Abstract
All organisms have to safeguard the integrity of their genome to prevent malfunctioning and oncogenic transformation. Sophisticated DNA damage response mechanisms have evolved to detect and repair genomic lesions. With the emergence of live-cell microscopy of individual cells, we now begin to appreciate the complex spatiotemporal kinetics of the DNA damage response and can address the causes and consequences of the heterogeneity in the responses of genetically identical cells. Here, we highlight key discoveries where live-cell imaging has provided unprecedented insights into how cells respond to DNA double-strand breaks and discuss the main challenges and promises in using this technique.
Keywords: live-cell imaging, single cell, DNA damage, fluorescence microscopy, dynamics
INTRODUCTION
The integrity of our genome is continually challenged by damaging agents such as reactive byproducts of cellular metabolism, background radiation and environmental mutagens. Of the various types of DNA lesions that arise within the cell, DNA double-strand breaks (DSBs) are particularly dangerous as their improper repair can cause chromosomal rearrangements that promote oncogenic transformation [1, 2]. DSBs activate a complex DNA damage response (DDR) cascade, which includes recognition of the damage and initiation of repair, as well as amplification of the damage signal and its transmission to checkpoints that temporarily halt the cell cycle and allow additional time for repair [3]. In case full repair is not achieved, the DDR directs damaged cells towards permanent cell cycle arrest or apoptosis. A functional DDR is therefore vital for preserving genomic integrity and many features of this response are highly conserved from yeast to humans [4, 5].
Modern medicine has exploited the DDR for cancer treatment [6]. Frontline therapies, such as radiation and chemotherapy, function by generating high levels of DNA damage, often in the form of DSBs. Cancer cells are more susceptible to the adverse effects of DNA damage compared with healthy cells, as they frequently harbor mutations that compromise the DDR and cell cycle checkpoints. However, the effectiveness of anti-cancer therapies depends on the repair capacity of the targeted cells, highlighting the importance of understanding the mechanisms and dynamics of the DDR in healthy and transformed cells.
A characteristic outcome in cancer therapy is ‘fractional killing’, where each round of therapy kills some but not all cells in a tumor [7]. Traditionally, this was thought to result from differences in the cells’ accessibility and sensitivity to the drug, the involvement of cancer stem cells or from genetic heterogeneity between cells of the same tumor [8–10]. However, much recent work has shown a large cell-to-cell variability in phenotypes and behaviors of genetically identical cells even when they are grown in the same conditions and treated uniformly [11, 12]. Such cellular heterogeneity among isogenic cells was previously shown to have implications in numerous cellular processes including the activation of T cells [13], the timing of apoptosis [14] and the differentiation of stem cells [15]. Similarly, it is possible that the incomplete killing of tumors in response to DNA-damaging drugs results from differences in the DDR of individual tumor cells arising from stochastic fluctuations in gene expression and protein levels, differences in cellular states (such as cell cycle phase) or an altered microenvironment. In order to gain deeper insights into factors that regulate the DDR and to accurately predict the cellular response to DNA damage, it is therefore essential to observe these responses at the single-cell level.
Historically, DNA repair has been investigated using molecular and biochemical approaches that are based on measurements of populations of cells. For example, early measurements of the kinetics of DSB repair were obtained from physical estimates of DNA size using pulsed-field gel electrophoresis that pooled DNA from a large number of damaged cells [16]. Single-cell methods such as the comet assay allowed detection of the levels of DNA damage in individual cells; however, it was difficult to relate these measurements to the exact number of breaks in a cell [17]. In addition, both methods investigated DNA repair outside the normal physiological context (i.e. outside the nucleus of a cell). The subsequent discovery that DNA repair proteins and modified histones organize into macromolecular structures (foci) around break sites, which can be easily visualized by immunofluorescence and light microscopy, provided a breakthrough advance in our ability to observe the number and location of DSBs in situ, in single cells [18–21]. Such analyses have been instrumental in providing key conceptual advances regarding the identity and function of various factors involved in the DDR and DSB repair [22–25].
While the visualization of damaged-induced foci by immunofluorescence allowed quantification of distinct DSBs in the nucleus, the ‘kinetics’ of the repair process was inferred by averaging across populations of cells fixed at specific times after damage. Such measurements may not be representative of the true kinetics in single cells (Figure 1). Moreover, cell fixation precludes insight into the precise dynamics and order of events at the sites of DNA damage. Live-cell imaging has recently emerged as a powerful technique for visualizing and quantifying the DNA repair process over time in single cells. This technique has been elegantly utilized for understanding the spatiotemporal aspects of the DDR such as the mobility of repair proteins [26, 27] and their assembly into foci at damage sites [28–30], as well as the movements of damaged DNA and chromatin [31–39]. In the following sections, we describe these key areas in which live-cell imaging has provided unprecedented insights into the DDR and outline the main experimental techniques used for these analyses. We also discuss the limitations of using live-cell reporters for studying cellular responses and conclude with open questions that have the potential to be addressed using live-cell imaging methods.
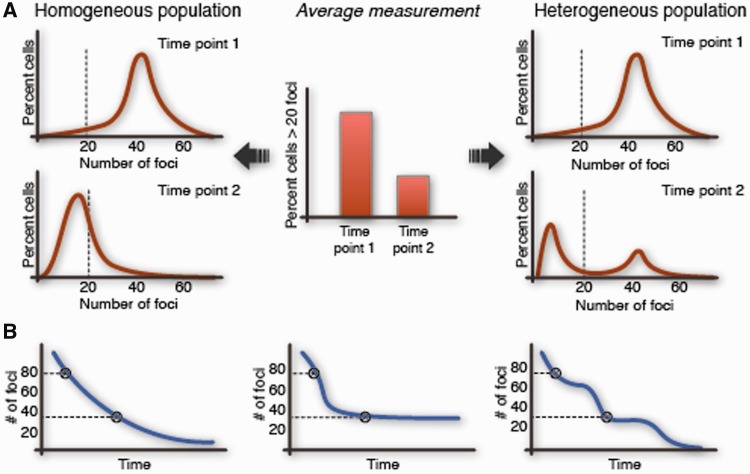
Figure 1:
Ensemble measurements can mask the variation in the kinetics of repair between cells. The kinetics of DNA DSB repair are popularly measured by averaging the number of foci in groups of fixed cells at specific times after damage. Such measurements do not capture the natural variation in the population and can mask the true kinetics of repair in single cells. (A) Left—a homogeneous population that repairs rather uniformly and right—a heterogeneous population that exhibits bimodality in the rates of repair; both yield the same average measurements at different times after damage. (B) The decay in the number of foci for three different cells is shown. Each of these cells has unique kinetics of repair, however, would yield the same number of foci if fixed at the indicated times.
INSIGHTS FROM SHORT-TERM LIVE-CELL IMAGING OF THE DDR
Imaging DNA repair, and more generally biological processes, in living cells has been revolutionized by the discovery of green fluorescent protein (GFP) and the subsequent development of its spectral variants [40–42]. These fluorescent proteins can be attached to cellular proteins by gene fusion and allow monitoring the levels and intracellular distributions of the tagged proteins by fluorescence microscopy at high temporal and spatial resolutions. Studies combining GFP-tagging of repair proteins with the induction of localized DSBs [43] and the use of dynamic imaging approaches, such as fluorescence recovery after photobleaching (FRAP, reviewed in [44–52]), fluorescence loss in photobleaching (FLIP; reviewed in [46, 49, 53]) and more recently in vivo fluorescence correlation spectroscopy (FCS; reviewed in [54–57]), have provided a wealth of knowledge about the spatiotemporal kinetics of repair proteins at DSBs.
FRAP is used to measure the diffusion and binding rates of fluorescently tagged proteins on short time-scales, typically seconds to minutes [58]. The fluorescence signal is irreversibly bleached in a small intracellular region of interest by exposure to a short pulse of high-intensity light (Figure 2A). The subsequent recovery of the fluorescence signal, which occurs by exchange of the bleached molecules with the unbleached fluorescent molecules from surrounding regions, is measured as a function of time. The rate at which the fluorescence recovers provides a measure of the mobility of the tagged protein, which is determined by its rate of diffusion and its in vivo binding interactions.
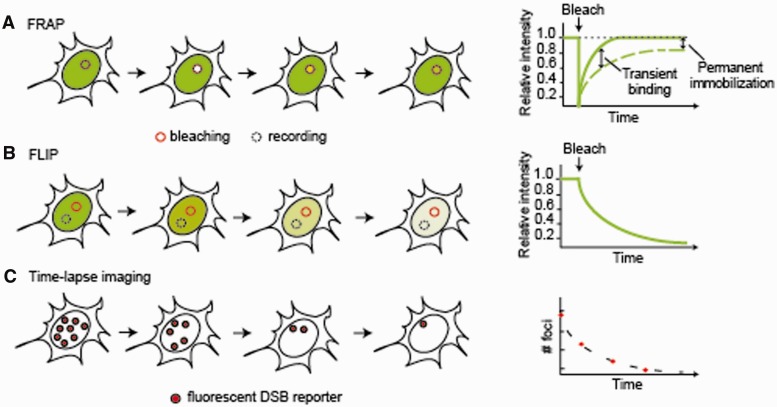
Figure 2:
Live-cell imaging techniques for studying DNA repair. (A) FRAP. A region of interest (solid circle) in the nucleus is photobleached with a strong laser pulse and the recovery of fluorescence intensity within the same region is monitored in time. The diffusion of the tagged protein and the kinetics of its binding interactions are measured from the rate of recovery (dashed line) in comparison to an inert protein of similar size (solid line). A delayed FRAP recovery indicates transient binding events and the fraction of recovery reflects the proportion of mobile molecules. In this example, ∼80% of tagged proteins in the bleached region are mobile. (B) FLIP. Bleaching within a region of interest (solid circle) is accompanied by measurement of fluorescence intensity in another region of the nucleus (dashed circle). The fluorescence decay in the recorded region indicates the extent of exchange between the two compartments. (C) Time-lapse microscopy. The expression and localization of the tagged proteins is monitored in time. In this example, the number of repair foci formed by the tagged protein is monitored to measure the rates of repair.
To assess if the mobility of the fusion protein is influenced by interactions with other proteins or cellular compartments, its FRAP recovery is compared with that of an inert, non-binding molecule, ideally of the same size. When the tagged protein has a slower FRAP recovery than the inert counterpart, transient binding is implicated and the degree of slowdown in the FRAP recovery provides a measure of binding kinetics. Furthermore, the proportion of the original fluorescence that is recovered over the measurement time indicates the mobile protein fraction in the bleached region. When the mobile fraction is less than 100%, some fluorescent molecules may be stably bound to a fixed substrate such as a DSB site. In FLIP, one region of the cell is photobleached, and the loss of fluorescence is recorded in another region of the cell (Figure 2B). Different species of the tagged protein migrate into the bleached region at different rates; hence, FLIP provides measurements of the steady state exchange of proteins between the two compartments as well as the binding status of the tagged protein and the relative proportions of its different states [59].
FRAP and FLIP have been instrumental in establishing that the cytologically stable repair foci are in fact highly dynamic structures, where many of the proteins involved are in constant dynamic exchange between the chromatin-bound fraction and the free nucleoplasmic pool. Different DDR factors are retained in foci for distinct periods of time depending on the availability of specific binding sites and interaction with other DDR factors. For example, the mediator protein Mdc1 is retained in foci at least 10 times longer than the sensor protein Nbs1 [29]. Mediator proteins such as Mdc1 and 53BP1 concentrate at the break site through a constant dynamic exchange between the bound and free fractions. In contrast, effector proteins such as the Chk2 kinase interact with the DSB site transiently and then disperse throughout the nucleus [28–30]. To determine the hierarchies in which different proteins are recruited to, and retained in foci, FRAP analysis was performed in cells where the expression of key DDR factors had been abrogated by specific small interfering RNAs [30]. Such studies provided insights into two key aspects of the DDR; first, that the dynamic exchange of factors at the break sites affords flexible access to the DNA ends and allows rapid adjustment of enzymatic activities at different stages of the repair. Second, they demonstrate how a localized damage signal generated by DSB is efficiently propagated throughout the nucleus by the disseminating effector proteins.
The most notable application of these techniques in advancing our knowledge of DNA repair came from the work of Essers et al. [26], in which FRAP was used to investigate how repair factors assemble at break sites. By GFP-tagging proteins of the Rad52 epistasis group that participate in the homologous recombination (HR) pathway, they elegantly demonstrated that these proteins are highly mobile and do not exist as pre-formed repair complexes in the free nucleoplasm. Rather, these proteins assemble into holocomplexes de novo at DSB sites. This allows repair proteins to rapidly diffuse through the entire nucleoplasm as individual molecules or small complexes and encounter aberrant DNA structures in a distributive fashion through random binding events. Through binding to their target sites, these proteins are transiently immobilized and organized into repair complexes before they again dissociate from the DSB and regain mobility. On-the-spot assembly of repair complexes allows rapid detection and easy access to break sites and greater flexibility in the composition of the repair complex as compared with large, pre-assembled holoenzymes. This is reminiscent of what has been observed for the nucleotide excision repair pathway that counteracts UV damage and appears to be a general phenomenon of DNA repair [60].
FCS complements FRAP and FLIP in detecting the mobility of proteins and their interactions in vivo. It measures fluctuations in photon number resulting from fluorescent particles diffusing in and out of a small sample volume (∼1 µm3) in the range of microseconds. These fluctuations reflect the average number of fluorescent molecules in the volume as well as the time of diffusion of each molecule across the measurement volume. FCS can also detect several diffusing species including the fraction of bound and unbound proteins, therefore providing both the concentration of the tagged protein and its diffusion and affinity constants [61]. Using FCS to study the behavior of GFP-tagged BRCA2 in living cells; Jeyasekharan et al. [27] demonstrated that these proteins exhibit restricted mobility under undamaged conditions. However, DNA damage triggers a redistribution of BRCA2 from slow moving forms to those with enhanced mobility and this change is accompanied by an increased binding between BRCA2 and Rad51.
INSIGHTS FROM LONG-TERM TIME-LAPSE IMAGING OF THE DDR IN SINGLE CELLS
Time-lapse imaging can capture the level and localization of fluorescently tagged proteins in live cells (Figure 2C). The duration of imaging may extend from a few milliseconds up to days, depending on the nature of the cellular process under observation. Additionally, multiple cellular proteins can be fused with distinct fluorescent tags and simultaneously monitored in the same cell.
Time-lapse imaging has been utilized for investigating the movement of broken DNA in a global chromatin context and to understand if efficient repair occurs at any location in the nucleus or is spatially restricted to selective nuclear compartments. By following two independent DNA lesions in yeast using fluorescent tagging near the break sites, it has been observed that DSBs are mobile and merge in to a common focus containing the HR repair machinery [31]. This suggests that in yeast, distinct dedicated repair centers exist as preferential sites for DSB repair and DSBs on separate chromosomes coalesce into these shared centers. Such merged repair centers may promote the tethering of broken DNA ends and facilitate the capture of homologous templates for HR repair. More recently, quantitative particle tracking of fluorescently tagged chromosomal loci was used to characterize the mobility of damaged DNA during the homology search process in the yeast nucleus. It was observed that homologous chromosomes reside far apart from each other and demonstrate limited movement in the undamaged nucleus [32]. Following DSB induction, the damaged chromosome moves in a ∼10-fold larger space to pair with its intact homologue. Increased mobility of the damaged chromosome requires the key resection and recombination enzymes Sae2, Rad51 and Rad54 as well as components of the DNA damage checkpoint—Mec1 (ATR in mammals) and Rad9 [32, 33]. In addition, the mobility of the damaged DNA is influenced by the nature of the DNA lesion. One-ended DSBs, which resemble collapsed replication forks and are tethered to their template for repair, demonstrate restricted movement compared with DSB ends that are not repaired or slowly repaired using templates other than the sister chromatid [33]. Furthermore, persistent DSBs in yeast migrate towards the nuclear periphery where they associate with the nuclear pore complex that might facilitate repair [34].
The picture emerging from observing the mobility of DSB ends in mammalian cells is more complicated. It has been reported that DSBs induced by γ-irradiation or restriction endonucleases are immobile and do not coalesce into shared repair centers [35]. These observations are consistent with findings from studies of global chromatin movement using fluorescently tagged histones which showed that in mammalian cells, chromatin regions damaged by irradiation undergo no or only limited motion in the nucleus [37]. These results have recently been challenged by experiments indicating that DSBs roam a 2- to 3-fold larger nuclear area compared with undamaged chromatin [36]. As in the case of yeast, the mobility of the damaged regions in mammalian cells may depend on the type of lesion, as for example breaks induced by topoisomerase poisons show increased mobility compared with DSBs induced by irradiation [36, 38]. DSBs induced by α-particles and deprotected telomeres have also been reported to show increased mobility [38, 39].
In a recent study from our group, we used long-term time-lapse imaging to investigate the cell-to-cell variability in the kinetics of DSB repair and the choice of repair pathway [62]. We found that heterogeneity in DSB repair is linked to the cell cycle states of individual cells. Asynchronously growing cells were engineered to simultaneously express fluorescent protein reporters for the total number of DSBs, HR and cell cycle. These cells were monitored for several hours after damage, and the rates of repair and proportion of HR were quantified for each cell. It was observed that the kinetics of DSB repair and the level of HR change with cell cycle stage (Figure 3). Specifically, cells damaged in mid-S attain the greatest proportions of HR and demonstrate the slowest repair. The balance between the alternate DSB repair pathways—non-homologous end-joining and HR—changes gradually as cells enter or exit S phase and maximal use of HR occurs at the peak of DNA replication. This suggests that in human cells, the extent of active replication, rather than the presence of a sister chromatid, influences the balance between the two repair pathways.
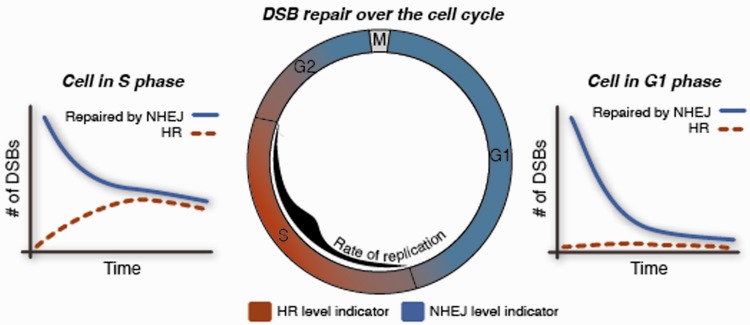
Figure 3:
The effect of cell cycle on the balance between alternate repair pathways. Cells that simultaneously expressed fluorescent protein reporters for DNA DSBs, HR and cell cycle were monitored by long-term time-lapse imaging. Analyses of the numbers of HR foci in single cells revealed that G1 cells repair exclusively by the non-homologous end-joining (NHEJ) pathway, while cells in S and G2 phase use both NHEJ and HR for repair. The level of HR changes gradually as cells enter or exit S phase and maximal use of HR occurs in mid-S at the peak of active DNA replication.
LIMITATIONS OF LIVE-CELL IMAGING
Although live-cell imaging methods present many advantages over their fixed cell counterparts, there are still important limitations to these techniques. Fluorescent reporter proteins are commonly generated by fusing the coding sequence for the fluorescent protein to either the N- or C-terminus of the protein of interest. Many proteins tolerate the addition of a fluorescent tag; however, control experiments are essential to confirm that the fusion protein faithfully represents the expression, sub-cellular localization and function of the endogenous protein. In addition, it is important to verify that the cellular process under study has not been perturbed by overexpressing one of its components. Ideally, the fluorescently tagged version of the protein replaces the endogenous one. This is best achieved by gene replacement, which preserves the chromatin context of the gene. Various strategies are available to integrate a fluorescent protein in the endogenous gene locus [63–66], and genome-scale libraries of fluorescently tagged strains are already available for both yeast and human cells [64, 67]. Alternately, the tagged protein could be introduced into a null background or the expression of the endogenous protein can be reduced using specific small interfering RNA or short hairpin RNA. In addition, some technical limitations also exist in performing live-cell imaging. Until recently, only up to three different fluorescent markers could be visualized simultaneously due to spectral limitations. Recent progress in identifying proteins that fluoresce in the near-infrared spectrum [68, 69] can now allow four channels to be monitored at the same time. Furthermore, computational analysis of time-lapse imaging data is still challenging. It requires sophisticated, automated data-processing procedures including cell segmentation, foci identification and single-particle tracking, which often need to be tailored to the specific experimental system [70].
PERSPECTIVES AND FUTURE DIRECTIONS
Live-cell imaging is rapidly evolving as a powerful technique for resolving the complex spatial and temporal interrelationships between various DDR factors that orchestrate the cellular response to DSBs. It allows an investigation of dynamical processes in ‘real time’ instead of giving a ‘snapshot’ of a cell’s current state. By observing cellular phenomena at the level of single cells, we can gain a unique perspective on biological processes that is distinct from the concepts gained by averaging data from populations of cells, which merges their diversity into a median trait and leads to loss of information.
At present, the main challenges in live-cell imaging include the time to construct the necessary biological tools and the expertise to analyze the image data. However, with the development of genome scale, tagged protein libraries and new computational tools, it is likely that live-cell imaging will be used more routinely and set the standard for measuring dynamic cellular processes at a high resolution. Furthermore, with current improvements in imaging technologies, it is becoming more feasible to measure individual cellular responses directly in vivo in the context of whole tissues and organisms.
Measurements using DNA damage and repair reporters in live cells will enable us to address additional long-standing questions in this field. For example, studies using reporters for different repair pathways in altered cell states can provide insights into their ability to compensate for each other under selective drug action and at different stages of the repair process. Similar analyses of different mediator and repair proteins will help determine the timing at which commitment to a specific repair pathway occurs and the factors leading to these decisions. Additionally, investigations combining reporters for repair and checkpoint regulators with indicators of cell fates will instruct us on how the rates and mechanisms of repair correlate with time through the checkpoints and with the execution of specific cellular outcomes. Such analyses afford an integrated, system-level understanding of the complex interrelationships between the myriad signaling and repair pathways that comprise the DDR.
Key points.
Measurements that average over a population of cells can mask the complexity and heterogeneity of biological processes in individual cells.
Live-cell imaging allows acquisition of quantitative dynamic information with high temporal and spatial resolution in individual cells.
Live-cell imaging has provided insights into the mobility of damaged DNA, the assembly of DNA repair complexes, and the cell-to-cell variability in the response to DNA double-strand breaks.
Current challenges in live-cell imaging include the construction and validation of fluorescent protein reporters and the need for sophisticated image analysis tools.
FUNDING
This work was supported by the National Institutes of Health [GM083303] and Hoffman-La Roche.
Acknowledgements
We thank all members of our laboratory for lively discussions and comments on this topic.
Biographies
Ketki Karanam is a PhD candidate at the Department of Systems Biology at Harvard Medical School (Boston, MA, USA). She uses quantitative live-cell imaging to study DNA double-strand break repair in single human cells.
Alexander Loewer is a group leader at the Max Delbruek Center (Berlin, Germany). His group uses live-cell time-lapse microscopy to understand signaling dynamics in single cells, focusing on the interplay of cellular networks.
Galit Lahav is an associate professor at the Department of Systems Biology at Harvard Medical School (Boston, MA, USA). Her group combines experimental and theoretical approaches to study the dynamics of signaling networks and cellular decision-making in single cells.
References
- 1.Lobrich M, Jeggo PA. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat Rev Cancer. 2007;7:861–9. doi: 10.1038/nrc2248. [DOI] [PubMed] [Google Scholar]
- 2.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–23. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 3.Su TT. Cellular responses to DNA damage: one signal, multiple choices. Ann Rev Genet. 2006;40:187–208. doi: 10.1146/annurev.genet.40.110405.090428. [DOI] [PubMed] [Google Scholar]
- 4.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–9. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 5.Harrison JC, Haber JE. Surviving the breakup: the DNA damage checkpoint. Ann Rev Genet. 2006;40:209–35. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- 6.Helleday T, Petermann E, Lundin C, et al. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 7.Berenbaum MC. In vivo determination of the fractional kill of human tumor cells by chemotherapeutic agents. Cancer Chemother Rep 1. 1972;56:563–71. [PubMed] [Google Scholar]
- 8.Chabner BA, Longo DL. Cancer Chemotherapy and Biotherapy: Principles and Practice. Philadelphia: Lippincott Williams & Wilkins; 2006. [Google Scholar]
- 9.Skeel RT. Handbook of Cancer Chemotherapy. Philadelphia: Lippincott Williams & Wilkins; 2003. [Google Scholar]
- 10.Reya T, Morrison SJ, Clarke MF, et al. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 11.Loewer A, Lahav G. We are all individuals: causes and consequences of non-genetic heterogeneity in mammalian cells. Curr Opin Genet Dev. 2011;21:753–8. doi: 10.1016/j.gde.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snijder B, Pelkmans L. Origins of regulated cell-to-cell variability. Nat. Rev. Mol Cell Biol. 2011;12:119–25. doi: 10.1038/nrm3044. [DOI] [PubMed] [Google Scholar]
- 13.Feinerman O, Veiga J, Dorfman JR, et al. Variability and robustness in T cell activation from regulated heterogeneity in protein levels. Science. 2008;321:1081–4. doi: 10.1126/science.1158013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spencer SL, Gaudet S, Albeck JG, et al. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459:428–32. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang HH, Hemberg M, Barahona M, et al. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–7. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iliakis GE, Cicilioni O, Metzger L. Measurement of DNA double-strand breaks in CHO cells at various stages of the cell cycle using pulsed field gel electrophoresis: calibration by means of 125I decay. Int J Radiation Biol. 1991;59:343–57. doi: 10.1080/09553009114550321. [DOI] [PubMed] [Google Scholar]
- 17.Shaposhnikov S, Frengen E, Collins AR. Increasing the resolution of the comet assay using fluorescent in situ hybridization–a review. Mutagenesis. 2009;24:383–9. doi: 10.1093/mutage/gep021. [DOI] [PubMed] [Google Scholar]
- 18.Maser RS, Monsen KJ, Nelms BE, et al. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol Cell Biol. 1997;17:6087–96. doi: 10.1128/mcb.17.10.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haaf T, Golub EI, Reddy G, et al. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc Natl Acad Sci USA. 1995;92:2298–302. doi: 10.1073/pnas.92.6.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scully R, Chen J, Ochs RL, et al. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 1997;90:425–35. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- 21.Lobrich M, Shibata A, Beucher A, et al. gammaH2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations and optimization. Cell Cycle. 2010;9:662–69. doi: 10.4161/cc.9.4.10764. [DOI] [PubMed] [Google Scholar]
- 22.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lisby M, Rothstein R. Choreography of recombination proteins during the DNA damage response. DNA Repair. 2009;8:1068–76. doi: 10.1016/j.dnarep.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25:409–33. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bekker-Jensen S, Mailand N. Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA Repair. 2010;9:1219–28. doi: 10.1016/j.dnarep.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 26.Essers J, Houtsmuller AB, van Veelen L, et al. Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J. 2002;21:2030–7. doi: 10.1093/emboj/21.8.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeyasekharan AD, Ayoub N, Mahen R, et al. DNA damage regulates the mobility of Brca2 within the nucleoplasm of living cells. Proc Natl Acad Sci USA. 2010;107:21937–42. doi: 10.1073/pnas.1009577107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lukas C, Falck J, Bartkova J, et al. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat Cell Biol. 2003;5:255–60. doi: 10.1038/ncb945. [DOI] [PubMed] [Google Scholar]
- 29.Lukas C, Melander F, Stucki M, et al. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J. 2004;23:2674–83. doi: 10.1038/sj.emboj.7600269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bekker-Jensen S, Lukas C, Melander F, et al. Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J Cell Biol. 2005;170:201–11. doi: 10.1083/jcb.200503043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lisby M, Mortensen UH, Rothstein R. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat Cell Biol. 2003;5:572–7. doi: 10.1038/ncb997. [DOI] [PubMed] [Google Scholar]
- 32.Mine-Hattab J, Rothstein R. Increased chromosome mobility facilitates homology search during recombination. Nat Cell Biol. 2012;14:510–7. doi: 10.1038/ncb2472. [DOI] [PubMed] [Google Scholar]
- 33.Dion V, Kalck V, Horigome C, et al. Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nat Cell Biol. 2012;14:502–9. doi: 10.1038/ncb2465. [DOI] [PubMed] [Google Scholar]
- 34.Nagai S, Dubrana K, Tsai-Pflugfelder M, et al. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soutoglou E, Dorn JF, Sengupta K, et al. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol. 2007;9:675–82. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krawczyk PM, Borovski T, Stap J, et al. Chromatin mobility is increased at sites of DNA double-strand breaks. J Cell Sci. 2012;125:2127–33. doi: 10.1242/jcs.089847. [DOI] [PubMed] [Google Scholar]
- 37.Kruhlak MJ, Celeste A, Dellaire G, et al. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol. 2006;172:823–34. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dimitrova N, Chen YC, Spector DL, et al. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456:524–8. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aten JA, Stap J, Krawczyk PM, et al. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science. 2004;303:92–5. doi: 10.1126/science.1088845. [DOI] [PubMed] [Google Scholar]
- 40.Tsien RY. The green fluorescent protein. Ann Rev Biochem. 1998;67:509–44. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 41.Patterson G, Davidson M, Manley S, et al. Superresolution imaging using single-molecule localization. Ann Rev Phys Chem. 2010;61:345–67. doi: 10.1146/annurev.physchem.012809.103444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stepanenko OV, Shcherbakova DM, Kuznetsova IM, et al. Modern fluorescent proteins: from chromophore formation to novel intracellular applications. BioTechniques. 2011;51:313–14. doi: 10.2144/000113765. 316, 318 passim. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagy Z, Soutoglou E. DNA repair: easy to visualize, difficult to elucidate. Trends Cell Biol. 2009;19:617–29. doi: 10.1016/j.tcb.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 44.Houtsmuller AB, Vermeulen W. Macromolecular dynamics in living cell nuclei revealed by fluorescence redistribution after photobleaching. Histochem Cell Biol. 2001;115:13–21. doi: 10.1007/s004180000234. [DOI] [PubMed] [Google Scholar]
- 45.Reits EA, Neefjes JJ. From fixed to FRAP: measuring protein mobility and activity in living cells. Nat Cell Biol. 2001;3:E145–7. doi: 10.1038/35078615. [DOI] [PubMed] [Google Scholar]
- 46.Dundr M, Misteli T. Measuring dynamics of nuclear proteins by photobleaching, Current protocols in cell biology/editorial board, Juan S. Bonifacino… [et al.] 2003 doi: 10.1002/0471143030.cb1305s18. Chapter 13: Unit 13 15. [DOI] [PubMed] [Google Scholar]
- 47.van Royen ME, Farla P, Mattern KA, et al. Fluorescence recovery after photobleaching (FRAP) to study nuclear protein dynamics in living cells. Methods Mol Biol. 2009;464:363–85. doi: 10.1007/978-1-60327-461-6_20. [DOI] [PubMed] [Google Scholar]
- 48.Mueller F, Mazza D, Stasevich TJ, et al. FRAP and kinetic modeling in the analysis of nuclear protein dynamics: what do we really know? Curr Opin Cell Biol. 2010;22:403–11. doi: 10.1016/j.ceb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lippincott-Schwartz J, Snapp E, Kenworthy A. Studying protein dynamics in living cells. Nat Rev Mol Cell Biol. 2001;2:444–56. doi: 10.1038/35073068. [DOI] [PubMed] [Google Scholar]
- 50.Sprague BL, McNally JG. FRAP analysis of binding: proper and fitting. Trends Cell Biol. 2005;15:84–91. doi: 10.1016/j.tcb.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 51.Carrero G, McDonald D, Crawford E, et al. Using FRAP and mathematical modeling to determine the in vivo kinetics of nuclear proteins. Methods. 2003;29:14–28. doi: 10.1016/s1046-2023(02)00288-8. [DOI] [PubMed] [Google Scholar]
- 52.Kimura H. Histone dynamics in living cells revealed by photobleaching. DNA Repair. 2005;4:939–50. doi: 10.1016/j.dnarep.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 53.Koster M, Frahm T, Hauser H. Nucleocytoplasmic shuttling revealed by FRAP and FLIP technologies. Curr Opin Biotechnol. 2005;16:28–34. doi: 10.1016/j.copbio.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 54.Maiti S, Haupts U, Webb WW. Fluorescence correlation spectroscopy: diagnostics for sparse molecules. Proc Natl Acad Sci USA. 1997;94:11753–7. doi: 10.1073/pnas.94.22.11753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Craenenbroeck E, Engelborghs Y. Fluorescence correlation spectroscopy: molecular recognition at the single molecule level. JMR. 2000;13:93–100. doi: 10.1002/(SICI)1099-1352(200003/04)13:2<93::AID-JMR492>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 56.Pramanik A. Ligand-receptor interactions in live cells by fluorescence correlation spectroscopy. Curr Pharmaceutical Biotechnol. 2004;5:205–12. doi: 10.2174/1389201043377002. [DOI] [PubMed] [Google Scholar]
- 57.Bacia K, Schwille P. A dynamic view of cellular processes by in vivo fluorescence auto- and cross-correlation spectroscopy. Methods. 2003;29:74–85. doi: 10.1016/s1046-2023(02)00291-8. [DOI] [PubMed] [Google Scholar]
- 58.Wachsmuth M, Caudron-Herger M, Rippe K. Genome organization: balancing stability and plasticity. Biochimica et Biophysica Acta. 2008;1783:2061–79. doi: 10.1016/j.bbamcr.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 59.Cole NB, Smith CL, Sciaky N, et al. Diffusional mobility of golgi proteins in membranes of living cells. Science. 1996;273:797–801. doi: 10.1126/science.273.5276.797. [DOI] [PubMed] [Google Scholar]
- 60.Gillet LC, Scharer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem Rev. 2006;106:253–76. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 61.Michelman-Ribeiro A, Mazza D, Rosales T, et al. Direct measurement of association and dissociation rates of DNA binding in live cells by fluorescence correlation spectroscopy. Biophys J. 2009;97:337–46. doi: 10.1016/j.bpj.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karanam K, Kafri R, Loewer A, et al. Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase. Mol Cell. 2012;47:320–9. doi: 10.1016/j.molcel.2012.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poser I, Sarov M, Hutchins JR, et al. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat Methods. 2008;5:409–15. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sigal A, Milo R, Cohen A, et al. Dynamic proteomics in individual human cells uncovers widespread cell-cycle dependence of nuclear proteins. Nat Methods. 2006;3:525–31. doi: 10.1038/nmeth892. [DOI] [PubMed] [Google Scholar]
- 65.Issaeva I, Cohen AA, Eden E, et al. Generation of double-labeled reporter cell lines for studying co-dynamics of endogenous proteins in individual human cells. PloS One. 2010;5:e13524. doi: 10.1371/journal.pone.0013524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li T, Huang S, Zhao X, et al. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 2011;39:6315–25. doi: 10.1093/nar/gkr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Howson R, Huh WK, Ghaemmaghami S, et al. Construction, verification and experimental use of two epitope-tagged collections of budding yeast strains. Compar Funct Genom. 2005;6:2–16. doi: 10.1002/cfg.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Filonov GS, Piatkevich KD, Ting LM, et al. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat Biotechnol. 2011;29:757–61. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shu X, Royant A, Lin MZ, et al. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science. 2009;324:804–07. doi: 10.1126/science.1168683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walter T, Shattuck DW, Baldock R, et al. Visualization of image data from cells to organisms. Nat Methods. 2010;7:S26–41. doi: 10.1038/nmeth.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]