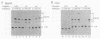Abstract
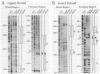
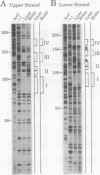
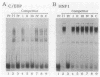
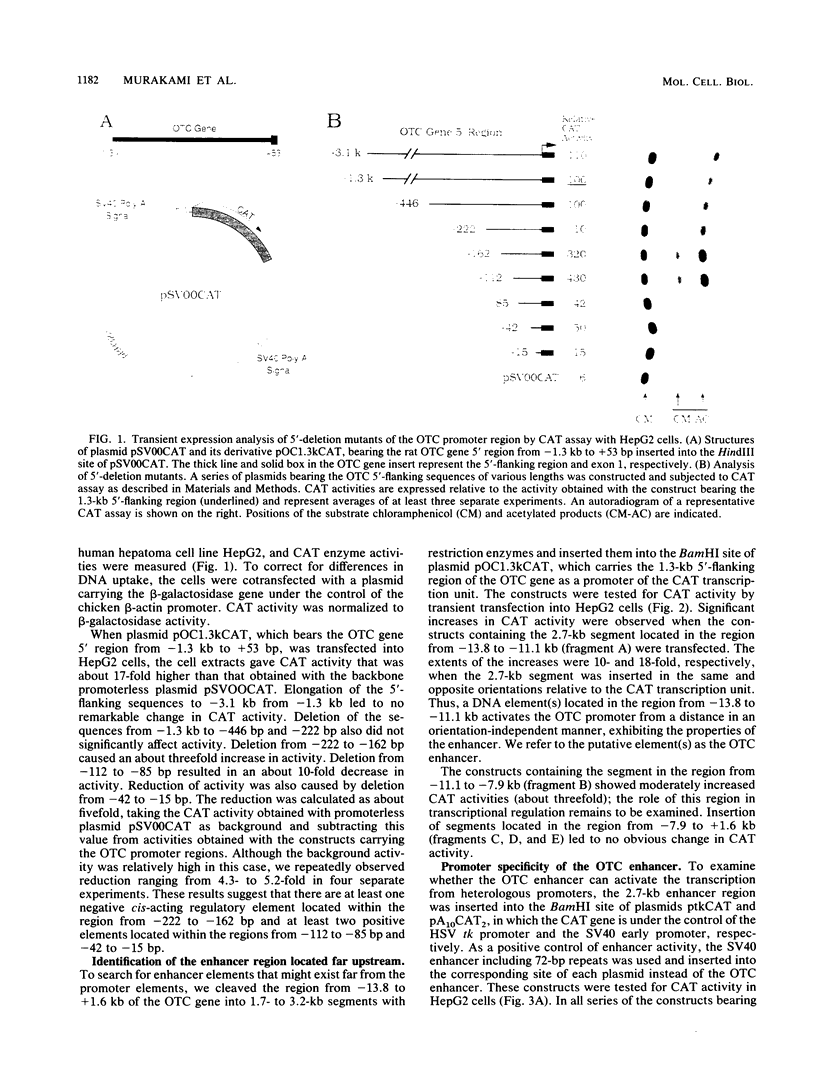
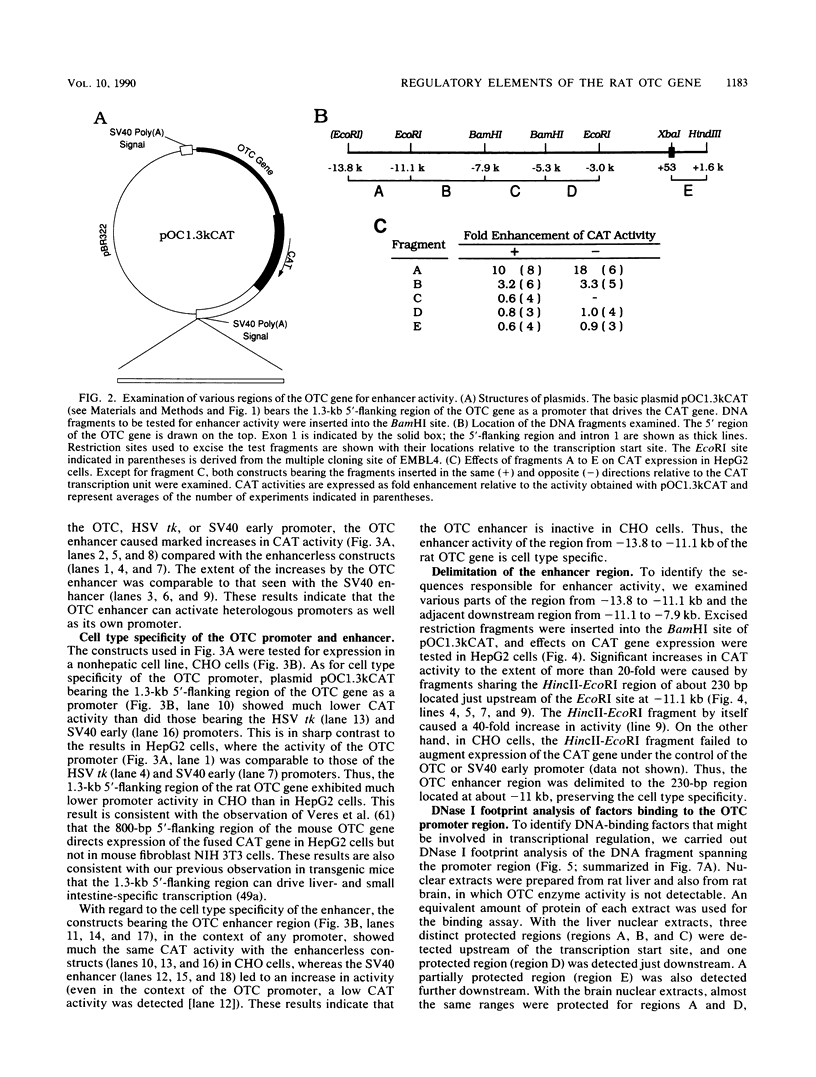
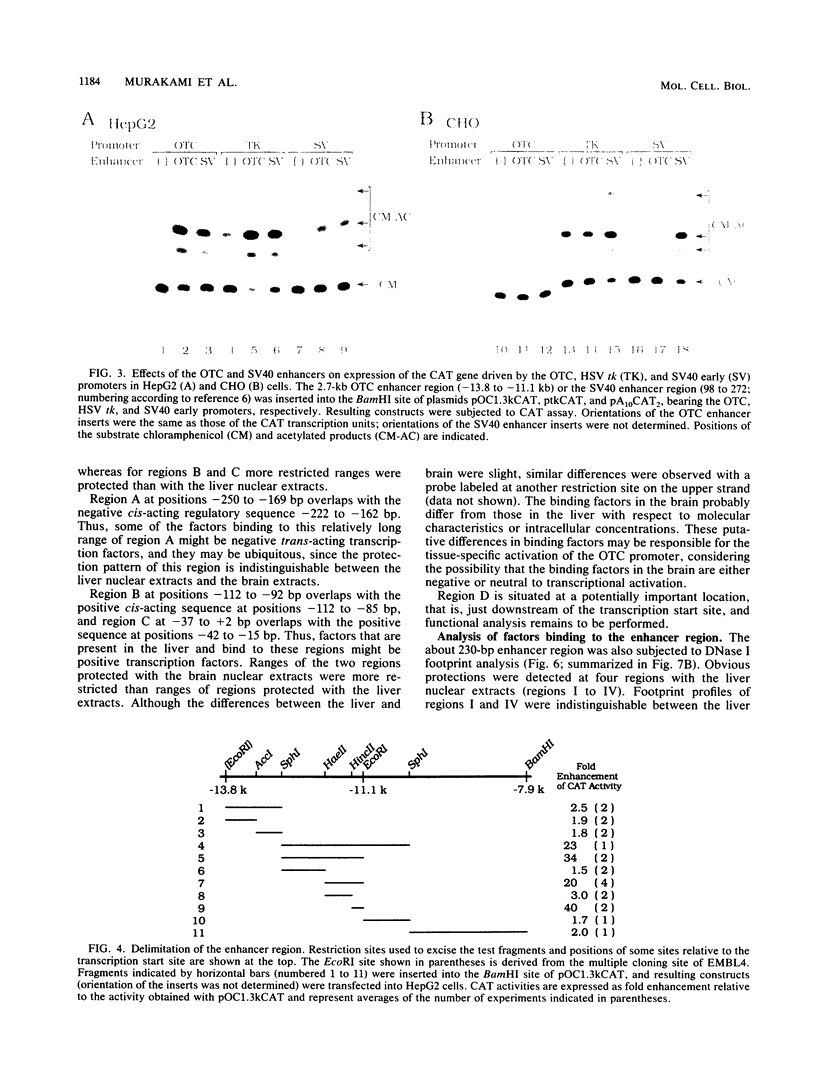
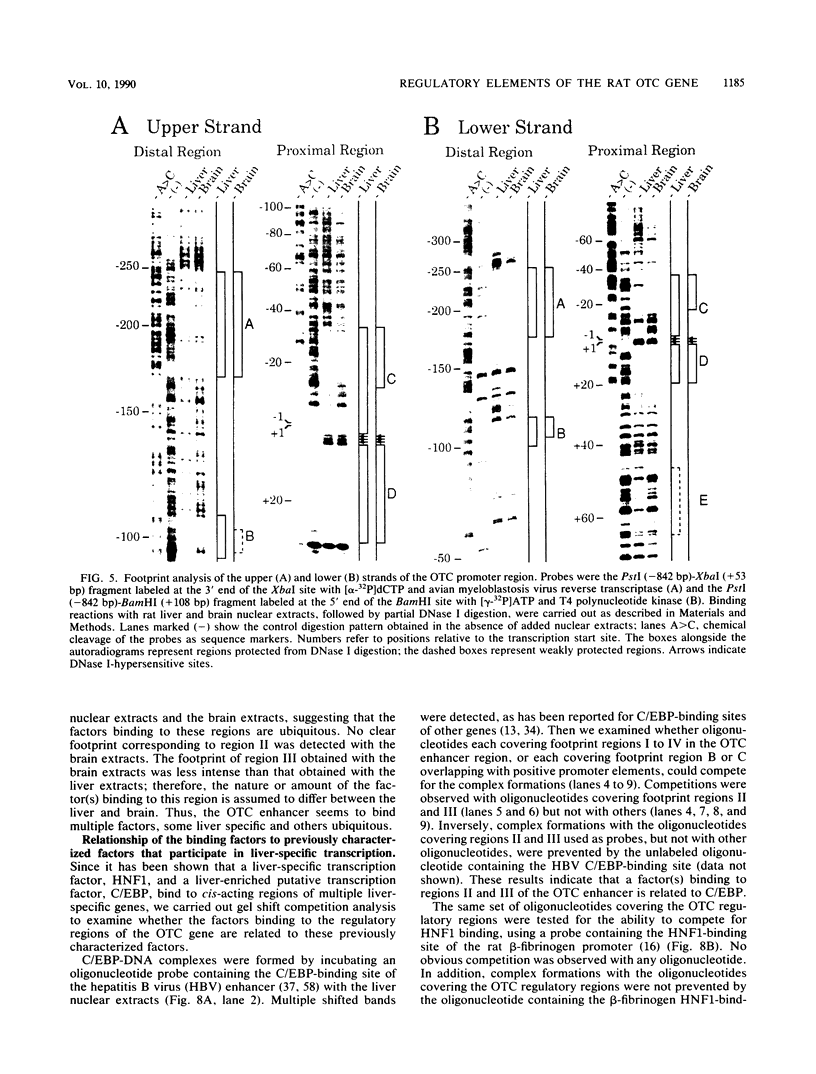
The gene for ornithine transcarbamylase (OTC; EC 2.1.3.3), a urea cycle enzyme, is expressed almost exclusively in the liver and small intestine. To identify DNA elements regulating transcription of the OTC gene in the liver, transient expression analysis was carried out by using hepatoma (HepG2) and nonhepatic (CHO) cell lines. The 1.3-kilobase 5'-flanking region of the rat OTC gene directed expression of the fused chloramphenicol acetyltransferase gene in HepG2 cells much more efficiently than in CHO cells. Analysis of deletion mutants of the 5'-flanking region in HepG2 cells revealed that there are at least one negative and two positive regulatory elements within the about 220-base-pair immediate 5'-flanking region. DNase I footprint analysis showed the presence of factors binding to these regulatory elements in nuclear extracts of rat liver and brain, and footprint profiles at the two positive elements exhibited liver-specific features. Transient expression analysis also revealed the existence of an enhancer region located 11 kilobases upstream of the transcription start site. The OTC enhancer was able to activate both its own and heterologous promoters in HepG2 but not in CHO cells. The enhancer was delimited to an about 230-base-pair region, and footprint analysis of this region revealed four protected areas. Footprint profiles at two of the four areas exhibited liver-specific features, and gel shift competition analysis showed that a factor(s) binding to the two liver-specific sites is related to C/EBP. These results suggest that both liver-specific promoter and enhancer elements regulate expression of the OTC gene through interaction with liver-specific factors binding to these elements.
Full text
PDF

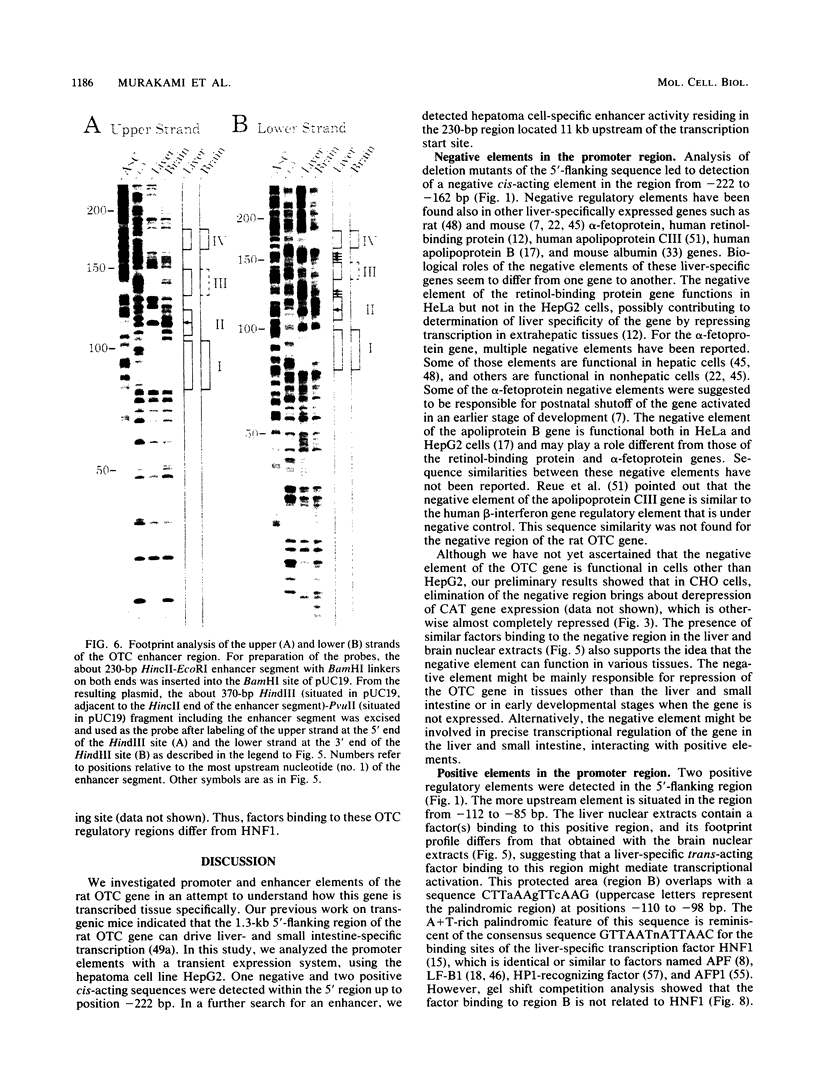
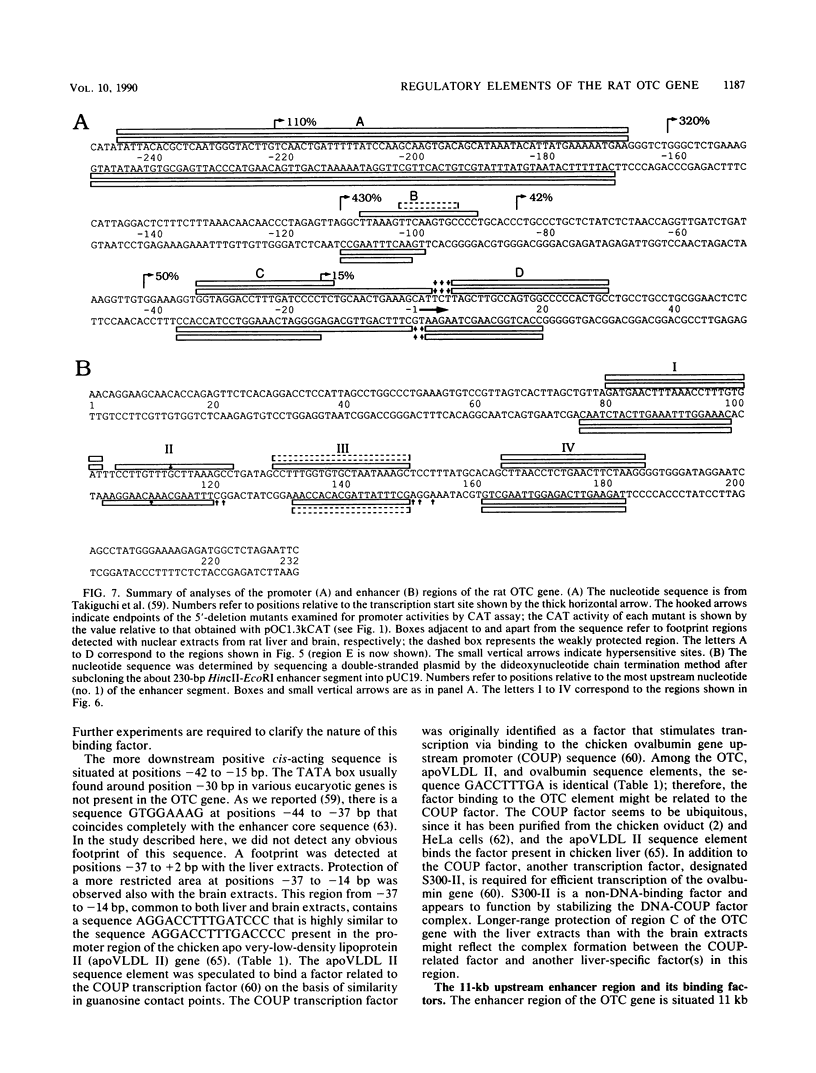




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Araki E., Shimada F., Shichiri M., Mori M., Ebina Y. pSV00CAT: low background CAT plasmid. Nucleic Acids Res. 1988 Feb 25;16(4):1627–1627. doi: 10.1093/nar/16.4.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagchi M. K., Tsai S. Y., Tsai M. J., O'Malley B. W. Purification and characterization of chicken ovalbumin gene upstream promoter transcription factor from homologous oviduct cells. Mol Cell Biol. 1987 Dec;7(12):4151–4158. doi: 10.1128/mcb.7.12.4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumhueter S., Courtois G., Crabtree G. R. A variant nuclear protein in dedifferentiated hepatoma cells binds to the same functional sequences in the beta fibrinogen gene promoter as HNF-1. EMBO J. 1988 Aug;7(8):2485–2493. doi: 10.1002/j.1460-2075.1988.tb03095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenmeier E. H., Gwynn B., Howard S., Jerry J., Gordon J. I., Landschulz W. H., McKnight S. L. Tissue-specific expression, developmental regulation, and genetic mapping of the gene encoding CCAAT/enhancer binding protein. Genes Dev. 1989 Aug;3(8):1146–1156. doi: 10.1101/gad.3.8.1146. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C. A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol. 1983;100:243–255. doi: 10.1016/0076-6879(83)00059-2. [DOI] [PubMed] [Google Scholar]
- Camper S. A., Tilghman S. M. Postnatal repression of the alpha-fetoprotein gene is enhancer independent. Genes Dev. 1989 Apr;3(4):537–546. doi: 10.1101/gad.3.4.537. [DOI] [PubMed] [Google Scholar]
- Cereghini S., Blumenfeld M., Yaniv M. A liver-specific factor essential for albumin transcription differs between differentiated and dedifferentiated rat hepatoma cells. Genes Dev. 1988 Aug;2(8):957–974. doi: 10.1101/gad.2.8.957. [DOI] [PubMed] [Google Scholar]
- Cereghini S., Raymondjean M., Carranca A. G., Herbomel P., Yaniv M. Factors involved in control of tissue-specific expression of albumin gene. Cell. 1987 Aug 14;50(4):627–638. doi: 10.1016/0092-8674(87)90036-5. [DOI] [PubMed] [Google Scholar]
- Christy R. J., Yang V. W., Ntambi J. M., Geiman D. E., Landschulz W. H., Friedman A. D., Nakabeppu Y., Kelly T. J., Lane M. D. Differentiation-induced gene expression in 3T3-L1 preadipocytes: CCAAT/enhancer binding protein interacts with and activates the promoters of two adipocyte-specific genes. Genes Dev. 1989 Sep;3(9):1323–1335. doi: 10.1101/gad.3.9.1323. [DOI] [PubMed] [Google Scholar]
- Ciliberto G., Dente L., Cortese R. Cell-specific expression of a transfected human alpha 1-antitrypsin gene. Cell. 1985 Jun;41(2):531–540. doi: 10.1016/s0092-8674(85)80026-x. [DOI] [PubMed] [Google Scholar]
- Colantuoni V., Pirozzi A., Blance C., Cortese R. Negative control of liver-specific gene expression: cloned human retinol-binding protein gene is repressed in HeLa cells. EMBO J. 1987 Mar;6(3):631–636. doi: 10.1002/j.1460-2075.1987.tb04801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa R. H., Grayson D. R., Xanthopoulos K. G., Darnell J. E., Jr A liver-specific DNA-binding protein recognizes multiple nucleotide sites in regulatory regions of transthyretin, alpha 1-antitrypsin, albumin, and simian virus 40 genes. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3840–3844. doi: 10.1073/pnas.85.11.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa R. H., Lai E., Grayson D. R., Darnell J. E., Jr The cell-specific enhancer of the mouse transthyretin (prealbumin) gene binds a common factor at one site and a liver-specific factor(s) at two other sites. Mol Cell Biol. 1988 Jan;8(1):81–90. doi: 10.1128/mcb.8.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois G., Baumhueter S., Crabtree G. R. Purified hepatocyte nuclear factor 1 interacts with a family of hepatocyte-specific promoters. Proc Natl Acad Sci U S A. 1988 Nov;85(21):7937–7941. doi: 10.1073/pnas.85.21.7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois G., Morgan J. G., Campbell L. A., Fourel G., Crabtree G. R. Interaction of a liver-specific nuclear factor with the fibrinogen and alpha 1-antitrypsin promoters. Science. 1987 Oct 30;238(4827):688–692. doi: 10.1126/science.3499668. [DOI] [PubMed] [Google Scholar]
- Das H. K., Leff T., Breslow J. L. Cell type-specific expression of the human apoB gene is controlled by two cis-acting regulatory regions. J Biol Chem. 1988 Aug 15;263(23):11452–11458. [PubMed] [Google Scholar]
- Frain M., Swart G., Monaci P., Nicosia A., Stämpfli S., Frank R., Cortese R. The liver-specific transcription factor LF-B1 contains a highly diverged homeobox DNA binding domain. Cell. 1989 Oct 6;59(1):145–157. doi: 10.1016/0092-8674(89)90877-5. [DOI] [PubMed] [Google Scholar]
- Friedman A. D., Landschulz W. H., McKnight S. L. CCAAT/enhancer binding protein activates the promoter of the serum albumin gene in cultured hepatoma cells. Genes Dev. 1989 Sep;3(9):1314–1322. doi: 10.1101/gad.3.9.1314. [DOI] [PubMed] [Google Scholar]
- Gerster T., Matthias P., Thali M., Jiricny J., Schaffner W. Cell type-specificity elements of the immunoglobulin heavy chain gene enhancer. EMBO J. 1987 May;6(5):1323–1330. doi: 10.1002/j.1460-2075.1987.tb02371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godbout R., Ingram R. S., Tilghman S. M. Fine-structure mapping of the three mouse alpha-fetoprotein gene enhancers. Mol Cell Biol. 1988 Mar;8(3):1169–1178. doi: 10.1128/mcb.8.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godbout R., Ingram R., Tilghman S. M. Multiple regulatory elements in the intergenic region between the alpha-fetoprotein and albumin genes. Mol Cell Biol. 1986 Feb;6(2):477–487. doi: 10.1128/mcb.6.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski K., Carneiro M., Schibler U. Tissue-specific in vitro transcription from the mouse albumin promoter. Cell. 1986 Dec 5;47(5):767–776. doi: 10.1016/0092-8674(86)90519-2. [DOI] [PubMed] [Google Scholar]
- Hammer R. E., Krumlauf R., Camper S. A., Brinster R. L., Tilghman S. M. Diversity of alpha-fetoprotein gene expression in mice is generated by a combination of separate enhancer elements. Science. 1987 Jan 2;235(4784):53–58. doi: 10.1126/science.2432657. [DOI] [PubMed] [Google Scholar]
- Hata A., Tsuzuki T., Shimada K., Takiguchi M., Mori M., Matsuda I. Structure of the human ornithine transcarbamylase gene. J Biochem. 1988 Feb;103(2):302–308. doi: 10.1093/oxfordjournals.jbchem.a122265. [DOI] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Hayashi S., Goto K., Okada T. S., Kondoh H. Lens-specific enhancer in the third intron regulates expression of the chicken delta 1-crystallin gene. Genes Dev. 1987 Oct;1(8):818–828. doi: 10.1101/gad.1.8.818. [DOI] [PubMed] [Google Scholar]
- Heard J. M., Herbomel P., Ott M. O., Mottura-Rollier A., Weiss M., Yaniv M. Determinants of rat albumin promoter tissue specificity analyzed by an improved transient expression system. Mol Cell Biol. 1987 Jul;7(7):2425–2434. doi: 10.1128/mcb.7.7.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbomel P., Bourachot B., Yaniv M. Two distinct enhancers with different cell specificities coexist in the regulatory region of polyoma. Cell. 1984 Dec;39(3 Pt 2):653–662. doi: 10.1016/0092-8674(84)90472-0. [DOI] [PubMed] [Google Scholar]
- Herbst R. S., Friedman N., Darnell J. E., Jr, Babiss L. E. Positive and negative regulatory elements in the mouse albumin enhancer. Proc Natl Acad Sci U S A. 1989 Mar;86(5):1553–1557. doi: 10.1073/pnas.86.5.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B. W., Lagacé M., Shore G. C. Activity of the carbamyl phosphate synthetase I promoter in liver nuclear extracts is dependent on a cis-acting C/EBP recognition element. Mol Cell Biol. 1989 Jul;9(7):2928–2933. doi: 10.1128/mcb.9.7.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. F., Landschulz W. H., Graves B. J., McKnight S. L. Identification of a rat liver nuclear protein that binds to the enhancer core element of three animal viruses. Genes Dev. 1987 Apr;1(2):133–146. doi: 10.1101/gad.1.2.133. [DOI] [PubMed] [Google Scholar]
- Laimins L. A., Khoury G., Gorman C., Howard B., Gruss P. Host-specific activation of transcription by tandem repeats from simian virus 40 and Moloney murine sarcoma virus. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6453–6457. doi: 10.1073/pnas.79.21.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., Adashi E. Y., Graves B. J., McKnight S. L. Isolation of a recombinant copy of the gene encoding C/EBP. Genes Dev. 1988 Jul;2(7):786–800. doi: 10.1101/gad.2.7.786. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Lenardo M., Pierce J. W., Baltimore D. Protein-binding sites in Ig gene enhancers determine transcriptional activity and inducibility. Science. 1987 Jun 19;236(4808):1573–1577. doi: 10.1126/science.3109035. [DOI] [PubMed] [Google Scholar]
- Li Y., Shen R. F., Tsai S. Y., Woo S. L. Multiple hepatic trans-acting factors are required for in vitro transcription of the human alpha-1-antitrypsin gene. Mol Cell Biol. 1988 Oct;8(10):4362–4369. doi: 10.1128/mcb.8.10.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtsteiner S., Schibler U. A glycosylated liver-specific transcription factor stimulates transcription of the albumin gene. Cell. 1989 Jun 30;57(7):1179–1187. doi: 10.1016/0092-8674(89)90055-x. [DOI] [PubMed] [Google Scholar]
- Lichtsteiner S., Wuarin J., Schibler U. The interplay of DNA-binding proteins on the promoter of the mouse albumin gene. Cell. 1987 Dec 24;51(6):963–973. doi: 10.1016/0092-8674(87)90583-6. [DOI] [PubMed] [Google Scholar]
- Lindgren V., de Martinville B., Horwich A. L., Rosenberg L. E., Francke U. Human ornithine transcarbamylase locus mapped to band Xp21.1 near the Duchenne muscular dystrophy locus. Science. 1984 Nov 9;226(4675):698–700. doi: 10.1126/science.6494904. [DOI] [PubMed] [Google Scholar]
- Miyazaki J., Takaki S., Araki K., Tashiro F., Tominaga A., Takatsu K., Yamamura K. Expression vector system based on the chicken beta-actin promoter directs efficient production of interleukin-5. Gene. 1989 Jul 15;79(2):269–277. doi: 10.1016/0378-1119(89)90209-6. [DOI] [PubMed] [Google Scholar]
- Molné M., Houart C., Szpirer J., Szpirer C. Combinatorial control of positive and negative, upstream and intragenic regulatory DNA domains of the mouse alpha 1-foetoprotein gene. Nucleic Acids Res. 1989 May 11;17(9):3447–3457. doi: 10.1093/nar/17.9.3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaci P., Nicosia A., Cortese R. Two different liver-specific factors stimulate in vitro transcription from the human alpha 1-antitrypsin promoter. EMBO J. 1988 Jul;7(7):2075–2087. doi: 10.1002/j.1460-2075.1988.tb03047.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris S. M., Jr, Kepka D. M., Sweeney W. E., Jr, Avner E. D. Abundance of mRNAs encoding urea cycle enzymes in fetal and neonatal mouse liver. Arch Biochem Biophys. 1989 Feb 15;269(1):175–180. doi: 10.1016/0003-9861(89)90097-0. [DOI] [PubMed] [Google Scholar]
- Muglia L., Rothman-Denes L. B. Cell type-specific negative regulatory element in the control region of the rat alpha-fetoprotein gene. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7653–7657. doi: 10.1073/pnas.83.20.7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins L. J., Veres G., Caskey C. T., Chapman V. Differential methylation of the ornithine carbamoyl transferase gene on active and inactive mouse X chromosomes. Mol Cell Biol. 1987 Nov;7(11):3916–3922. doi: 10.1128/mcb.7.11.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T., Takiguchi M., Inomoto T., Yamamura K., Mori M. Tissue- and developmental stage-specific expression of the rat ornithine carbamoyltransferase gene in transgenic mice. Dev Genet. 1989;10(5):393–401. doi: 10.1002/dvg.1020100507. [DOI] [PubMed] [Google Scholar]
- Pinkert C. A., Ornitz D. M., Brinster R. L., Palmiter R. D. An albumin enhancer located 10 kb upstream functions along with its promoter to direct efficient, liver-specific expression in transgenic mice. Genes Dev. 1987 May;1(3):268–276. doi: 10.1101/gad.1.3.268. [DOI] [PubMed] [Google Scholar]
- Reue K., Leff T., Breslow J. L. Human apolipoprotein CIII gene expression is regulated by positive and negative cis-acting elements and tissue-specific protein factors. J Biol Chem. 1988 May 15;263(14):6857–6864. [PubMed] [Google Scholar]
- Ryall J. C., Quantz M. A., Shore G. C. Rat liver and intestinal mucosa differ in the developmental pattern and hormonal regulation of carbamoyl-phosphate synthetase I and ornithine carbamoyl transferase gene expression. Eur J Biochem. 1986 May 2;156(3):453–458. doi: 10.1111/j.1432-1033.1986.tb09603.x. [DOI] [PubMed] [Google Scholar]
- Ryall J., Nguyen M., Bendayan M., Shore G. C. Expression of nuclear genes encoding the urea cycle enzymes, carbamoyl-phosphate synthetase I and ornithine carbamoyl transferase, in rat liver and intestinal mucosa. Eur J Biochem. 1985 Oct 15;152(2):287–292. doi: 10.1111/j.1432-1033.1985.tb09196.x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawadaishi K., Morinaga T., Tamaoki T. Interaction of a hepatoma-specific nuclear factor with transcription-regulatory sequences of the human alpha-fetoprotein and albumin genes. Mol Cell Biol. 1988 Dec;8(12):5179–5187. doi: 10.1128/mcb.8.12.5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S. E., Veres G., Caskey C. T. The genetic structure of mouse ornithine transcarbamylase. Nucleic Acids Res. 1988 Feb 25;16(4):1593–1601. doi: 10.1093/nar/16.4.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorpp M., Kugler W., Wagner U., Ryffel G. U. Hepatocyte-specific promoter element HP1 of the Xenopus albumin gene interacts with transcriptional factors of mammalian hepatocytes. J Mol Biol. 1988 Jul 20;202(2):307–320. doi: 10.1016/0022-2836(88)90460-3. [DOI] [PubMed] [Google Scholar]
- Shaul Y., Ben-Levy R. Multiple nuclear proteins in liver cells are bound to hepatitis B virus enhancer element and its upstream sequences. EMBO J. 1987 Jul;6(7):1913–1920. doi: 10.1002/j.1460-2075.1987.tb02451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takiguchi M., Murakami T., Miura S., Mori M. Structure of the rat ornithine carbamoyltransferase gene, a large, X chromosome-linked gene with an atypical promoter. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6136–6140. doi: 10.1073/pnas.84.17.6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S. Y., Sagami I., Wang H., Tsai M. J., O'Malley B. W. Interactions between a DNA-binding transcription factor (COUP) and a non-DNA binding factor (S300-II). Cell. 1987 Aug 28;50(5):701–709. doi: 10.1016/0092-8674(87)90328-x. [DOI] [PubMed] [Google Scholar]
- Veres G., Craigen W. J., Caskey C. T. The 5' flanking region of the ornithine transcarbamylase gene contains DNA sequences regulating tissue-specific expression. J Biol Chem. 1986 Jun 15;261(17):7588–7591. [PubMed] [Google Scholar]
- Wang L. H., Tsai S. Y., Sagami I., Tsai M. J., O'Malley B. W. Purification and characterization of chicken ovalbumin upstream promoter transcription factor from HeLa cells. J Biol Chem. 1987 Nov 25;262(33):16080–16086. [PubMed] [Google Scholar]
- Weiher H., König M., Gruss P. Multiple point mutations affecting the simian virus 40 enhancer. Science. 1983 Feb 11;219(4585):626–631. doi: 10.1126/science.6297005. [DOI] [PubMed] [Google Scholar]
- Widen S. G., Papaconstantinou J. Liver-specific expression of the mouse alpha-fetoprotein gene is mediated by cis-acting DNA elements. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8196–8200. doi: 10.1073/pnas.83.21.8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnholds J., Philipsen J. N., Ab G. Tissue-specific and steroid-dependent interaction of transcription factors with the oestrogen-inducible apoVLDL II promoter in vivo. EMBO J. 1988 Sep;7(9):2757–2763. doi: 10.1002/j.1460-2075.1988.tb03130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wraight C., Lingelbach K., Hoogenraad N. Comparison of ornithine transcarbamylase from rat liver and intestine. Evidence for differential regulation of enzyme levels. Eur J Biochem. 1985 Dec 2;153(2):239–242. doi: 10.1111/j.1432-1033.1985.tb09292.x. [DOI] [PubMed] [Google Scholar]
- Xanthopoulos K. G., Mirkovitch J., Decker T., Kuo C. F., Darnell J. E., Jr Cell-specific transcriptional control of the mouse DNA-binding protein mC/EBP. Proc Natl Acad Sci U S A. 1989 Jun;86(11):4117–4121. doi: 10.1073/pnas.86.11.4117. [DOI] [PMC free article] [PubMed] [Google Scholar]