in the Additional file 1: Section 5.1, we get
| (36) |
as N→∞. Therefore, a difference between the 10th and 12th reaction terms is approximated in terms of
| (37) |
which converges to from (34). Therefore, we get
| (38) |
as N→∞.
Now, from the equations for and , we get
| (39a) |
| (39b) |
Using the law of large numbers of Poisson processes in (27), the reaction terms in (39a) and (39b) are asymptotically equal to
| (40a) |
| (40b) |
Using (40a), (40b), and (38), the limiting equations of (39a) and (39b) are given as
| (41) |
In (41), note that since X9(0)=0 as given in Table 1.
Since and balance conditions are satisfied, in the finite time interval. Since γ8=2,
| (42) |
Using (38) and (42), is averaged as
| (43) |
From (33) and (43), we get
| (44) |
For more details used in (43) and (44), see Lemma 1.5 and Theorem 2.1 in [15]. Finally, we get the limiting equation of as
Theorem 1
For γ=0, converges to the solution of (3) for . For γ=1, converges to the solution of (4) for . In (3), is a discrete process, while is a deterministic process in (4). For γ=2, converges to the solution of (5) for .
Conditional equilibrium distributions
In the previous section, we derived limiting models in three different time scales. Except for the subset of species in the limiting model, the remaining species are approximated as constants in the first time scale, since their natural time scale exponents (γi) are larger than γ=0, i.e., species with γi>γ=0 did not start to fluctuate at these times yet. In the second and third time scales, there are subsets of species whose natural time scale exponents are smaller than γ=1 and 2, respectively. Normalized species numbers with γi<γfluctuate very rapidly at these times and their averaged behavior is approximated in terms of other variables which converge to a nondegenerate limit. For those species, the normalized species numbers do not converge to a limit in a functional sense, but still we can find a limit in a probabilistic sense (i.e. convergence in distribution) and their distribution. Conditioned on the normalized species numbers which converge to a nondegenerate limit in the time scale of interest, we can find the conditional equilibrium (or the local averaging) distributions of species numbers whose natural time scale exponents are smaller than the time scale exponents of interests. Conditioning on the normalized species numbers which converge to a nondegenerate limit is similar to fixing slowly-moving variables and describing behavior of the fast-fluctuating variables in terms of slowly-moving variables treating them as constants. In the next remark, we give a conditional equilibrium distribution of the subset of species with natural time scale exponents smaller than γ=1 and γ=2.
Remark 2
For γ=1, for each t>0, converges in distribution to such that conditioned on has a binomial distribution with parameter
respectively, that is,
For γ=2, for each t>0, converges in distribution to where and are independent Poisson distributed random variables with parameters
and
The detailed method to compute conditional equilibrium distributions is given in Section 6 in [[9]].
Mean value of the random variable with a binomial distribution, B(n,p), is equal to np. Therefore, for γ=1, we treat as constant and get a limit of the averaged values for and as
Mean value of the random variable with a Poisson distribution, Pois(λ), is equal to λ, and we obtain a limit of the averaged values for and as the parameters given in Remark 2.
Simulation results
Recall that the normalized species numbers after a time change are defined as
Using the limiting models in the three time scales given in (3)-(5), we approximate the species numbers in the full model by unnormalizing the species numbers and applying time change backward as
using a real value N0=100 for the parameter. In Figures 2, 3, 4 and Figure 5(a)-(d), the panels located in the left column give mean and standard deviation from the mean of stochastic simulation for Xi(t) and the panels located in the right column give mean and standard deviation from the mean of simulation for using the limiting models. The mean and standard deviation of species numbers are computed from 3000 realizations of the sample path of the stochastic simulation.
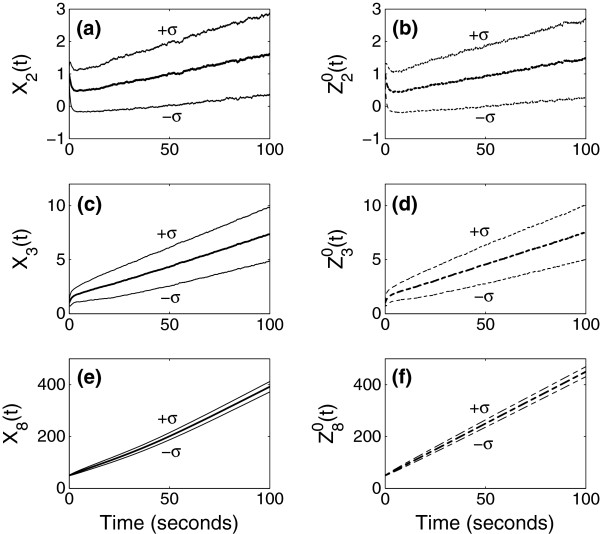
Figure 2.
Simulation results when γ = 0. Simulation of the full model (left) and that of approximation using the limiting model (right) when the time is of order (=1).
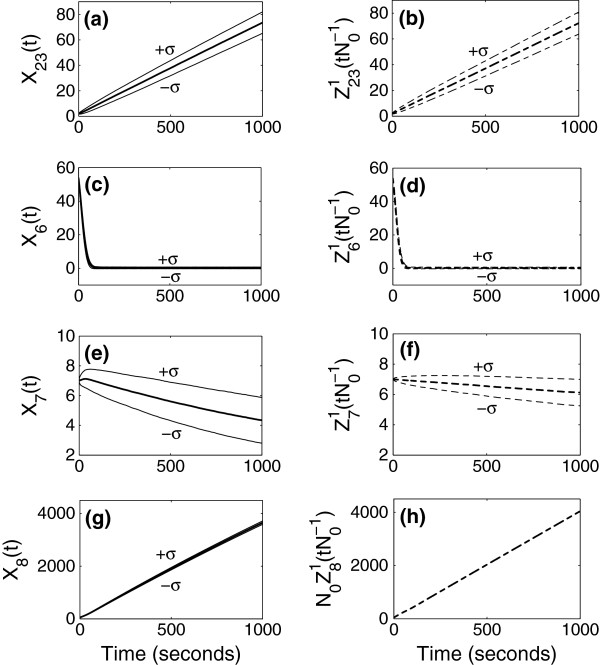
Figure 3.
Simulation results when γ = 1. Simulation of the full model (left) and that of approximation using the limiting model (right) when the time is of order N0(=100).
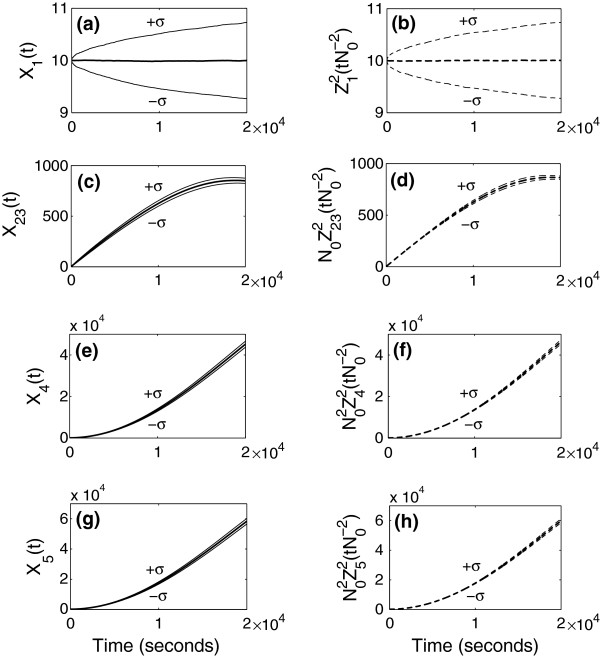
Figure 4.
Simulation results when γ = 2. Simulation of the full model (left) and that of approximation using the limiting model (right) when the time is of order (=10000).
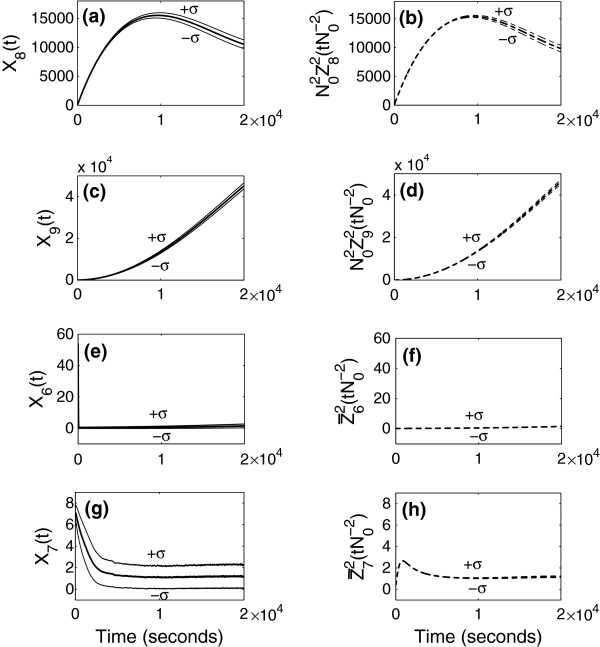
Figure 5.
Simulation results when γ = 2 (continued). Simulation of the full model (left) and that of approximation using the limiting model (right) when the time is of order . Figures (e), (f), (g), and (h) are simulation results for species 6 and 7. The graphs (f) and (h) give approximation of the averaged species numbers of S6and S7.
In Figure 2, we compare the simulation for the full model and for the approximation using the limiting model in the first scaling. The first scaling (γ=0) is for the times of order , and we look at the evolution of mean and standard deviation of the species numbers up to 100 sec. The full model and the limiting model for γ=0 are stochastic, and the limiting model approximates the evolution of statistics of the species numbers quite precisely. As shown in Figure 2(f) overestimates X8(t), since the limiting model does not include reactions consuming S8. Therefore, consumption of S8may not be captured well in the approximation.
In Figure 3, we compare the simulation for the full model and for the approximation using the limiting model in the second scaling. Since the second scaling (γ=1) is for the times of order , we observe the evolution of the species numbers up to 1000 sec. In this time scale, the evolution of S8shown in Figure 3(h) is approximated by a deterministic variable. The evolution of the species number of S8 in the full model given in Figure 3(g) is stochastic, but its standard deviation is very small. As in the previous time scale, slightly overestimates X8(t), since the limiting model does not include any consumptions of S8. The remaining three species, S23, S6, and S7are approximated by stochastic variables. The increasing species number of S23 in time and the rapid decrease in species number of S6are well captured by the limiting model. The species numbers of S7are described by stochastic variables both in the full model and in the limiting model. The behavior of S7in two models is not exactly the same, and discrepancy of the mean species numbers of S7 comes from the approximation of X4(t) in terms of its initial value. In the limiting model, S7is approximated as a stochastic process decreasing by 1 with the propensity proportional to . However, X4(t) increases during the times in [0,1000] sec in the full model, and this difference gives slower decreasing rate of the mean number of S7 in the limiting model than that in the full model.
In Figure 4 and Figure 5(a)-(d), we compare the simulation for the full model and for the approximation obtained from the limiting model in the third scaling. Since the third scaling (γ=2) is for the times of order , we look at the simulation up to 20,000 sec. In this time scale, the limiting model is stochastic. The species number of S1 in the limiting model is approximated by a stochastic discrete variable increasing and decreasing by 1, and the remaining species numbers in the limiting model satisfy stochastic equations driven by the stochastic discrete variable . As we have seen in the proof of Theorem 1 in the Additional file 1: Section 5.1, the processes for S1 in the full model and in the limiting model are exactly the same. Therefore, we use a same series of random numbers, when we simulate the full and limiting models. In Figure 4(b), the process for S1 is random, but its standard deviation is very small. Therefore, in one realization of simulation of the limiting model, behavior of S1appears as constant. Since all the remaining variables in the limiting model are governed by the variable for S1 and they satisfy the stochastic differential equations, evolution of one sample path of the species numbers for S23, S4, S5, S8, and S9 in the limiting model looks like a solution of the system of ordinary differential equations.
In Figure 5, (e)-(h) are the species numbers for S6and S7in the full model and their averaged values in the limiting model. Note that the species numbers for S6 and S7 do not appear in the limiting model, since their values are approximated in terms of other species numbers. Therefore, the difference between mean species numbers for S6and S7in the full model and those in the approximation does not affect the error directly. For γ=2, and are asymptotically averaged out by the variables in the limiting model as given in Remark 2. Since the averaged value for S6 plays an important role in the evolution of in the limiting model and since the averaged value for S7 gives the conditional mean value for S7 in the limiting model, we compare the species numbers of S6and S7 in the full model and the approximated averaged values in the limiting model. In Figure 5(f) and (h), we plot the mean and standard deviation from the mean for
in time. They are stochastic variables determined by the ones in the limiting model with very small fluctuations. Since and describe averaged behavior of S6and S7, X6(t) and X7(t) in Figure 5(e) and (g) have more fluctuations than the averaged species numbers in Figure 5(f) and (h).
In Figure 5(e)-(h) there is a discrepancy between the species numbers and their averaged values in the very early time, and the discrepancy comes from a disagreement in initial values of the species numbers in the full model and those of the averaged values in the limiting model. The integrated species numbers for S6 and S7 up to times of order 10,000 are supposed to be approximated by the integrated averaged values over the time interval, and the initial difference is due to a boundary layer phenomenon.
Error estimates
In the previous sections, we scaled species numbers and derived their limit to approximate temporal behavior of the species numbers in the full network. Among three limiting models given in (3)-(5), the first two are systems with discrete variables (except for ) which change by integer values. On the other hand, the last one is a hybrid system with both discrete and continuous variables. A discrete variable increases or decreases by one and stochasticity of all other variables comes from how much fluctuates. Since rarely changes at the times of our interest, the rest of the variables in (5) behaves such as a solution of systems of ordinary differential equations. Our choice of the scaling parameter value, N0=100, is not very large and it is possible that the limiting model does not contain enough fluctuations as much as the full network actually has due to our assumption that N0is replaced by a large parameter N.
In this section, we estimate an error between the normalized species numbers and their limit given in (5) at the times of 10,000 sec. Define
and denote U(t)=(U23(t),U4(t),U5(t),U8(t),U9(t))T as a limit of UN(t) as N goes to infinity. Note that we do not consider an error between and , since they are exactly the same processes. In the next remark, we show that UN(t) converges to U(t) in the probabilistic sense and thus the error between and is approximately of order . Since U(t) gives an explicit form of the error, we have better approximation of Xi(t) for γ=2 as
for S23, S4, S5, S8, and S9.
Remark 3
For γ=2, for each t>0, UN(t) converges in distribution to U(t) which is a solution of
where W(t) is a standard Brownian motion and
The detailed method to compute an error using the central limit theorem is derived in [14].
Estimating order of magnitude of an error is an analogue of that in van Kampen’s system size expansion [16]. A difference is that in the system size expansion, the system state representing the species numbers is scaled by the system size Ω and noise between the scaled process and its deterministic value is approximated as a random variable of order Ω−1/2. In our approach N is not a system size but a parameter for scaling, and species numbers are scaled by powers of N. Though the limiting model for γ=2 is not deterministic, it is still possible to estimate an error analytically due to the fact that which produces stochasticity in the limiting model is an exact process equal to . Another difference between our approach and van Kampen’s system size expansion is that a subset of species numbers is averaged in terms of other species numbers which appear in the limiting model for γ=2 due to the various scales involved.
Our estimates of the error is also different from diffusion approximations. In the diffusion approximations, the reaction terms centered by their propensities in the stochastic equations for discrete variables of species numbers are approximated in terms of time-changed Brownian motion. On the other hand, the noise term in the error estimates is determined by both the centered reaction terms in the equations for discrete variables and a difference between the discrete variables for the normalized species number and their continuous limit.
To find the asymptotic order of magnitude of , we show convergence of to a nonzero finite limit for some rN. Among the species S23, S4, S5, S8, and S9, the species number of S23is scaled with the smallest exponent, and thus noise in the limit of is determined dominantly by the component . Since is the species number scaled by N, we expect that rN=N1/2and the error between the scaled species numbers and their limit is of order . For a detailed approach to derive rNand U(t), see more about the central limit theorem in [14]. The fact that all components but the first one in the diffusion term in the equation for U(t) are zero supports the idea that noise is dominantly determined by the error between and . A sketch of the proof of Remark 3 is given in the Additional file 1: Section 6.
Conclusions
We considered a stochastic model for a well-stirred biochemical network with small numbers of molecules for some species. As the biochemical network consists of more species and reactions, network topology becomes more complex and it is harder to analyze. Therefore, how to reduce the biochemical network while preserving its important biochemical features is a very important issue.
In this paper, we applied the multiscale approximation method introduced by Ball et al. [8] and extended by Kang and Kurtz [9] to a heat shock response model of E. coli developed by Srivastava et al. [11]. Using the fact that the species numbers and the reaction rate constants in the model vary over several orders of magnitude, we scaled them using a scaling parameter with different exponents both of which contribute to determining the time scales of species. We derived balance conditions for each species and for a subset of linear combinations of species explicitly in this model, and chose appropriate values for the scaling exponents satisfying the balance conditions. Assuming that initial values of the species numbers are positive, satisfying the balance conditions is required to get a nondegenerate limiting model. We assumed that the reaction rate constants do not change in time, while we may use several sets of scaling exponents for the species numbers due to rapid changes in some species numbers in time. In this analysis, we chose three sets of scaling exponents, and they are used to derive limiting models in different time scales.
In each time scale we derived a limiting model, and used it to approximate the species numbers in the full network. In the limiting model, species numbers whose scaling exponents are larger than those of all rates of reactions involving the species are treated as constants, since changes of the species numbers due to the reactions are not noticeable at these times. When the scaling exponent of the species number is smaller than the scaling exponents of the rates of some productions and consumptions of the species and in case the scaling exponents for both kinds of reactions are equal, the scaled species number is averaged out and is approximated in terms of other variables. Therefore, the limiting model includes a subset of species and reactions and network topology in it becomes simpler. We derived the conditional equilibrium distributions of the fast-fluctuating species numbers and studied errors between the scaled species numbers and their limits in the third time scale.
Using the limiting models, we approximated the temporal evolution of species numbers in three time scales. By comparing stochastic simulation of the full model and approximations using the limiting models, we see that the main features of evolution of species numbers are well captured by the limiting models.
Competing interests
The author(s) declare that they have no competing interests.
Authors’ contributions
Based on the model of heat shock response of E. coli developed in [11], the author applied the multiscale approximation method introduced in [9] to the model. The author derived limiting models, showed convergence of the scaled species numbers to their limits, and estimated errors analytically. The author simulated the full network model and approximate processes using the limiting models and compared the results.
Supplementary Material
Supplementary material for “A multiscale approximation in a heat shock response model of E. coli.” This is a supplementary material of the paper including calculations and tables.
Acknowledgements
The author would like to greatly thank Thomas G. Kurtz for his continuous support and many helpful discussion. This work is an extension of the author’s Ph.D work at the University of Wisconsin, is proceeded while the author held a postdoctoral appointment under Hans G. Othmer at the University of Minnesota, and is completed while the author held a postdoctoral appointment in the Mathematical Biosciences Institute at the Ohio State University. The support provided by three appointments is acknowledged. This research has been supported in part by the National Science Foundation under grant DMS 05-53687, 08-05793, and 09-31642 and the Mathematical Biosciences Institute.
References
- Kærn M, Elston T, Blakem W, Collins J. Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet. 2005;6(6):451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- Gillespie D. A general method for numerically simulating the stochastic time evolution of coupled chemical reactions. J Comput Phys. 1976;22(4):403–434. [Google Scholar]
- Gillespie D. Exact stochastic simulation of coupled chemical reactions. J Phys Chem. 1977;81(25):2340–2361. [Google Scholar]
- Rao C, Arkin A. Stochastic chemical kinetics and the quasi-steady-state assumption: application to the Gillespie algorithm. J Chem Phys. 2003;118(11):4999–5010. [Google Scholar]
- Haseltine E, Rawlings J. Approximate simulation of coupled fast and slow reactions for stochastic chemical kinetics. J Chemi Phys. 2002;117(15):6959–6969. [Google Scholar]
- Cao Y, Gillespie D, Petzold L. Multiscale stochastic simulation algorithm with stochastic partial equilibrium assumption for chemically reacting systems. J Comput Phys. 2005;206(2):395–411. [Google Scholar]
- Pahle J. Biochemical simulations: stochastic, approximate stochastic and hybrid approaches. Briefings Bioinf. 2009;10(1):53–64. doi: 10.1093/bib/bbn050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball K, Kurtz T, Popovic L, Rempala G. Asymptotic analysis of multiscale approximations to reaction networks. Ann Appl Probability. 2006;16(4):1925–1961. [Google Scholar]
- Kang HW, Kurtz T. Separation of time-scales and model reduction for stochastic reaction networks. 2012. arXiv preprint arXiv:1011.1672, to appear in Annals of Applied Probability.
- Crudu A, Debussche A, Radulescu O. Hybrid stochastic simplifications for multiscale gene networks. BMC Syst Biol. 2009;3:89. doi: 10.1186/1752-0509-3-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R, Peterson M, Bentley W. Stochastic kinetic analysis of the Escherichia coli stress circuit using σ32-targeted antisense. Biotechnol Bioeng. 2001;75(1):120–129. doi: 10.1002/bit.1171. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Kaizu K, Hu B, Tomita M. A multi-algorithm, multi-timescale method for cell simulation. Bioinformatics. 2004;20(4):538–546. doi: 10.1093/bioinformatics/btg442. [DOI] [PubMed] [Google Scholar]
- Weinan E, Vanden-Eijnden E. Nested stochastic simulation algorithm for chemical kinetic systems with disparate rates. J Chem Phys. 2005;123(19):194107. doi: 10.1063/1.2109987. [DOI] [PubMed] [Google Scholar]
- Kang HW, Popovic L, Kurtz T. Central limit theorems and diffusion approximations for multiscale Markov chain models. 2012. arXiv preprint arXiv:1208.3783, submitted.
- Kurtz T. Averaging for martingale problems and stochastic approximation. Appl Stochastic Anal. 1992. pp. 186–209.
- Van Kampen NG. Stochastic processes in physics and chemistry (North-Holland Personal Library) Elsevier; 2007. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for “A multiscale approximation in a heat shock response model of E. coli.” This is a supplementary material of the paper including calculations and tables.