Abstract
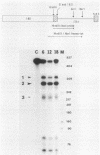
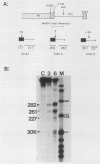
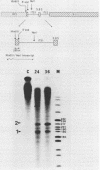
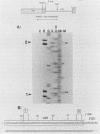
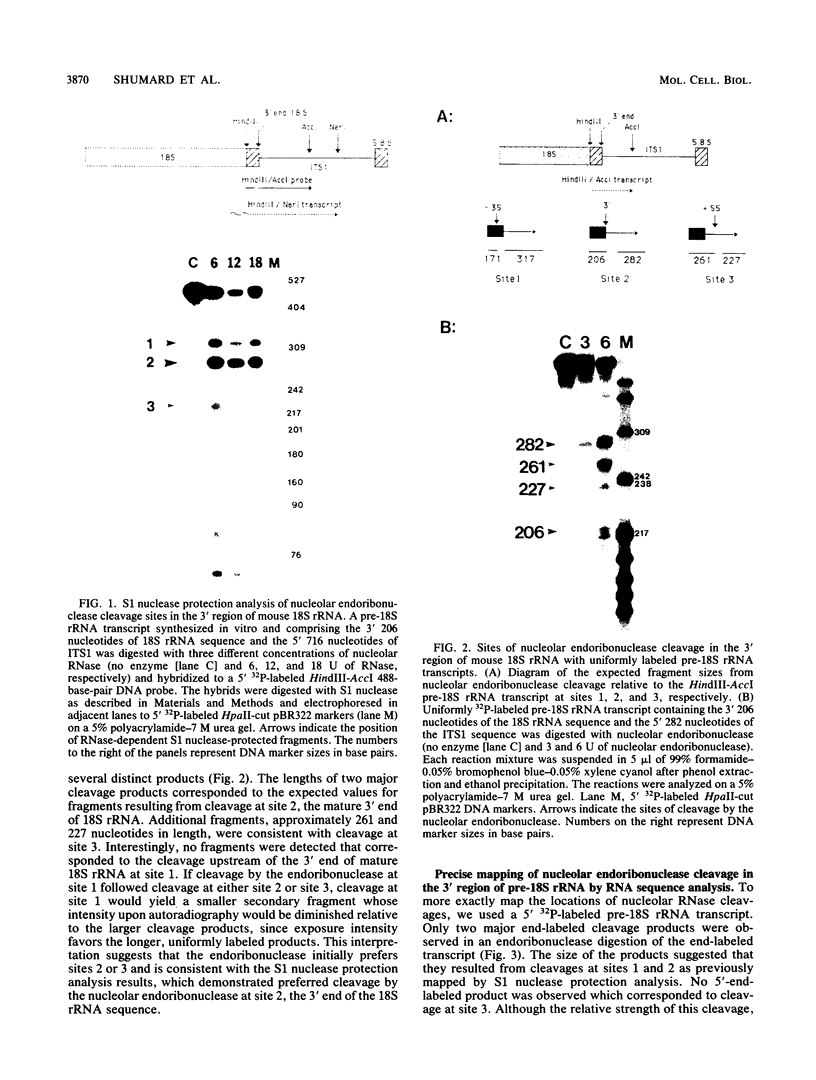
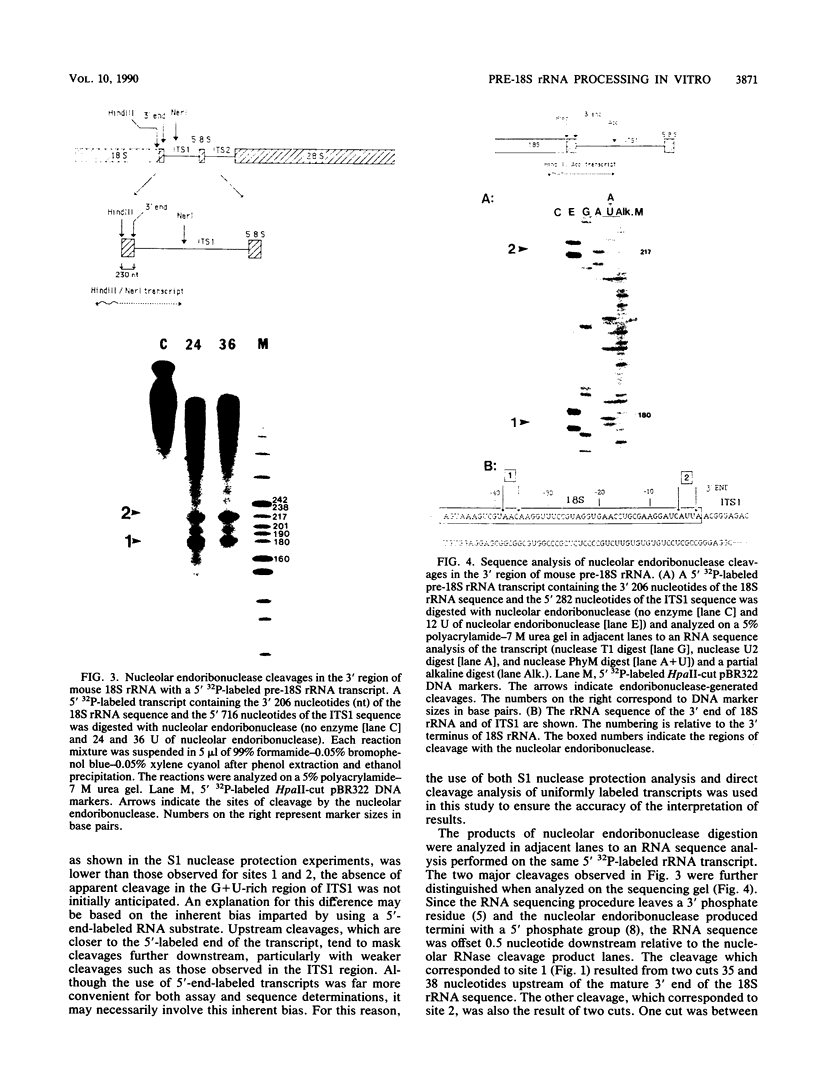
In an investigation of the possible involvement of a highly purified nucleolar endoribonuclease in processing of pre-rRNA at the 3' end of the 18S rRNA sequence, an in vitro synthesized pre-18S rRNA transcript containing the 3' end region of 18S rRNA and the 5' region of the first internal transcribed spacer (ITS1) was used as a substrate for the enzyme. Cleavages generated by the nucleolar RNase were localized by S1 nuclease protection analysis and by the direct release of labeled rRNA products. Precise determination of the specificity of cleavage was achieved by RNA sequence analysis with end-labeled rRNA transcripts. These data demonstrated that the purified nucleolar RNase cleaved the pre-18S rRNA transcript at three specific sites relative to the 3' region of 18S rRNA. The first two sites included the mature 3'-end 18S rRNA sequence and a site approximately 55 nucleotides downstream of the 3'-end 18S rRNA sequence, both of which corresponded directly to recent results (Raziuddin, R. D. Little, T. Labella, and D. Schlessinger, Mol. Cell. Biol. 9:1667-1671, 1989) obtained with transfected mouse rDNA in hamster cells. The other cleavage occurred approximately 35 nucleotides upstream from the mature 3' end in the 18S rRNA sequence. The results from this study mimic the results obtained from in vivo studies for processing in the 3' region of pre-18S rRNA, supporting the proposed involvement of this nucleolar endoribonuclease in rRNA maturation.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachellerie J. P., Michot B., Raynal F. Recognition signals for mouse pre-rRNA processing. A potential role for U3 nucleolar RNA. Mol Biol Rep. 1983 May;9(1-2):79–86. doi: 10.1007/BF00777477. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Bowman L. H., Goldman W. E., Goldberg G. I., Hebert M. B., Schlessinger D. Location of the initial cleavage sites in mouse pre-rRNA. Mol Cell Biol. 1983 Aug;3(8):1501–1510. doi: 10.1128/mcb.3.8.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman L. H., Rabin B., Schlessinger D. Multiple ribosomal RNA cleavage pathways in mammalian cells. Nucleic Acids Res. 1981 Oct 10;9(19):4951–4966. doi: 10.1093/nar/9.19.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudov K. P., Dabeva M. D., Hadjiolov A. A., Todorov B. N. Processing and migration of ribosomal ribonculeic acids in the nucleolus and nucleoplasm of rat liver nuclei. Biochem J. 1978 May 1;171(2):375–383. doi: 10.1042/bj1710375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler D. C., Eales S. J. Isolation and characterization of a single-stranded specific endoribonuclease from Ehrlich cell nucleoli. J Biol Chem. 1982 Dec 10;257(23):14384–14389. [PubMed] [Google Scholar]
- Eichler D. C., Eales S. J. Purification and properties of a novel nucleolar exoribonuclease capable of degrading both single-stranded and double-stranded RNA. Biochemistry. 1985 Jan 29;24(3):686–691. doi: 10.1021/bi00324a022. [DOI] [PubMed] [Google Scholar]
- Eichler D. C., Tatar T. F. Properties of a purified nucleolar ribonuclease from Ehrlich ascites carcinoma cells. Biochemistry. 1980 Jun 24;19(13):3016–3022. doi: 10.1021/bi00554a028. [DOI] [PubMed] [Google Scholar]
- Hadjiolov A. A., Milchev G. I. Synthesis and maturation of ribosomal ribonucleic acids in isolated HeLa cell nuclei. A tracer study on the topology of the 45S precursor of ribosomal ribonucleic acids. Biochem J. 1974 Aug;142(2):263–272. doi: 10.1042/bj1420263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjiolov A. A., Nikolaev N. Maturation of ribosomal ribonucleic acids and the biogenesis of ribosomes. Prog Biophys Mol Biol. 1976;31(2):95–144. doi: 10.1016/0079-6107(78)90006-8. [DOI] [PubMed] [Google Scholar]
- Hadjiolova K. V., Georgiev O. I., Nosikov V. V., Hadjiolov A. A. Localization and structure of endonuclease cleavage sites involved in the processing of the rat 32S precursor to ribosomal RNA. Biochem J. 1984 May 15;220(1):105–116. doi: 10.1042/bj2200105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjiolova K. V., Georgiev O. I., Nosikov V. V., Hadjiolov A. A. Mapping of the major early endonuclease cleavage site of the rat precursor to rRNA within the internal transcribed spacer sequence of rDNA. Biochim Biophys Acta. 1984 Jun 16;782(2):195–201. doi: 10.1016/0167-4781(84)90024-1. [DOI] [PubMed] [Google Scholar]
- Kass S., Craig N., Sollner-Webb B. Primary processing of mammalian rRNA involves two adjacent cleavages and is not species specific. Mol Cell Biol. 1987 Aug;7(8):2891–2898. doi: 10.1128/mcb.7.8.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasater L. S., Eichler D. C. Isolation and properties of a single-strand 5'----3' exoribonuclease from Ehrlich ascites tumor cell nucleoli. Biochemistry. 1984 Sep 11;23(19):4367–4373. doi: 10.1021/bi00314a019. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raziuddin, Little R. D., Labella T., Schlessinger D. Transcription and processing of RNA from mouse ribosomal DNA transfected into hamster cells. Mol Cell Biol. 1989 Apr;9(4):1667–1671. doi: 10.1128/mcb.9.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumard C. M., Eichler D. C. Ribosomal RNA processing. Limited cleavages of mouse preribosomal RNA by a nucleolar endoribonuclease include the early +650 processing site. J Biol Chem. 1988 Dec 25;263(36):19346–19352. [PubMed] [Google Scholar]
- Tiemeier D. C., Tilghman S. M., Leder P. Purification and cloning of a mouse ribosomal gene fragment in coliphage lambda. Gene. 1977;2(3-4):173–191. doi: 10.1016/0378-1119(77)90016-6. [DOI] [PubMed] [Google Scholar]