Abstract
Creutzfeldt-Jakob disease (CJD) is a rare transmissible neurodegenerative disorder. The etiology of sporadic form of CJD remains unsolved. In addition to the codon 129 polymorphism, polymorphisms in the non-coding region of PRNP are considered as important factors in sCJD development. To assess a possible association between PRNP 1368 SNP and sCJD, we compared the genotype, allele and haplotype frequencies of the 1368 SNP among 46 sCJD patients of Dutch origin with the respective frequencies in healthy controls. We detected a significant association between sCJD and 1368T/T genotype. A significant difference was also observed in 1368 alleles’ distribution. In the haplotype analysis, haplotype 1368C-129G was associated with decreased risk of sCJD in Dutch population. Our findings support the hypothesis that genetic variations in the regulatory region of the PRNP gene may influence the pathogenesis of sCJD.
Keywords: Creutzfeldt-Jakob disease, genetic susceptibility, polymorphisms, prion disease, prion protein gene
Introduction
Creutzfeldt-Jakob disease (CJD) belongs to transmissible spongiform encephalopathies (TSE) or prion diseases and occurs as an acquired, inherited (familial) or sporadic form. A central role in the pathogenesis of TSE is an accumulation of an abnormal isoform of prion protein (PrPSc) in the affected brains. PrPSc is derived through a posttranslational conformational conversion of the cellular prion protein isoform (PrPC). Mutations in the prion protein gene (PRNP) are linked to the occurrence of familial form of CJD (fCJD).1 The majority of CJD cases (more than 90%) occurs sporadically (sCJD) and the etiology of these cases remains obscure. A common polymorphism of codon 129 of PRNP gene was recognized as a risk factor. Homozygosity for methionine or valine at codon 129 increases the risk of sporadic and iatrogenic CJD and modifies the phenotype of fCJD.2-4 The genotype at codon 129 was demonstrated to influence the clinical and neuropathological phenotype of disease.5,6 Although the polymorphism in codon 129 was shown to be associated with TSE it cannot be the sole genetic factor predisposing to the disease. Therefore, subsequent studies have been focused on the existence of additional genetic factors located outside of the coding region of PRNP.
Polymorphisms in the promoter region of the prion protein gene were found to influence susceptibility to prion diseases in both cattle and mice.7-9 It was shown that incubation period of disease following scrapie challenge of transgenic mice expressing hamster PrP gene is shorter in animals with higher expression level of the transgene.10 Heterozygous null PrP mice demonstrated lower PrP expression level and had longer incubation time after infection with scrapie compared with wild type mice.11 Moreover, transgenic mice harboring high copy numbers of wild-type PrP transgenes developed a lethal neuromuscular disorder with necrotizing myopathy of skeletal muscle, a demyelinating polyneuropathy and vacuolation in the central nervous system. The incubation time of disease was dependent on the transgene dosage.10 Since animal studies have shown the relationship between the level of PrP gene expression and incubation time of disease, it is likely that humans with higher PrP expression levels are more susceptible to the disease development or may have shorter incubation period. For this reason genetic variations in the regulatory region of PRNP are considered as CJD susceptibility factors. In recent years, several SNPs were found in PRNP upstream of exon 1 and intronic regions (Fig. 1). Fifty-six polymorphic sites located within 25kb region flanking PRNP gene were found by Mead et al.12 An association between -101, +310, as well as +385 SNPs and sCJD was reported by McCormack and colleagues.13 A single nucleotide polymorphism 1368 upstream of PRNP exon 1 has been shown to be associated with sporadic CJD in British population. However, no difference in age at onset of disease or in disease duration between the three genotypes at SNP 1368 was noticed.12
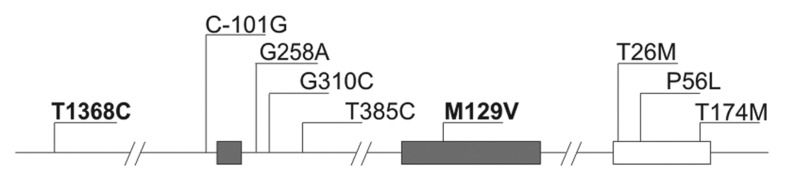
Figure 1. The polymorphisms in PRNP and PRND genes. PRNP exons are shown as gray boxes and PRND is shown as open box. Polymorphisms genotyped in the present study are indicated in bold.
The purpose of the current study was to investigate the allele, genotype, and haplotype frequencies of 1368 PRNP polymorphic position and codon 129 polymorphism among the Dutch sCJD patients and controls to determine if SNP 1368 modulates susceptibility to sCJD in the Dutch population.
Results
To assess the correlation between SNP 1368 of PRNP and susceptibility to sporadic form of CJD, we examined the genotype and allele frequencies of this polymorphism among 46 Dutch sCJD patients and 138 healthy controls. The genotype distribution of PRNP at codon 129 and position 1368 were in Hardy-Weinberg equilibrium (HWE) for both groups. As expected, an over-representation of 129Met/Met homozygosity was noticed among sCJD patients compared with controls (OR 3.411, 95% CI 1.512–7.758; p = 0.0013). As shown in Table 1, a significant association between sCJD and 1368T/T homozygosity (OR 0.381, 95% CI 0.177–0.820; p = 0.0183 for T/C vs. T/T 1368 genotype) was observed. The association of 1368T/T genotype with sCJD was also apparent when T/T vs. T/C-C/C (dominant model) genotypes frequencies were compared between groups (OR 2.472, 95% CI 1.165–5.252; p = 0.015). There were also significant differences in allele frequencies between patients and controls (OR 0.603, 95% CI 0.372–0.980; p = 0.0412 for C vs. T 1368 allele). These results imply that 1368T/T homozygosity acts as a sCJD risk factor whereas heterozygosity at this locus protects against sCJD. Further investigation of the association between various genotypes and sCJD revealed that double heterozygosity at 1368 and 129 has strong protective effect (OR 0.093, 95% CI 0.023–0.372; p = 0.0003). The frequency of 1368T/C-129A/G genotype was nearly six times higher in controls than in patients. The slight negative association with sCJD was found for genotypes 1368C/C-129A/G (OR 0.110, 95% CI 0.013–0.963; p = 0.033) and 1368T/C-129G/G (p = 0.036).
Table 1. Genotype and allele frequencies of the 1368 and 129 PRNP polymorphisms among sCJD patients and controls.
| |
sCJD n (%) |
Controls n (%) |
OR |
95% CI |
p value* |
|---|---|---|---|---|---|
| 1368 genotype | |||||
| TT |
21(45.6) |
35 (25.4) |
reference |
|
|
| TC |
16 (34.8) |
70 (50.7) |
0.381 |
0.177–0.820 |
0.018 |
| CC | 9 (19.6) | 33 (23.9) | 0.455 | 0.182–1.134 | 0.121 |
| 1368 allele | |||||
|---|---|---|---|---|---|
| T |
58 (63.0) |
140 (50.7) |
reference |
|
|
| C | 34 (37.0) | 136 (49.3) | 0.603 | 0.372–0.980 | 0.041 |
| 129 genotype | |||||
|---|---|---|---|---|---|
| AA |
24 (52.2) |
38 (27.5) |
3.411 |
1.512–7.758 |
0.0013 |
| AG |
15 (32.6) |
81 (58.7) |
reference |
|
|
| GG | 7 (15.2) | 19 (13.8) | 1.989 | 0.630–6.177 | 0.248 |
| 129 allele | |||||
|---|---|---|---|---|---|
| A |
63 (68.5) |
157 (56.9) |
reference |
|
|
| G | 29 (31.5) | 119 (43.1) | 1.647 | 0.971–2.802 | 0.051 |
| Genotype 1368–129 |
|||||
|---|---|---|---|---|---|
| TT-AA |
3 (6.5) |
8 (5.8) |
0.580 |
0.126–2.672 |
0.713 |
| TC-AA |
13 (28.2) |
11 (8) |
1.826 |
0.605–5.512 |
0.403 |
| CC-AA |
8 (17.4) |
19 (13.8) |
0.651 |
0.212–1.997 |
0.573 |
| TT-AG |
11 (24.0) |
17 (12.3) |
reference |
|
|
| TC-AG |
3 (6.5) |
50 (36.2) |
0.093 |
0.023–0.372 |
0.0003 |
| CC-AG |
1 (2.2) |
14 (10.1) |
0.110 |
0.013–0.963 |
0.033 |
| TT-GG |
7 (15.2) |
10(7.2) |
1.082 |
0.317–3.694 |
1.000 |
| TC-GG | 0 | 9 (6.5) | 0.036 |
Two-tailed Fisher’s exact test; OR, odds ratio; CI, confidence interval.
To investigate whether SNP 1368 and codon 129 SNP were in linkage disequilibrium, D’ and r2 were calculated among controls. A SNP 1368 was found to be in incomplete linkage disequilibrium with codon 129 polymorphism (D’ = 0.545; r2 = 0.171). The analysis of the haplotype frequencies in Dutch sCJD and controls revealed that four haplotypes were constructed by 1368 and codon 129 polymorphisms. Haplotype 1368C-129G was shown to be associated with decreased risk of sCJD in Dutch population (OR 0.09, 95% CI 0.01–0.72; p = 0.024) (Table 2).
Table 2. Haplotype frequencies of the 1368 and 129 PRNP polymorphisms among sCJD patients and controls.
| Haplotype |
Frequency in: |
OR (95% CI) |
p value |
|
|---|---|---|---|---|
| sCJD | controls | |||
| C- A |
0.358 |
0.384 |
1.000 |
|
| T-G |
0.303 |
0.323 |
0.93 (0.51–1.69) |
0.81 |
| T-A |
0.323 |
0.185 |
1.76 (0.97–3.19) |
0.065 |
| C-G | 0.012 | 0.109 | 0.09 (0.01–0.72) | 0.024 |
Discussion
The identification of genetic risk factors in sCJD is important for understanding the mechanisms of pathogenesis and susceptibility to disease. Association of codon 129 polymorphism with sCJD has been already well documented in many populations.2-6 Subsequent studies undertaken in the last years revealed a number of SNPs in upstream and intronic regions of PRNP and in the prion-like protein gene (PRND).13,14
In the current study, we detected a significant association of 1368T allele and T/T genotype with sCJD in the Dutch population. Homozygous 1368T/T genotype was nearly twice more common among patients than in controls (45.6 vs. 25.4%). We found that the presence of 1368T/C genotype significantly decreased the risk of sCJD development (Table 1). Additinally, protective effect of the C allele was noticed when the number of allele C carriers was compared with the number of T/T homozygous individuals (OR 0.405, 95% CI 0.190–0.858; p = 0.015). Results presented in this article are in inconsistency with some of the earlier studies performed accross other populations. PRNP T1368C polymorphism was found to be associated with sCJD in UK (p = 0.003) and among 129Met/Met homozygotes in German study (p = 0.049).12,15 However, no differences in genotype and allele frequencies between sCJD patients and controls were found in Korean populations.16 This discrepancy may be related to the differencies in the ethnic background of the investigated populations or to different demographic structure. It may be also explained by differences in the linkage disequilibrium for SNP 1368 and nearby locus involved in CJD across studied populations. It is especially interesting that our results are not concordant with an earlier Dutch study performed on 46 sCJD and 248 controls by Croes et al. who found no association between SNP 1368 and sCJD.17 The differences between both studies may be due to the relatively small sample sizes. Therefore, further studies on larger population should be undertaken to avoid random fluctuations.
Furthermore, our analysis of differences in distribution of double 1368 and codon129 genotypes revealed that TC-AG genotype acts as strong protective factor. The weaker effect of decreasing susceptibility to sCJD was shown for CC-AG and TC-GG genotypes (Table 1). These results in part confirm the previous study undertaken in British population by Mead et al. who found decreased frequency of TC-AG genotype in sCJD compared with controls (5.4% vs. 22.3%).12
Haplotype-based association analysis revealed that the distribution of C-G haplotype was significantly different between sCJD patients and controls. Our results are in contrast to the results reported by Jeong et al. who showed no association of C-G containing haplotype (ht6) with CJD in Korean population.16
The notion that prion protein is involved in development and intensity of oxidative stress and, thereby, contributes to the neurodegeneration is widely accepted. Contribution of prion protein in neurodegenerative disorders is also considered. However, while codon 129 polymorphism was found to be related with other neurological diseases,18,19 no association between SNP 1368 and occurrence of other neurodegenerative disorders was detected.20,21
Polymorphisms of non-coding region of prion protein gene could act through enhanced expression of PRNP causing increase of prion protein concentration and making PrP conversion more likely. Differences between studied groups may reflect the influence of additional factors such a PrPSc strain type, environmental factors, or coincidence of variations in other genes. Further investigations including larger cohorts study and functional analysis as well as identification of the other genes contributing to the disease process should be undertaken to solve a mechanism of susceptibility to prion disease.
Material and Methods
We studied 46 neuropathologically confirmed sCJD cases of Dutch origin and 138 control individuals with no history of neurodegenerative disorders. Sequencing of the entire PRNP open reading frame of all CJD patients was performed to exclude the cases with mutations in the PRNP gene.
Genomic DNA was extracted post mortem from brain tissue of CJD cases and from blood leukocytes of controls following standard procedures. PRNP 1368 genotyping was performed according to the procedure described elsewhere.16 Briefly, DNA was amplified using PCR with J-1 (GAGAAAACCTTGCGTCAGCA) and J-2 (AAGGTGCAGAAAAGATGGGC) primers to obtain a 586bp product. The PCR cycling conditions were 94°C for 2 min for initial denaturation followed by 35 cycles at 94°C for 45 s, 56°C for 45 s, 72°C for 1 min 30 s, and 1 cycle at 72°C for 10 min. Then PCR products were digested at 37°C for 1 h with five units of PvuII (Fermentas Life Sciences). Restriction products were visualized in UV light after electrophoresis in 1.5% agarose gel with ethidium bromide. Codon 129 polymorphism was determined during sequencing of PRNP open reading frame.
Genotype and alleles frequencies of PRNP 1368 position and codon 129 were tested for HWE using chi-square test. Allele and genotype differences between patients and controls were tested by Fisher exact test. Haplotype frequencies were calculated and compared between sCJD cases and controls using SNPStats program (http://bioinfo.iconcologia.net/SNPstats).22 Odds ratio (OR) and their corresponding 95% confidence intervals (CI) were calculated by logistic regression for the allele, genotype, and haplotype frequencies. To evaluate the non-random association of SNPs, pairwise linkage disequilibrium (LD) statistic D’ and correlation coefficient r2 were calculated using Haploview v4.2.23
Acknowledgments
The study was supported by Ministry of Scientific Research and Information Technology grant no. PBZ-KBN-124/P05/2004. We thank Piotr Kruszyński for his technical assistance in this project.
Glossary
Abbreviations:
- CJD
Creutzfeldt-Jakob disease
- PrPC
cellular isoform of prion protein
- PrPSc
disease associated isoform of prion protein
- PRNP
human prion protein gene
- PRND
prion-like protein gene
- TSE
transmissible spongiform encephalopathy
- SNP
single nucleotide polymorphism
Disclosure of Potential Conflicts of Interest
The authors certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.
Footnotes
Previously published online: www.landesbioscience.com/journals/prion/article/21773
References
- 1.Lloyd S, Mead S, Collinge J. Genetics of prion disease. Top Curr Chem. 2011;305:1–22. doi: 10.1007/128_2011_157. [DOI] [PubMed] [Google Scholar]
- 2.Palmer MS, Dryden AJ, Hughes JT, Collinge J. Homozygous prion protein genotype predisposes to sporadic Creutzfeldt-Jakob disease. Nature. 1991;352:340–2. doi: 10.1038/352340a0. [DOI] [PubMed] [Google Scholar]
- 3.Alperovitch A, Zerr I, Pocchiari M, Mitrova E, de Pedro Cuesta J, Hegyi I, et al. Codon 129 prion protein genotype and sporadic Creutzfeldt-Jakob disease. Lancet. 1999;353:1673–4. doi: 10.1016/S0140-6736(99)01342-2. [DOI] [PubMed] [Google Scholar]
- 4.Collinge J, Palmer MS, Dryden AJ. Genetic predisposition to iatrogenic Creutzfeldt-Jakob disease. Lancet. 1991;337:1441–2. doi: 10.1016/0140-6736(91)93128-V. [DOI] [PubMed] [Google Scholar]
- 5.Parchi P, Giese A, Capellari S, Brown P, Schulz-Schaeffer W, Windl O, et al. Classification of sporadic Creutzfeldt-Jakob disease based on molecular and phenotypic analysis of 300 subjects. Ann Neurol. 1999;46:224–33. doi: 10.1002/1531-8249(199908)46:2<224::AID-ANA12>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 6.Parchi P, Strammiello R, Giese A, Kretzschmar H. Phenotypic variability of sporadic human prion disease and its molecular basis: past, present, and future. Acta Neuropathol. 2011;121:91–112. doi: 10.1007/s00401-010-0779-6. [DOI] [PubMed] [Google Scholar]
- 7.Sander P, Hamann H, Pfeiffer I, Wemheuer W, Brenig B, Groschup MH, et al. Analysis of sequence variability of the bovine prion protein gene (PRNP) in German cattle breeds. Neurogenetics. 2004;5:19–25. doi: 10.1007/s10048-003-0171-y. [DOI] [PubMed] [Google Scholar]
- 8.Collinge J, Palmer MS, Sidle KC, Hill AF, Gowland I, Meads J, et al. Unaltered susceptibility to BSE in transgenic mice expressing human prion protein. Nature. 1995;378:779–83. doi: 10.1038/378779a0. [DOI] [PubMed] [Google Scholar]
- 9.Kashkevich K, Humeny A, Ziegler U, Groschup MH, Nicken P, Leeb T, et al. Functional relevance of DNA polymorphisms within the promoter region of the prion protein gene and their association to BSE infection. FASEB J. 2007;21:1547–55. doi: 10.1096/fj.06-7522com. [DOI] [PubMed] [Google Scholar]
- 10.Westaway D, Mirenda CA, Foster D, Zebarjadian Y, Scott M, Torchia M, et al. Paradoxical shortening of scrapie incubation times by expression of prion protein transgenes derived from long incubation period mice. Neuron. 1991;7:59–68. doi: 10.1016/0896-6273(91)90074-A. [DOI] [PubMed] [Google Scholar]
- 11.Büeler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, Aguet M, et al. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–47. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 12.Mead S, Mahal SP, Beck J, Campbell T, Farrall M, Fisher E, et al. Sporadic--but not variant--Creutzfeldt-Jakob disease is associated with polymorphisms upstream of PRNP exon 1. Am J Hum Genet. 2001;69:1225–35. doi: 10.1086/324710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCormack JE, Baybutt HN, Everington D, Will RG, Ironside JW, Manson JC. PRNP contains both intronic and upstream regulatory regions that may influence susceptibility to Creutzfeldt-Jakob Disease. Gene. 2002;288:139–46. doi: 10.1016/S0378-1119(02)00466-3. [DOI] [PubMed] [Google Scholar]
- 14.Peoc’h K, Guérin C, Brandel JP, Launay JM, Laplanche JL. First report of polymorphisms in the prion-like protein gene (PRND): implications for human prion diseases. Neurosci Lett. 2000;286:144–8. doi: 10.1016/S0304-3940(00)01100-9. [DOI] [PubMed] [Google Scholar]
- 15.Vollmert C, Windl O, Xiang W, Rosenberger A, Zerr I, Wichmann HE, et al. KORA group Significant association of a M129V independent polymorphism in the 5′ UTR of the PRNP gene with sporadic Creutzfeldt-Jakob disease in a large German case-control study. J Med Genet. 2006;43:e53. doi: 10.1136/jmg.2006.040931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeong BH, Lee KH, Lee YJ, Kim YH, Cho YS, Carp RI, et al. PRNP 1368 polymorphism is not associated with sporadic Creutzfeldt-Jakob disease in the Korean population. Eur J Neurol. 2008;15:846–50. doi: 10.1111/j.1468-1331.2008.02203.x. [DOI] [PubMed] [Google Scholar]
- 17.Croes EA, Alizadeh BZ, Bertoli-Avella AM, Rademaker T, Vergeer-Drop J, Dermaut B, et al. Polymorphisms in the prion protein gene and in the doppel gene increase susceptibility for Creutzfeldt-Jakob disease. Eur J Hum Genet. 2004;12:389–94. doi: 10.1038/sj.ejhg.5201161. [DOI] [PubMed] [Google Scholar]
- 18.Golanska E, Hulas-Bigoszewska K, Rutkiewicz E, Styczynska M, Peplonska B, Barcikowska M, et al. Polymorphisms within the prion (PrP) and prion-like protein (Doppel) genes in AD. Neurology. 2004;62:313–5. doi: 10.1212/01.WNL.0000103290.74549.DC. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Rowland LP, Mitsumoto H, Przedborski S, Bird TD, Schellenberg GD, et al. Prion protein codon 129 genotype prevalence is altered in primary progressive aphasia. Ann Neurol. 2005;58:858–64. doi: 10.1002/ana.20646. [DOI] [PubMed] [Google Scholar]
- 20.Rohrer JD, Mead S, Omar R, Poulter M, Warren JD, Collinge J, et al. Prion protein (PRNP) genotypes in frontotemporal lobar degeneration syndromes. Ann Neurol. 2006;60:616–, author reply 617. doi: 10.1002/ana.20931. [DOI] [PubMed] [Google Scholar]
- 21.Jeong BH, Lee KH, Lee YJ, Kim YJ, Choi EK, Kim YH, et al. Lack of association between PRNP 1368 polymorphism and Alzheimer’s disease or vascular dementia. BMC Med Genet. 2009;10:32. doi: 10.1186/1471-2350-10-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Solé X, Guinó E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–9. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 23.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]


