Abstract
Nanostructure-initiator mass spectrometry (NIMS) is a recently developed matrix-free laser desorption/ionization technique that has shown promise for peptide analyses. It is also useful in mass spectrometric imaging (MSI) studies of small molecule drugs, metabolites, and lipids, minimizing analyte diffusion caused by matrix application. In this study, NIMS and matrix-assisted laser desorption/ionization (MALDI) MSI of a crustacean model organism Cancer borealis brain were compared. MALDI was found to perform better than NIMS in these neuropeptide imaging experiments. Twelve neuropeptides were identified in MALDI MSI experiments whereas none were identified in NIMS MSI experiments. In addition, lipid profiles were compared using each ionization method. Both techniques provided similar lipid profiles in the m/z range 700 – 900.
Introduction
Neuropeptides represent a diverse class of signaling molecules that are challenging to analyze in neuronal tissues due to their low abundance and chemical complexity. Mass spectrometric imaging (MSI) is an emerging bioanalytical technique that facilitates direct measurement of many analytes’ locations in a tissue section within one acquisition. Research has demonstrated that MALDI MSI can be successfully applied to studies of neuropeptides in a variety of neural tissues.1–7 In these experiments, MALDI matrix is dissolved in a solvent and applied to tissue as a fine spray. The analytes from the tissue are extracted into a thin matrix layer, enabling the MALDI process. The application of a wet matrix to tissue can cause analyte migration, especially for more hydrophilic neuropeptide molecules, which decreases localization accuracy. Furthermore, the matrix crystal size could also have an impact on the resulting spatial resolution of the image. The recent development of the nanostructure-initiator mass spectrometry (NIMS) improved upon the desorption/ionization on porous silicon (DIOS) matrix-free method with results comparable to MALDI and ESI for the analysis of biological molecules.8–18 NIMS enhances the DIOS approach by incorporating a perfluorinated initiator compound into the surface’s nanopores. NIMS initiators do not absorb energy at UV laser wavelengths, do not ionize, and do not cocrystallize with analytes like MALDI matrices do.8, 19, 20 Rather, NIMS initiators adsorb analytes to their surface and only assist in the desorption process. It is hypothesized that the ionization process in NIMS is a product of the surface’s acidity resulting from the hydrofluoric acid etching process.9
NIMS has shown great performance for the detection of analytes with a m/z below 1800.8, 9 This matrix-free technique has also been applied successfully to imaging experiments of drugs, metabolites, and lipids.8, 21, 22 Since the majority of neuropeptides in crustacean species are below m/z 180023 and analyte delocalization is a concern in MALDI MSI analysis, this work examines the utility of NIMS for MSI-based studies of neuropeptides in C. borealis brain tissue.
Materials and Methods
All chemicals were purchased from Thermo Fisher Scientific (Waltham, MA) unless otherwise noted.
Animals
C. borealis (Jonah crabs) were purchased from The Fresh Lobster Company (Gloucester, MA) and stored in an artificial seawater aquarium at 12–13 °C. Prior to use, crabs were cold anesthetized in packed ice for 30 min. Details of animal treatment and dissection were described previously by Kutz et al.24 In this experiment, the brain was dissected in chilled (4 °C) physiological saline (composition: 440 mM NaCl; 11 mM KCl; 13 mM CaCl2; 26 mM MgCl2; 10 mM HEPES acid; pH 7.45). Animals were housed, treated and sacrificed following the animal care protocol approved by the institution’s Animal Care and Usage Committee.
Preparation of NIMS surfaces
A detailed protocol for NIMS chip preparation has been previously documented.9 Single side-polished, 100 mm silicon wafers (P/Boron, 〈100 〉 orientation, 525 ± 25 μm thick, 0.01–0.02 Ω-cm resistivity) were purchased from Silicon Quest International (Santa Clara, CA) and sliced into 36 x 36 mm squares. Cut chips were cleaned through submersion in Piranha solution (2:1 H2SO4:H2O2) for 30 min. Chips were then rinsed with deionized water and blown dry with high-purity nitrogen gas. Etching took place inside of a custom-made Teflon chamber using 25% hydrofluoric acid in ethanol under 0.30 A for 30 min. Chips were cleaned with methanol, dried, and surface-coated with 50 μL of 1,3-bis(heptadecafluoro-1,1,2,2-tetrahydrodecyl)tetramethyldisiloxane (BisF17) purchased from Gelest (Morrisville, PA).
Sample Preparation
Immediately after dissection, C. borealis brain was briefly rinsed in deionized water to reduce salt content, embedded in gelatin (100 mg/mL), and snap-frozen in an ethanol/dry-ice bath. The embedded brain was attached to a cryostat chuck with a minimal amount of optimal-cutting temperature polymer, allowed to chill to chamber temperature (−20 °C), and serially sectioned into 12 μm slices for MALDI and 2 – 4 μm slices for NIMS using a Thermo Scientific Microm HM 505E cryostat equipped with a Shandon MB22 microtome blade (Walldorf, Germany). Sliced tissue was then thaw-mounted onto either MALDI or NIMS surfaces. Mounted samples were dehydrated in a desiccator at −20 °C for 30 min and allowed to warm to room temperature for 1 hr prior to either matrix application for MALDI analysis or NIMS MS analysis. For MALDI experiments, multiple thin coats of 2,5-dihydroxybenzoic acid (DHB, 100 mg/mL in 50% methanol, v/v) were applied to the tissue using a Paasche airbrush (Chicago, IL). Five coats were applied to the tissue each for 20 s with 1 min dry time between each coat.
Data Acquisition and Processing
An Applied Biosystems (Framingham, MA) 4800 MALDI TOF/TOF mass spectrometer equipped with a 200 Hz, 355 nm Nd:YAG laser (spot diameter of 75 μm) was used for all MS analyses. Instrument parameters were optimized using the 4000 Series Explorer Software (Applied Biosystems). All data were acquired in positive reflectron mode. Peptides FMRFa (m/z 599.31), [Arg8] Vasopressin (AVP, CYFQNCPRGa, m/z 1084.45), and neurotensin (pyroELYENKPRRPYIL, m/z 1672.92) (American Peptide Company, Sunnyvale, CA) were used as external calibrants for mass spectrometric analysis. Imaging experiments were performed using the 4800 Imaging application (Novartis, Basel, Switzerland) available through the MALDI MSI web site (www.maldi-msi.org). Tissue MSI data were collected over the range m/z 550 – 1800 at 100 μm intervals. At each acquisition pixel either 200 laser shots (MALDI) or 20 laser shots (NIMS) were averaged to generate a mass spectrum for each pixel. Mass spectra were externally calibrated using peptide standards applied directly to the MALDI or NIMS target. TissueView software (Applied Biosystems) was used to process the MSI data into MSI images. Tandem mass spectra (MS/MS) were acquired using 2 kV collision induced dissociation (CID) method with air as the collision gas. One thousand laser shots were averaged to generate each mass spectrum, and sequence interpretation was performed manually.
Results and Discussion
NIMS and MALDI imaging of brain sections workflow
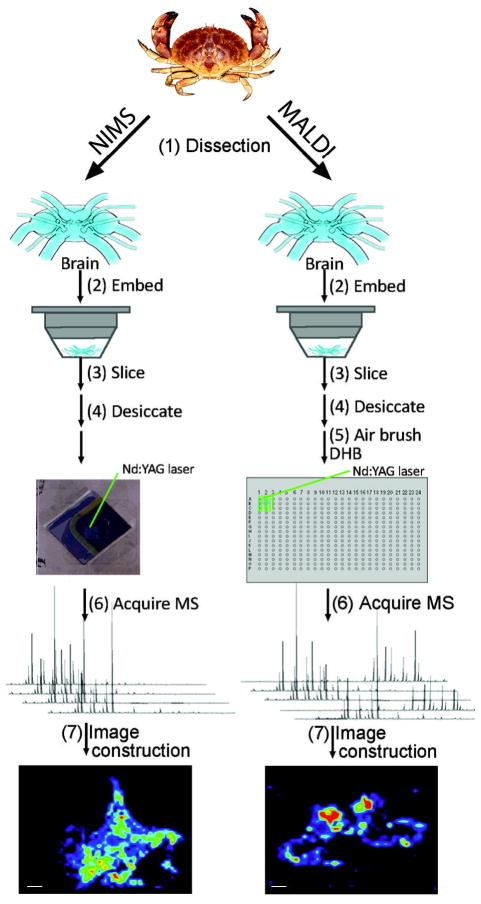
All NIMS imaging experiments were performed using 1,3-bis(heptadecafluoro-1,1,2,2-tetrahydrodecyl)tetramethyldisiloxane (BisF17) as this initiator. BisF17 was chosen as the initiator because it has worked well for peptide analyses and has been utilized in imaging experiments.8, 9, 21, 22 Initially NIMS imaging was performed using 12 μm thick tissue but required a high energy ablation step prior to mass analysis.8 Yanes et al. demonstrated that tissue sections cut at 2 – 4 μm thickness could directly be analyzed with this technique.22 Therefore, for NIMS experiments, C. borealis brain sections were cut at 2 – 4 μm thickness, and for MALDI experiments, tissue was cut at 12 μm thickness based on prior optimization results.3, 25 Experimental workflows for NIMS and MALDI MSI experiments are shown in Figure 1. In both experimental workflows, crabs were cold anesthetized in packed ice prior to dissection. The animals’ brain was placed in a small volume of liquid gelatin (100 mg/mL) and snap frozen. In NIMS experiments, the snap frozen tissue was placed on a cryostat chuck using a minimal amount of OCT and sliced into 2 – 4 μm thick sections at −20 °C. It should be noted that it was difficult to serially collect intact sections of 2 – 4 μm thickness. Out of three serial sections, usually only one was suitable for an imaging experiment and subsequently thaw mounted to a NIMS chip. Cryostat temperatures from −10 °C to −30 °C were tested for crustacean brain sectioning. The most consistent, intact, non-chattered brain slices were collected at −20 °C. For sections that were used in NIMS MSI experiments, the tissue mounted NIMS chips were placed in a desiccator at −20 °C for 30 min. Prior to MS analysis, the desiccator was removed from −20 °C and allowed to sit at room temperature for 1 hr. The NIMS plate was mounted to a stainless steel MALDI target using tape and placed into the mass spectrometer vacuum chamber for analysis. During data acquisition, 20 laser shots were averaged per pixel to generate a mass spectrum. It was observed that when > 20 laser shots were averaged per spectrum in NIMS experiments the average mass spectrum signal decreased (data not shown). In NIMS, the desorption process occurs underneath the tissue at the silicon surface where compounds that have interacted with the initiator are ionized into the mass analyzer.26 Therefore, once the NIMS surface is deconstructed by the laser energy, subsequent laser shots do not yield analyte signal thereby decreasing averaged peak intensity in a NIMS spectrum.
Figure 1.
MSI workflow. Brain tissue was dissected from C. borealis (1), embedded in gelatin, and snap frozen (2). For NIMS analysis, tissue was sliced at 2–4 μm thickness, mounted onto a BisF17 NIMS surface (3), and stored in a desiccator (4) until mass spectral analysis. Mass spectra are acquired across the tissue surface at 20 laser shots per pixel, 100 μm increments (6). Collected mass spectra were reassembled into a molecular ion distribution map using TissueView software. For MALDI analysis, tissue was sliced at 12 μm thickness, mounted onto a stainless steel target (3), and stored in a desiccator (4) until matrix application (5). Mass spectra were acquired across the tissue surface at 200 laser shots per pixel, 100 μm increments (6). Collected mass spectra were reassembled into a molecular ion distribution map using TissueView software. (Scale bar = 1 mm)
The MALDI MSI workflow is nearly identical to the NIMS MSI workflow, except for three aspects: tissue section thickness, application of MALDI matrix to the tissue, and laser shots per pixel. In the MALDI workflow, 12 μm sections are collected rather than 2 – 4 μm sections because these sections are easier to collect and handle. In addition, MALDI MSI does not rely on analytes desorbing from the mounting surface. In MALDI MSI, a UV-light absorbing weak organic acid is deposited/sprayed on the tissue and allowed to co-crystallize with it. When the laser irradiates the tissue, the MALDI matrix absorbs the energy and aids in the desorption/ionization of the analyte into the gas-phase. In NIMS MSI, a couple dozen laser shots at a single pixel results in the entire ablation of the tissue and deconstruction of the nanostructured surface. With MALDI, several hundred laser shots can be used on a single pixel without ablating all the tissue. During data acquisition in a MALDI MSI experiment 200 laser shots were averaged per pixel to generate a mass spectrum.
Although there are sample manipulation (tissue section thickness, MALDI matrix) and data acquisition differences (20 versus 200 laser shots per pixel) between the two desorption/ionization techniques, the authors do not believe that they will affect the results of this study since both methods were designed using optimized conditions that provide the best signal for each method.
Neuropeptide imaging
Four C. borealis brains with at least five slices for NIMS (n = 30 slices) and five slices for MALDI MS (n = 30 slices) analysis were used in this work. A total of eight NIMS chips were used during method development and analysis. Not all tissue slices were imaged. Some slices were used for optimization of tissue thickness, laser intensity, and laser shot evaluation while others only had direct tissue profiling analyses performed on them. It was determined that C. borealis brain should be sliced between 2 – 4 μm based on lipid ion signal intensities. This thickness is in agreement with previously published drug/metabolite NIMS MSI experiments in a mouse model.22 The laser intensity had to be greatly increased for tissue thicker than 5 μm in order to observe strong lipid ion peaks. The resulting spectra from tissue thicker than 5 μm lacked the analyte information obtained when using thinner tissue (data not shown). For NIMS analysis, the minimal laser intensity that would ablate tissue and produce lipid signal from tissue was used for imaging experiments. When performing non-tissue NIMS experiments where a peptide mixture was applied using the “Z-touch” method9, the laser intensity used for optimum sensitivity/resolution acquisition was lower than that used for MALDI experiments using alpha-cyano-4-hydroxy-cinnamic acid (CHCA) or DHB matrices. Supplemental Figure 2 shows the ion response of three neuropeptides (1 μM) applied to a NIMS surface using the “Z-touch” method. When performing tissue NIMS with 2 – 4 μm thick tissue, the laser intensity was increased to intensity similar to what would be used for a MALDI DHB matrix experiment. As mentioned previously, twenty laser shots were used to generate the best mass spectra for tissue NIMS. This result is logical since the desorption process causes the initiator to explode out of the silicon nanopores. After a certain number of laser shots (twenty in this case), the area under irradiation lacked initiator/analyte molecules so any further acquired mass spectra only contained background ions. Averaging background spectra with the spectra containing analyte would decrease the average mass spectrum’s analyte signal intensity.
Using the optimal data acquisition settings, spectra were collected from m/z 550 to 1800. Peptides FMRFa, Arg-vasopressin (AVP), and neurotensin were used to externally calibrate NIMS and MALDI mass spectra. Figure 2a shows the results from MALDI and NIMS neuropeptide imaging experiments. The neuropeptide rich region was m/z 900 to 1700. The MALDI spectra contained many neuropeptide ion peaks in this m/z region. MALDI molecular ion image distributions of three identified neuropeptides are shown in Figure 2b. In total, twelve neuropeptides were identified in this MALDI MSI experiment (Table 1). In contrast, the NIMS spectra did not contain peaks corresponding to any identified neuropeptide ions. A peak at m/z 1117.96 was observed in the NIMS analysis within this m/z region, but the ion peak was not observed in MALDI analysis and does not match the m/z of any known neuropeptides.
Figure 2.
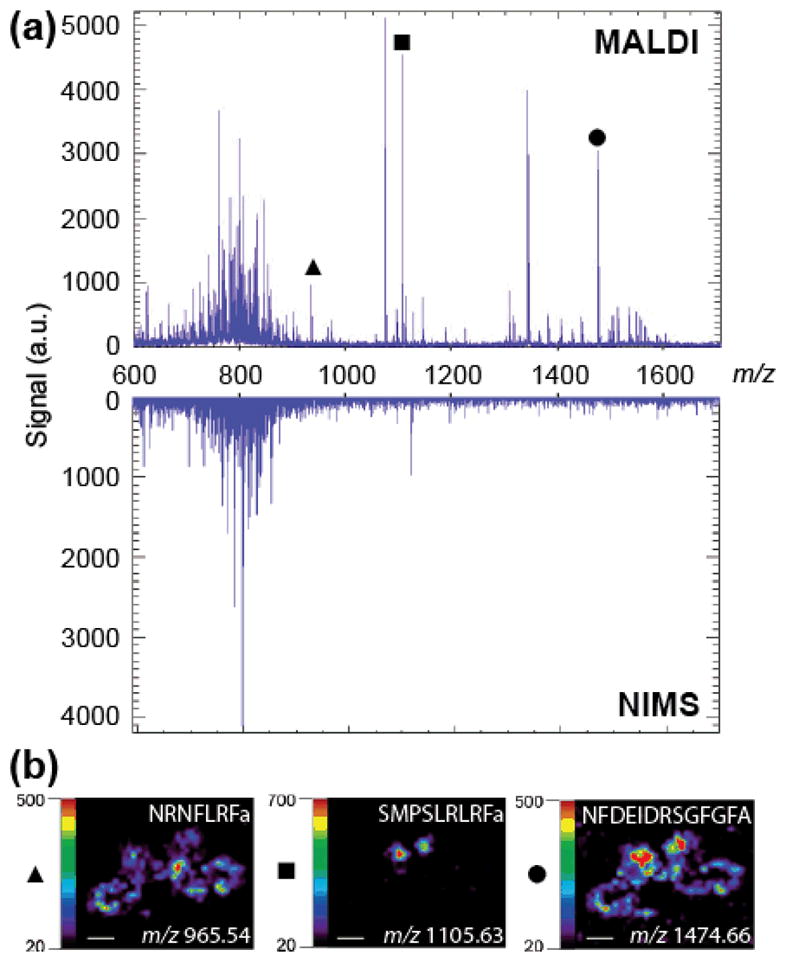
Neuropeptide NIMS and MALDI MSI of C. borealis brain. (a) Top mass spectrum shows a MALDI spectrum collected at a specific pixel of a MSI experiment showing rich lipid and neuropeptide content. Bottom mass spectrum displays a NIMS spectrum collected at a specific pixel of a MSI experiment showing little neuropeptide content but lipid ion intensities similar to MALDI. (b) Molecular ion distributions of three identified neuropeptides from a MALDI MSI experiment: NRNFLRFa (m/z 965.54), SMPSLRLRFa (m/z 1105.63), and NFDEIDRSGFGFA (m/z 1474.66). (Scale bar = 1 mm)
Table 1.
Neuropeptides identified in a single MALDI MSI experiment of C. borealis brain.
| Neuropeptide Sequence | Measured m/z | Theoretical m/z |
|---|---|---|
| RFamides | ||
| NRNFLRFa | 965.54 | 965.54 |
| SMPSLRLRFa | 1105.64 | 1105.63 |
| APQRNFLRFa | 1147.66 | 1147.65 |
| DVRTPALRLRFa | 1342.80 | 1342.81 |
| Orcokinins | ||
| NFDEIDRSGFGFA | 1474.65 | 1474.66 |
| NFDEIDRSGFGFV | 1502.69 | 1502.69 |
| NFDEIDRSSFGFV | 1532.68 | 1532.70 |
| NFDEIDRSSFGFN | 1547.69 | 1547.68 |
| NFDEIDRTGFGFH | 1554.72 | 1554.70 |
| SIFamide | ||
| GYRKPPFNGSIFa | 1381.74 | 1381.74 |
| C. borealis tachykinin related peptide | ||
| APSGFLGMRa | 934.50 | 934.49 |
| Others | ||
| HIGSLYRa | 844.48 | 844.48 |
The results suggest that NIMS is not compatible with direct imaging of neuropeptides in C. borealis brain tissue. To test whether these observations result from the lack of neuropeptides at the tissue/NIMS surface junction, one μM of AVP was deposited across the entire NIMS surface prior to thaw-mounting brain tissue. AVP ion signal was observed at surface locations adjacent to mounted tissue but was not observed when performing direct tissue analysis of the mounted tissue (Supplemental Figure 1). In an additional experiment, a droplet containing 1 μM of AVP was deposited on top of the mounted tissue. Once again, ion signals from the neuropeptide analytes were not observed, supporting the concept that the NIMS process occurs beneath the tissue.
Lipid imaging
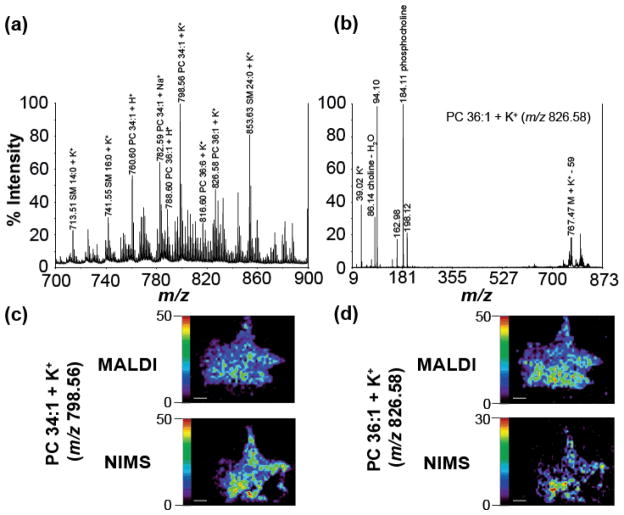
Although neuropeptide signals in the NIMS imaging experiments were not observed, the mass spectra in Figure 2a demonstrate that NIMS may be useful for lipid imaging experiments. Figure 3a shows a zoomed in view of ion peaks from m/z 700 to 900 from a MALDI MSI experiment. Using accurate mass in combination with MS/MS fragmentation (example of MS/MS, PC 36:1 + K+ shown in Figure 3b) several lipids in C. borealis brain tissue were identified. Figure 3c and 3d compare the MALDI and NIMS MSI distributions of PC 34:1 + K+ and PC 36:1 + K+, respectively. The MALDI and NIMS molecular ion distributions in these two sets of images matched well. In both sets of images, the lipid analytes appear to be more concentrated in the deutocerebrum region of the tissue. The NIMS slices are torn in the lower right part of the brain due to the fragile nature and difficulty of thaw-mounting inherent to 2 – 4 μm tissue slices.
Figure 3.
Lipid profiles and distributions from direct tissue and MSI NIMS/MALDI experiments of C. borealis brain. (a) Zoomed-in lipid rich area (m/z 700 – 900) of MALDI direct tissue mass spectrum. (b) MALDI MS/MS spectra of PC 36:1 + K+ (m/z 826.58). (c) MALDI and NIMS distribution of PC 34:1 + K+ (m/z 798.56). (d) MALDI and NIMS distribution of PC 36:1 + K+ (m/z 826.58). (Scale bar = 1 mm)
The observation that NIMS can be successfully utilized for lipid imaging in C. borealis brain complements previous results of using this technique in mammalian tissues.8, 21 It is important to note that the MALDI images in Figure 3c and 3d were collected by averaging 200 laser shots whereas the NIMS images only required 20 shots. The lipid ion intensities from NIMS and MALDI acquisitions are similar to each other in Figures 2, 3c, and 3d. The 10x decrease in laser shots required per pixel in NIMS has its advantages with respect to MSI throughput. For lipid distribution studies using tissue, NIMS MSI appears to be a viable, time-saving alternative to MALDI MSI, meriting further investigation.
Conclusions
Although NIMS MSI and MALDI MSI generate comparable results for lipid analytes (m/z 700 – 900), it does not appear that NIMS MSI can be successfully used for neuropeptide distribution studies in C. borealis brain at this time. The lack of neuropeptide chemical information generated in NIMS versus MALDI experiments may be due to the desorption/ionization differences between the two ionization methods, causing differences in sensitivity for low-level peptide analysis. In MALDI, tissue analytes are co-crystallized with a large molar excess of matrix, generating many opportunities for an analyte to become ionized in the gas plume. In contrast, it is hypothesized that the acidic conditions of the HF etched silicon surface is responsible for analyte ionization in NIMS. To increase the ionization efficiency of molecules in positive mode NIMS, researchers are investigating the use of photoacid initiators and different nanostructured surfaces.26 The development of matrix-free methods for neuropeptide MSI will mitigate spatial resolution limits and potential analyte delocalization associated with analyte diffusion in MALDI MSI experiments. It is possible that these advances will someday facilitate matrix-free analysis of neuropeptides in tissue, but MALDI currently remains the preferred method for neuropeptide localization and in situ measurement experiments in C. borealis brain tissue.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Hin-Koon Woo and Dr. Oscar Yanes in the Siuzdak lab at Scripps Center for Mass Spectrometry for their help in teaching the authors how to prepare NIMS surfaces. This work is supported in part by the National Science Foundation grant (CHE-0957784), and National Institutes of Health grant (1R01DK071801). L. L. acknowledges an H.I. Romnes Faculty Research Fellowship. R.M.S. acknowledges the NIH-supported Clinical Neuroengineering Training Program Predoctoral Fellowship (NRSA T32 EB011434).
References
- 1.Altelaar A, Taban IM, McDonnell LA, Verhaert PDEM, de Lange RPJ, Adan RAH, Mooi WJ, Heeren R, Piersma SR. Int J Mass Spectrom. 2007;260:203–211. [Google Scholar]
- 2.Chen R, Hui L, Sturm RM, Li L. J Am Soc Mass Spectrom. 2009;20:1068–1077. doi: 10.1016/j.jasms.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeKeyser SS, Kutz-Naber KK, Schmidt JJ, Barrett-Wilt GA, Li L. J Proteome Res. 2007;6:1782–1791. doi: 10.1021/pr060603v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jarecki JL, Andersen K, Konop CJ, Knickelbine JJ, Vestling MM, Stretton AO. ACS Chem Neurosci. 2010;1:505–519. doi: 10.1021/cn1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taban IM, Altelaar AFM, van der Burgt YEM, McDonnell LA, Heeren RMA, Fuchser J, Baykut G. J Am Soc Mass Spectrom. 2007;18:145–151. doi: 10.1016/j.jasms.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 6.Verhaert PDEM, Pinkse MWH, Strupat K, Conaway MCP. Methods Mol Biol. 2010;656:433–449. doi: 10.1007/978-1-60761-746-4_25. [DOI] [PubMed] [Google Scholar]
- 7.Zimmerman TA, Rubakhin SS, Romanova EV, Tucker KR, Sweedler JV. Anal Chem. 2009;81:9402–9409. doi: 10.1021/ac901820v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Northen TR, Yanes O, Northen MT, Marrinucci D, Uritboonthai W, Apon J, Golledge SL, Nom A, Siuzdak G. Nature. 2007;449:1033–1036. doi: 10.1038/nature06195. [DOI] [PubMed] [Google Scholar]
- 9.Woo HK, Northen TR, Yanes O, Siuzdak G. Nat Protoc. 2008;3:1341–1349. doi: 10.1038/nprot.2008.110. [DOI] [PubMed] [Google Scholar]
- 10.Wei J, Buriak JM, Siuzdak G. Nature. 1999;399:243–246. doi: 10.1038/20400. [DOI] [PubMed] [Google Scholar]
- 11.Amantonico A, Flamigni L, Glaus R, Zenobi R. Metabolomics. 2009;5:346–353. [Google Scholar]
- 12.Colantonio S, Simpson JT, Fisher RJ, Yavlovich A, Belanger JM, Puri A, Blumenthal R. Lipids. 2011;46:469–477. doi: 10.1007/s11745-010-3493-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng K, George KW, Reindl W, Keasling JD, Adams PD, Lee TS, Singh AK, Northen TR. Rapid Commun Mass Spectrom. 2012;26:611–615. doi: 10.1002/rcm.6134. [DOI] [PubMed] [Google Scholar]
- 14.Pavlásková Ki, Strnadová M, Strohalm M, Havlícek Vr, Sulc M, Volný M. Anal Chem. 2011;83:5661–5665. doi: 10.1021/ac200801t. [DOI] [PubMed] [Google Scholar]
- 15.Reindl W, Deng K, Gladden JM, Cheng G, Wong A, Singer SW, Singh S, Lee JC, Yao CH, Hazen TC, Singh AK, Simmons BA, Adams PD, Northen TR. Energy Environ Sci. 2011;4:2884. [Google Scholar]
- 16.Reindl W, Northen TR. Anal Chem. 2010;82:3751–3755. doi: 10.1021/ac100159y. [DOI] [PubMed] [Google Scholar]
- 17.Wyatt MF, Ding S, Stein BK, Brenton AG, Daniels RH. J Am Soc Mass Spectrom. 2010;21:1256–1259. doi: 10.1016/j.jasms.2010.03.038. [DOI] [PubMed] [Google Scholar]
- 18.Liu Q, Xiao Y, Pagan-Miranda C, Chiu YM, He L. J Am Soc Mass Spectrom. 2009;20:80–88. doi: 10.1016/j.jasms.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka K, Waki H, Ido Y, Akita S, Yoshida Y, Yoshida T, Matsuo T. Rapid Commun Mass Spectrom. 1988;2:151–153. [Google Scholar]
- 20.Karas M, Hillenkamp F. Anal Chem. 1988;60:2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- 21.Lee DY, Platt V, Bowen B, Louie K, Canaria CA, McMurray CT, Northen T. Integr Biol. 2012;4:693–699. doi: 10.1039/c2ib20043k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yanes O, Woo HK, Northen TR, Oppenheimer SR, Shriver L, Apon J, Estrada MN, Potchoiba MJ, Steenwyk R, Manchester M, Siuzdak G. Anal Chem. 2009;81:2969–2975. doi: 10.1021/ac802576q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L, Sweedler JV. Annu Rev Anal Chem. 2008;1:451–483. doi: 10.1146/annurev.anchem.1.031207.113053. [DOI] [PubMed] [Google Scholar]
- 24.Kutz KK, Schmidt JJ, Li L. Anal Chem. 2004;76:5630–5640. doi: 10.1021/ac049255b. [DOI] [PubMed] [Google Scholar]
- 25.Sugiura Y, Shimma S, Setou M. J Mass Spectrom Soc Jpn. 2006;54:45–48. [Google Scholar]
- 26.Calavia R, Fatima A, Correig X, Yanes O. J Proteomics. 2012;16:5061–5068. doi: 10.1016/j.jprot.2012.05.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.