Abstract
Photodynamic therapy (PDT) is one of the most promising and noninvasive methods for clinical treatment of different malignant diseases. Here, we present a novel strategy of designing an aptamer-based DNA nanocircuit capable of the selective recognition of cancer cells, controllable activation of photosensitizer and amplification of photodynamic therapeutic effect. The aptamers can selectively recognize target cancer cells and bind to the specific proteins on cell membranes. Then the overhanging catalyst sequence on aptamer can trigger a toehold-mediated catalytic strand displacement to activate photosensitizer and achieve amplified therapeutic effect. The specific binding-induced activation allows the DNA circuit to distinguish diseased cells from healthy cells, reducing damage to nearby healthy cells. Moreover, the catalytic amplification reaction will only take place close to the target cancer cells, resulting in a high local concentration of singlet oxygen to selectively kill the target cells. The principle employed in this study demonstrated the feasibility of assembling a DNA circuit on cell membranes and could further broaden the utility of DNA circuits for applications in biology, biotechnology, and biomedicine.
Keywords: DNA nanocircuit, aptamer, photosensitizer, cancer therapy
As a minimally invasive therapeutic modality, photodynamic therapy (PDT) is already greatly used in clinical treatment of cancers. PDT can destroy cancer cells when light irradiates a photosensitizer (PS), generating reactive singlet oxygen (1O2).1 Briefly, PDT involves a two-step process, whereby a nontoxic PS is delivered to an organism and then activated by an appropriate light source. However, because the 1O2 has a limited lifetime and diffusion distance, efficient and reliable PDT depends on generating 1O2 with methods that offer the greatest selectivity.2
PDT selectivity is usually controlled at two levels. The first level controls the spatial localization of PS reagents. This approach has been actively pursued by specifically delivering PS to the tumor site with regional light shining, which has effectively improved PDT selectivity and efficiency. 3, 4 But the tendency to cause damage to surrounding normal tissues still exists. To achieve greater selectivity, a molecular activation layer is added to further control the specificity of the PS. At this level, the probe initially stays in the nontoxic state and can only be activated when it interacts with its corresponding trigger at the tumor site. For example, we and others have developed activatable PDT methods which can be triggered by biomarkers, including membrane proteins5 and extracellular proteases,6 as well as cellular environments (e.g., pH)7 or other external stimuli, including artificial molecular switches.8–10 Molecular activation allows the PS to distinguish diseased from healthy cells, thus greatly improving the selectivity of PDT.
To achieve higher oncolytic efficacy in tumors, a sufficient dosage of drugs should be administered at the tumor site. However, greater selectivity is typically achieved by introducing multiple activation processes, but at the cost of decreasing the active drug amounts at the tumor site. For example, the triggers of activatable PDT are usually biomarkers present inside or outside the cells. However, the limited amount of trigger elements in the disease cells coupled with low activation efficiency may dramatically decrease the activation and killing effects of the PS. Selective amplification would effectively solve this problem. Researchers have applied some enzymes, such as protease whose overexpression is correlated with specific diseases, to continually catalyze PS activation, thereby amplifying the PDT effects.11 However, the application of enzymes is often limited by their microenvironment, including pH and temperature, in turn reducing the applications of enzyme-activatable PDT. Thus, to achieve more robust PDT with selectivity and amplification effect, a suitable medium is required that can both recognize the target cell and amplify the therapeutic effect.
As carriers of genetic information with well-regulated and predictable structures, nucleic acids are promising materials for signal amplification based on their nanometer size and programmability. Recent advances in the field of nucleic acids have generated nucleic-acid based circuits, in which enzyme-free signal amplification can be achieved by simple nucleic acid hybridization, such as hybridization chain reaction,12,13 entropy-triggered hybridization catalysis14 and DNA hairpin fuel catalysis.15,16 These methods show promise in amplifying PDT with such properties as high amplification efficiency, environmental robustness and ability to interact with other naturally occurring molecules. Meanwhile, the exploration and development of special single-stranded oligonucleotides, well known as aptamers,17–19 have extended the recognition capabilities of nucleic acids from Watson-Crick base-pairing to interactions with various targets, such as small molecules,20 proteins,21 and cells,22 via the aptamers’ unique secondary or tertiary structures. We recently developed an effective method to generate aptamer-based molecular probes for the specific recognition and targeting of cancer cells.22–25 Therefore, by combining the recognition and amplification abilities of these oligonucleotides, more efficient and specific PDT methods can be developed.
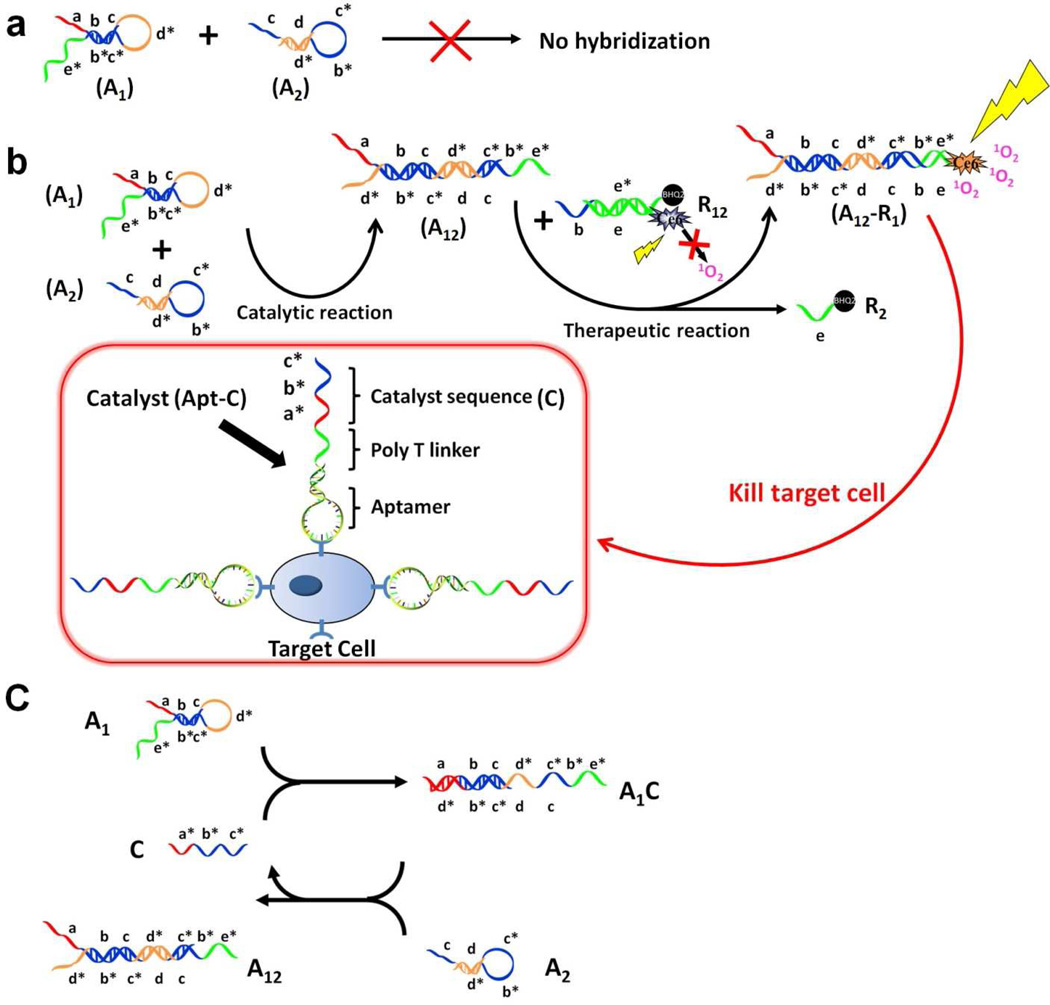
In this paper, we report the design of an aptamer-based DNA circuit capable of the selective recognition of cancer cells, controllable activation of PS and amplification of therapeutic effect. In particular, the amplification circuit motif comprises two DNA hairpins developed by Yin15 et al. and Li et al.16 In principle, two DNA hairpin structures, A1 and A2 initially do not hybridize with each other because of the effective block created by complementary domains. However, in the presence of another ssDNA sequence, termed catalyst (C), A1 and A2 can form a stable duplex without consuming C. As shown in Figure 1, A1, A2 and C contain a few functional domains labeled in lowercase letters. Complementarity between lettered domains is denoted by an asterisk. Initially, C can hybridize with the exposed toehold domain a of A1 and gradually open the stem of A1 to form intermediate A1C, but A1C has an exposed ssDNA domain c* able to hybridize with the exposed domain c in A2. Hence, after hybridization of c and c*, the sequence dc*b*d will undergo branch migration and displace the C sequence (c*b*a*) to form the A12 duplex. Importantly, the released C triggers further hybridizations of A1 and A2 in repeating cycles, thus providing the multiple-trigger effect absent in previous models. In this example, C catalyzes the formation of duplex A12 from A1 and A2 through a prescribed reaction pathway. The overall reaction is driven by a decrease in enthalpy resulting from the formation of A12 with a greater number of base pairs.
Figure 1. Working Scheme of DNA aptamer circuit on cell membrane.
(a). Scheme of the circuit without catalyst. (b). Scheme of the circuit on cell membrane. The circuit involves two individual steps. In the catalytic step, target cell labeled with Apt -C catalyzes DNA hairpins A1 and A2 to form duplex A12. In the therapeutic step, A12 can open duplex R12 and displace quencher-labeled single strand R2 to form A12–R1. Subsequently, Ce6-labeled R1 generates singlet oxygen (1O2) to kill cancer cells by irradiation at 404 nm. (c). Scheme of detailed reaction of DNA hairpins A1 and A2 catalyzed by C sequence. Different domains are labeled with different colors. All x domains are complementary to x*.
Selectivity is achieved by encoding the catalyst sequence C (a*b*c*) into an aptamer sequence (Apt-C) that targets cancer cells. To avoid forming undesired secondary structures, 17 poly T bases are used to separate the aptamer sequence and C. Under these conditions, the aptamer part can bind to the receptor on the target cancer cell membrane with a tail (C) exposed for the catalytic hybridizations of A1 and A2. Thus, one aptamer binding event can induce multiple hybridization events between A1 and A2 to form dsDNA A12, and the all-important amplification step essentially derives from the catalytic reaction.
To apply this cell-catalyzed hairpin amplification circuit to PDT therapy, dsDNA sequences denoted as R12 are employed to carry the photodynamic therapeutic reagents. Because of its high photosensitizing efficacy and low dark toxicity, Chlorin e6 (Ce6), a second-generation and easily modifiable photosensitizer, is modified on ssDNA R1, and a quencher, BHQ2, is conjugated on ssDNA R2 to quench the generation of 1O2 by Ce6 when no target cell is present. This design has several advantages. First, specific binding-induced activation allows the DNA circuit to distinguish diseased cells from healthy cells, reducing damage to nearby healthy cells which otherwise might be destroyed during PDT with conventional photosensitizers. Second, since the circulatory system in vivo can flush away the unbound aptamers and the catalyst C with aptamer is only present on the target cell membrane, the catalytic reaction will only take place close to the target cancer cells, resulting in a high local concentration of 1O2 to selectively kill the target cells. Third, traditional aptamer-based PDT has suffered from the drawback of insufficient killing effects from the 1:1 binding-induced singlet oxygen generation (SOG). While, in this new design, by incorporating the catalyst sequences of the hairpin amplification circuit on cell membranes, numerous binding-induced SOG events can be realized on each cell membrane. In addition, the uncatalyzed background of the circuit is nearly undetectable, resulting in fewer side effects to other healthy cells. Finally, to our best knowledge, this is the first design using the target cancer cell as the trigger to drive the DNA hybridizations. As such, this method may provide a universal strategy for signal amplification on cell membranes.
RESULTS AND DISCUSSION
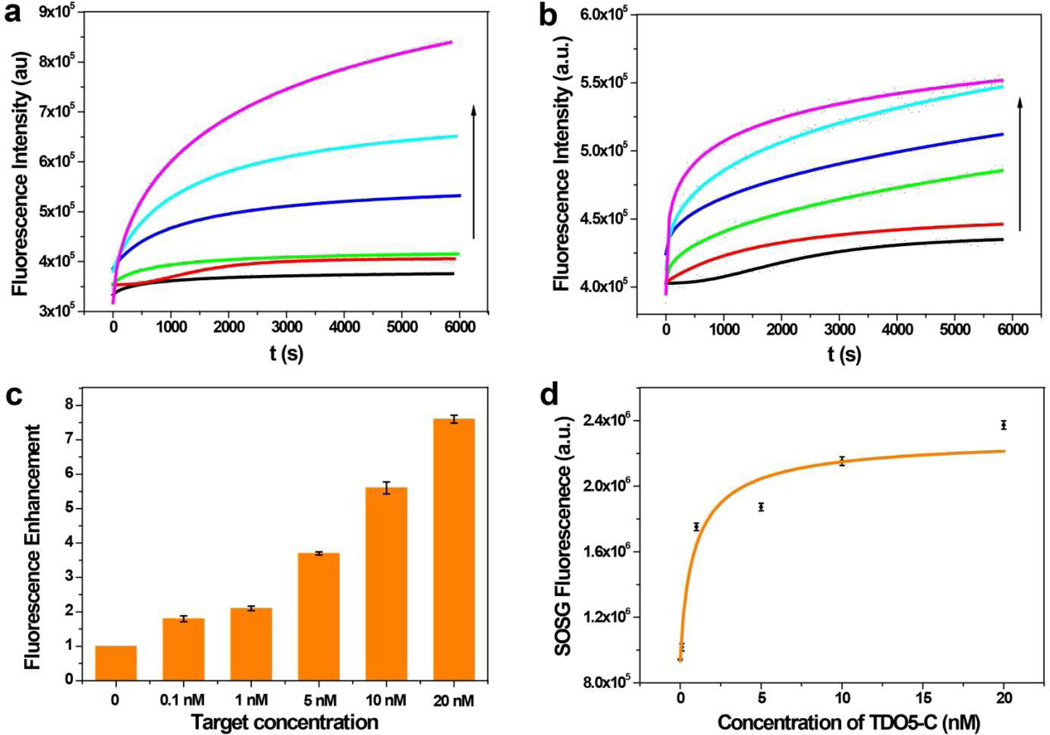
To demonstrate the effectiveness of the C (a*b*c*) in catalyzing the A1 and A2 hybridization, native gel electrophoresis was used. As shown in Figure S1 (Supporting Information), without C, A1 and A2 can be present stably without hybridization. However, when C is added, A1 and A2 hybridize with each other to form A12 with a yield even higher than that achieved by annealing of A1 and A2. To further study the amplification efficiency of the hairpin circuit, a FRET-based dsDNA R12 was designed with a stable fluorophore (FAM-R1) and quencher (DABCYL-R2) pair. To improve the thermostability and anti-enzymatic digestion ability of the R12 duplex, we incorporated 4 LNA (Locked nucleic acid) nucleotides into FAM-labeled R1.26 C was linked to an aptamer sequence TDO5, which targets acute lymphoblastic leukemia B-cells (Kd=74.7 nM) via a poly-T linker.23 Thus, TDO5-C was used as catalyst to initiate the A1/A2 hybridization, and the fluorescence was monitored. In the presence of different concentrations of TDO5-C (0–20 nM), dramatic signal enhancement was observed (Fig. 2a), indicating the effective catalytic effect of TDO5-C. In addition, the signals approached a maximum in 2 hr, indicating rapid kinetics of the catalytic hybridization.
Figure 2.
(a). Kinetics of DNA circuit containing A1, A2 and R12 with different concentrations of TDO5-C (fluorescence intensities corresponding to F1:n) monitored by FAM fluorescence. The colored lines represent 0, 0.1 nM, 1 nM, 5 nM, 10 nM and 20 nM of TDO5-C, respectively. (b). Kinetics of dsDNA R12 with different concentrations of A12 (fluorescence intensities corresponding to F1:1) monitored by FAM fluorescence. The colored lines represent 0, 0.1 nM, 1 nM, 5 nM, 10 nM and 20 nM of A12, respectively. (c). Comparison of the fluorescence enhancement fold of the catalytic DNA circuit and 1:1 displacement. The calculation is based on the equation Fold = , where fold is the fluorescence enhancement ratio of 1:n method to 1:1 method, B is the background fluorescence. The fluorescence intensities at 6000 s are used to plot against different target concentrations. Each bar presents the mean and standard deviation derived from three independent experiments. (d). The SOSG signal plotted as the function of TDO5-C concentration. The SOG was triggered by irradiation at 404 nm, the maximum absorption of Ce6, for 10 minutes. The SOSG fluorescence was obtained with excitation at 494 nm and emission from 500 nm to 600 nm. Each bar presents the mean and standard deviation derived from three independent experiments.
We also studied the fluorescence kinetics of the 1:1 displacement reaction (Fig. 2b). In particular, A12 was prepared by annealing equal concentrations of A1 and A2 in advance. Then, different concentrations of A12 were added to displace R12 in buffer solution. In Figure 2c, the fluorescence enhancement ratio between catalytic amplification circuit (1:n) and displacement reaction (1:1) is 8-fold at the target concentration of 20 nM after about 2 hr, indicating high amplification efficiency of this circuit.
For therapeutic applications with the DNA circuit, A1, A2 and R12 will be present together in the deactivated forms around cells, and small leakage hybridizations may occur. Therefore, we tested the leakage hybridization rate by measuring the fluorescence of buffer solution containing A1, A2 and R without TDO5-C for 8 hr (Fig. S2). Although a small leakage did occur, the result indicated that the second-order rate constant of uncatalyzed reaction could be estimated to be <10 M−1s−1, which is almost negligible for PDT applications.
Next, the photosensitizer Ce6 was conjugated to the ssDNA R1, and the BHQ-2 quencher was modified with ssDNA R2. Because of the close proximity between Ce6 and BHQ-2, up to 95% quenching efficiency of Ce6 was observed by our previous studies.27 Herein, the DNA hairpin circuit had significant fluorescence enhancement upon the addition of different concentrations of TDO5-C. This was illustrated by the Ce6 fluorescence which increased up to 10-fold with 20 nM TDO5-C in buffer (Fig. S3, Supporting Information). To evaluate the effect of different concentrations of TDO5-C on the amount of 1O2 generated by Ce6-modified R1, singlet oxygen sensor green (SOSG) was added, and its fluorescence enhancement was measured before and after irradiation at 404 nm. As shown in Figure 2d, SOSG fluorescence increased 3-fold with the introduction of 20 nM TDO5-C in the span of 1 hr, indicating that SOG could be mediated by TDO5-C.
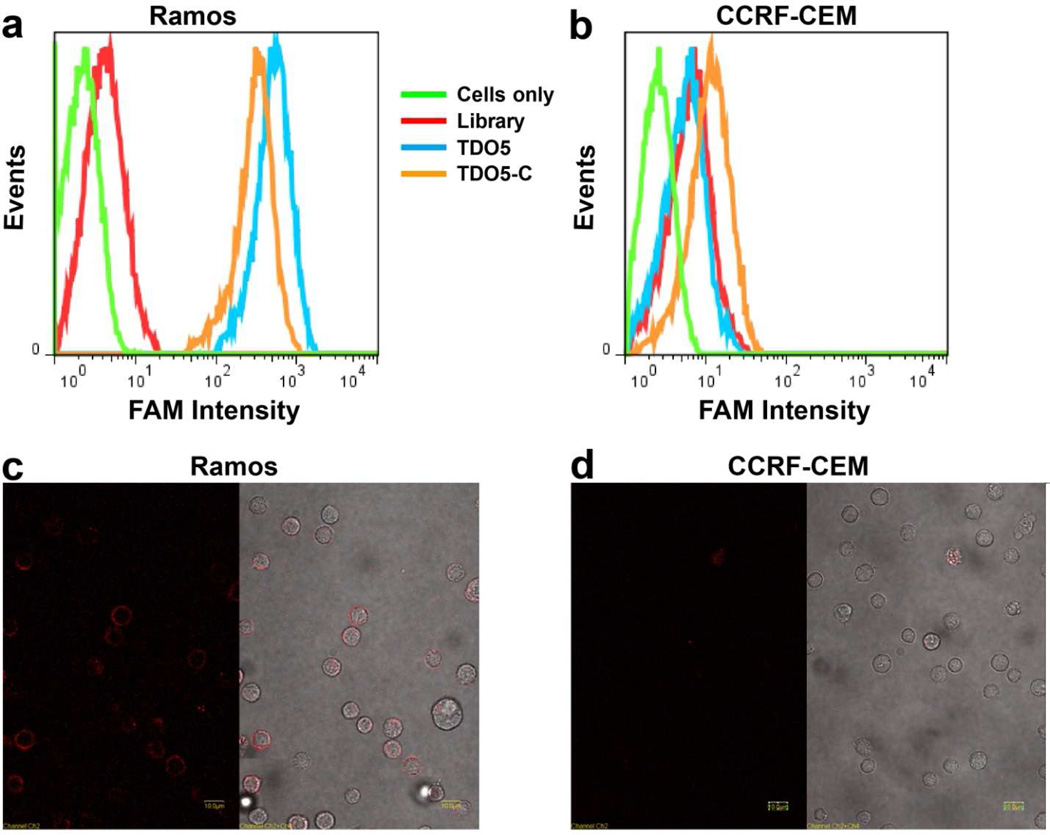
For proof of concept, a leukemia cell line was chosen as the target. Compared with solid tumor cells, leukemia cells are widespread in the circulatory system and are surrounded by normal blood cells. Under these circumstances, any nonspecific cytotoxin would also destroy the normal blood cells. Therefore, a therapeutic method which can selectively recognize and kill the target leukemia cells is highly desirable. As mentioned above, aptamer TDO5, which binds to the cancer cell membrane protein IgM with high affinity and selectivity, was used in our study with target cancer cell Ramos (acute lymphoblastic leukemia B-cells) and negative control cell CCRF-CEM (acute lymphoblastic leukemia T-cells). Therefore, if TDO5-C is present, it recognizes the target cancer cells and, importantly, also catalyzes the DNA hairpin hybridization to trigger SOG around cells. First, the selective binding of TDO5-C to Ramos cells was demonstrated by flow cytometry, as shown in Figure 3 a and b. Herein, aptamer TDO5 was used as positive control. Compared with TDO5, TDO5-C showed almost equally strong binding affinity to Ramos cells at 4°C, indicating that the cell membranes were partially covered by TDO5-C. However, both TDO5 and TDO5-C exhibited weak affinity to the control CCRF-CEM cells, as evidenced by only small fluorescence peak shifts. In addition, to confirm that TDO5-C was bound to the cell membrane surface without internalization, confocal microscopy images were taken with TMR-labeled TDO5-C incubated with Ramos and CCRF-CEM cells (Fig. 3 c and d). Since only the cell membrane surface was labeled with fluorescence, the strong binding and low uptake efficiency of TDO5-C makes it suitable for the catalysis of the hairpin circuit in close proximity to the target cancer cells.
Figure 3.
a and b, Flow cytometry results of FAM-labeled TDO5-C binding with Ramos (a) and CCRF-CEM (b). Aptamer TDO5 was used as positive control to show the maximum binding affinity. Cells: 200 k/sample; aptamer concentration: 250 nM. c and d, Confocal imaging of Ramos cells (c) and CCRF-CEM cells (d) incubated with 250 nM TMR-modified TDO5-C at 4°C. Fluorescence image (left). Overlap of optical image and fluorescence image (right). The scale bar is 10 µm.
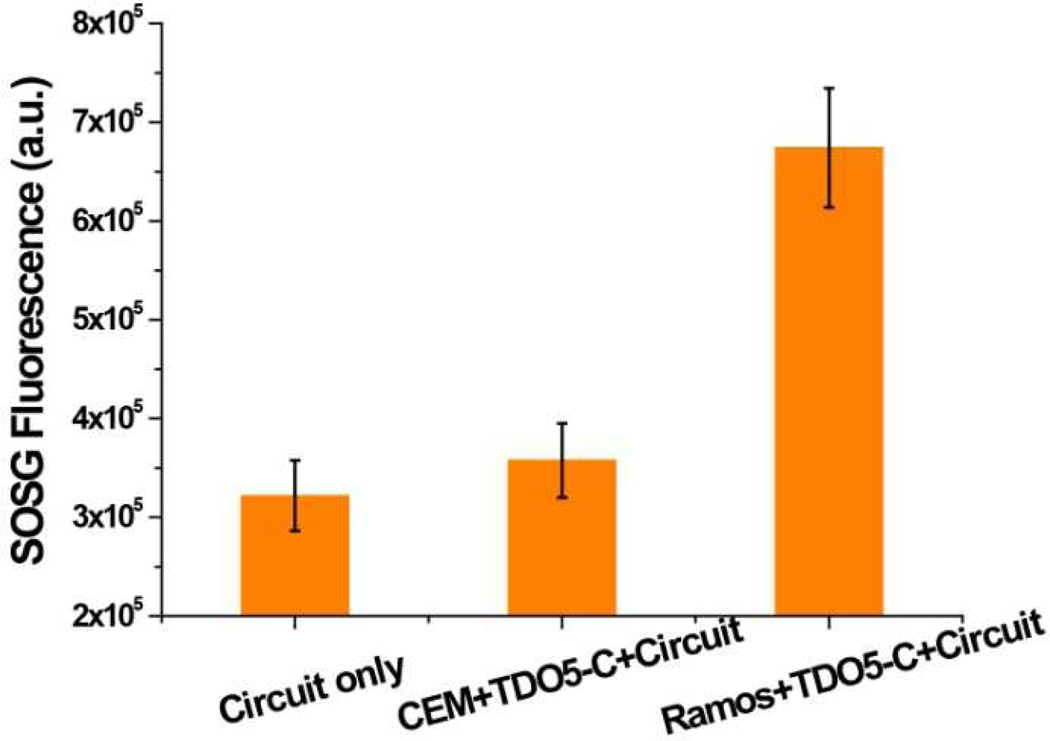
To determine whether the amplification effect of TDO5-C remains active on the cell membrane, SOG was evaluated by incubating TDO5-C-labeled cells with the circuit (A1, A2 and R12) in PBS buffer, followed by adding SOSG sensor and monitoring the fluorescence. As shown in Figure 4, obvious SOSG fluorescence enhancement was observed when incubating the circuit with TDO5-C-labeled target cells (Ramos), which can be attributed to the catalytic effect of TDO5-C on the target cell membranes. As a control, the SOG triggered by TDO5-C-labeled CCRF-CEM cells was studied. Little SOSG fluorescence enhancement was observed compared to the circuit only (A1, A2 and R12), indicating that the selective binding of TDO5-C can induce selective SOG.
Figure 4.
SOSG fluorescence of DNA circuit (A1, A2 and R12) incubated with buffer, TDO5-C-labeled CCEF-CEM cells (control) and TDO5-C-labeled Ramos cells (target). Each bar presents the mean and standard deviation derived from three independent experiments. (Cells: 200k/sample; λex=494 nm and λem=532 nm).
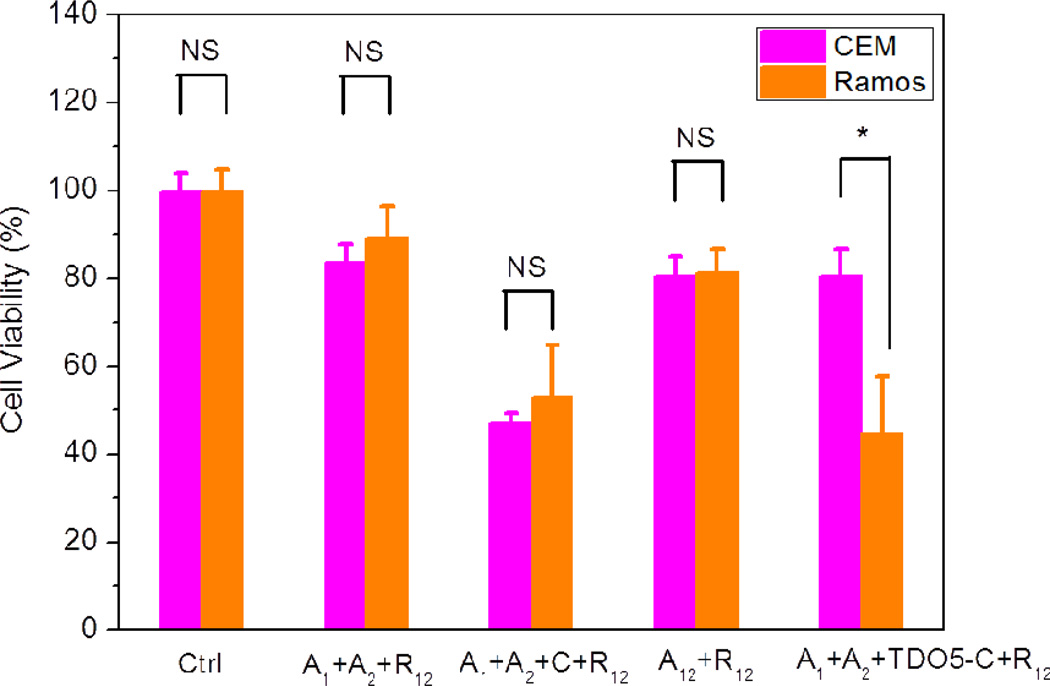
Cell destruction by PDT was studied by irradiation with white light. As indicated by the SOSG studies, 1O2 is produced after the binding of TDO5-C with Ramos followed by the hybridization reaction of A1 and A2 to form A12 and the displacement of R1 from R12 by A12. Therefore, the phototoxicity of the cell-surface circuit to cancer cells was studied by MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] assay. After 3 hr of light irradiation, the target cells (Ramos) and the control cells (CCRF-CEM) were cultured for 36 hr before evaluating cell viability with MTS reagent. Figure 5 shows the MTS data expressed as the mean viability (standard deviation). The statistical differences were assessed by Student's t-test. When A1, A2 and R12 were combined with nonlabeled cells and irradiated with white light, very little damage was observed to either target or control cells (cell viability of around 85%). These data are consistent with the previous SOG resulting from the weak leakage hybridizations of A1 and A2. However, when 100 nM free C was incubated with both cell lines, no statistical difference was evident (P > 0.30), and both cell types showed low cell viabilities of about 50%. Herein, because the free C sequence did not selectively recognize target cells, it catalyzed the amplification reaction without selectivity and caused relatively equal cell death for both target cells and control cells. On the other hand, when each of the cell lines was first incubated with TDO5-C, rinsed, and then incubated with A1, A2 and R12, high phototoxicity was observed for the target Ramos cells (45%), compared to 80% for the control cells (CCEF-CEM) with P < 0.02, indicating that TDO5-C can catalyze the hairpin circuit on specific cell membranes. In addition, we also found that the statistical toxicity difference between (A1+A2 + C+ R12) group and (A1+A2 + TDO5-C+ R12) group is evident (P < 0.001) for CEM cells, but not for Ramos cells (P > 0.3) as the consequence of catalytic selectivity of TDO5-C. Finally, to compare the cytotoxicity of our method with the 1:1 displacement method, we incubated the preannealed A12 (1 µ) and R12 with cells and found a much higher cell viability of 79% for both cell lines. Under these conditions, the 1:1 displacement method did not show any selectivity to the two cell lines based on the lack of recognition element (P > 0.30). These comparisons demonstrate the selective and amplified therapeutic effect of our method.
Figure 5.
Cell viability result using MTS assay. The in vitro cytotoxicity was measured after 36 hr of incubation in cell medium with 3 hr of white light irradiation. Cells: 500k/sample. Each bar presents the mean and standard deviation derived from three independent experiments. P values were calculated by Student's t-test: ns, nonsignificance: P > 0.30 and * for P < 0.02, n = 3.
CONCLUSION
In conclusion, our results demonstrate the feasibility of assembling a DNA circuit on cell membranes to achieve amplified and targeted photodynamic therapy. The DNA circuit, composed of four functional modules (A1, A2, R12 and TDO5-C) totally made of DNA, can greatly amplify the singlet oxygen generation and selectively kill cancer cells. In particular, the DNA hairpin amplification circuit can be catalyzed by specifically designed nucleic acid sequences. Many nucleic acids, including messenger RNA, microRNAs and small interfering RNAs, are important biomarkers for various diseases.28–30 If sequences for these biomolecules are available, the amplification hairpin DNA circuit can be designed to perform other biological and biomedical functions inside targeted disease cells with effective delivery methods. Second, increasing numbers of aptamers have been developed to target the membranes of a variety of cancer cell lines, thus establishing the universality of this DNA hairpin circuit for targeted and amplified therapy. Finally, as an application of DNA circuit to biological cells, the prototype DNA circuit demonstrated here has the potential to enhance DNA technology with new insights and will broaden the utility of DNA circuits for applications in biology, biotechnology, and biomedicine.
MATERIALS AND METHODS
Cell Culture
Ramos (CRL-1596, B-cell line, human Burkitt's lymphoma) and CCRF-CEM (CCL-119, T-cell line, human acute lymphoblastic leukemia) were cultured in RPMI 1640 medium (American Type Culture Collection) with 10% fetal bovine serum (FBS, Invitrogen, Carlsbad, CA, USA) and 0.5 mg/mL penicillin-streptomycin (American Type Culture Collection) at 37°C under a 5% CO2 atmosphere. Cells were washed before and after incubation with washing buffer [4.5 g/L glucose and 5 mM MgCl2 in Dulbecco's PBS with calcium chloride and magnesium chloride (Sigma-Aldrich)]. Binding buffer used for selection was prepared by adding yeast tRNA (0.1 mg/mL; Sigma-Aldrich) and BSA (1 mg/mL; Fisher Scientific) to the wash buffer to reduce background binding.
Ce6-modified DNA synthesis
An amino group (Glen Research Corp.) was incorporated at the 5’-end using the synthesis protocol specified by the company. After removing the MMT protection group on the 5’-amino of the sequence on machine, the CPG beads were washed with acetonitrile (ACN) 10 times and dried with nitrogen for off-machine coupling of Ce6. Each Ce6 molecule has three carboxyl groups for conjugation with the amino group. To improve the coupling efficiency and reduce the multiple coupling products, the amount of Ce6 was 10 times more than DNA product in the coupling reaction. Ten µmole Ce6 was mixed with an equal molecular amount of N,N’-dicyclohexylcarbodiimide (DCC, Sigma-Aldrich, Inc.) and N-hydroxysuccinimide (NHS, Sigma-Aldrich, Inc.) and dissolved in 250 µL N,N-Dimethylformamide (DMF) for the activation reaction with 1 hr stirring. The coupling reaction was performed with vigorous stirring overnight, followed by washing with ACN. Then the DNA product was purified by HPLC.
DNA purification
Native PAGE was applied to purify the A1, A2 hairpin strands to remove excess strands and avoid undesired system leakage. A1 and A2 were annealed at concentrations of around 50 µM in 1× TAE-Mg buffer (40 mM Tris-Acetate-EDTA, pH 8.0, 12.5 mM Mg(Ac)2) and cooled to room temperature. Native PAGE gels (12%) in 1× TAE-Mg buffer were run at 110 V for 90 minutes at 4°C and stained with GelRed stain solution (Biotium, Inc., Hayward, CA). Only the sharp bands were cut from the gels, chopped into small pieces, and soaked in 1× TAE-Mg2+ buffer for 24 hr. The buffer was extracted and concentrated with centrifugal filter devices (Millipore, Billerica, MA). Finally, the purified DNA sequences were quantified by UV spectrometry and kept in buffer for future use.
Fluorescence kinetics of the DNA hairpin circuit in buffer
All fluorescence measurements were performed using a Fluorolog (Jobin Yvon Horiba) with a 100 µL macro cuvette. DNA sequences A1, A2 were separately refolded in the Fluo buffer (20mM Tris, pH 7.5; 140mM NaCl; 5mM KCl). This and other refolding reactions involved heating to 90°C for 1 min, followed by slowly decreasing the temperature to 25°C at a rate of 0.1°C s−1. After purification by gel electrophoresis, the annealed sequences were stocked in Fluo buffer for later use. An amount of 10 µM stock of R was prepared by annealing 10 µM FAM-R1 and 15 µM DABCYL-R2 in Fluo buffer. An excess of R2 ensures efficient quenching of R1, but does not interfere with the readout of A1 and A2. A mixture of 100 nM A1, 100 nM A2 and 150 nM R12 was prepared in 1×Fluo buffer. The fluorescence at 518 nm was monitored at 25 °C after adding different amounts of TDO5-C. To evaluate the amplification effect of the circuit, a 1:1 displacement reaction was performed under the same conditions by mixing 10 µM A1 and A2 in 1×Fluo buffer and heating to 90°C for 3 minutes, followed by slowly decreasing the temperature to 25 °C to form the stable duplex A12. A 150 nM sample of R12 was incubated in 1×Fluo buffer, followed by adding different concentrations of A12 and monitoring the fluorescence.
Test of the fluorescence response and SOG of the DNA hairpin circuit
For these experiments, Ce6-modifed R1 and BHQ2-modified R2 were used to form duplex R12. To study the Ce6 fluorescence response to different concentrations of TDO5-C, 100 nM A1, 100 nM A2 and 150 nM R12 were mixed in 1×Fluo buffer. The excitation wavelength was set at 404 nm with emission scanned from 600 nm to 800 nm. When testing the SOG, the concentration of SOSG probe was set at 2 µM together with 100 nM A1, 100 nM A2 and 150 nM R12 in 1×Fluo buffer. To extend the lifetime of 1O2 and increase the sensitivity of SOG assay, all buffers and samples were prepared using deuterium oxide. The SOG was triggered by irradiation at 404 nm, the maximum absorption of Ce6, for 10 minutes. The SOSG fluorescence was obtained with excitation at 494 nm and emission from 500 nm to 600 nm.
Test of the SOG response of the DNA hairpin circuit to cancer cells
3×105 of Ramos and CCRF-CEM cells were prepared in 100 µL of washing buffer separately. Fifty pmol of nonlabeled TDO5-C were added and incubated for 30 minutes. After washing the cells twice with washing buffer, the two different cell types were resuspended in 100 µL of washing buffer. Then 100 nM A1, 100 nM A2 and 150 nM R12 were incubated with cells for 1 hr. To extend the lifetime of 1O2 and increase the sensitivity of SOG assay, all buffers and samples were prepared using deuterium oxide. Two micromolar SOSG sensors were added to the cell medium, and SOG was triggered by irradiation at 404 nm, the maximum absorption of Ce6, for 10 minutes. The fluorescence was monitored at 25 °C with the excitation wavelength at 494 nm and emission from 500 nm to 600 nm.
Flow Cytometric Analysis
In flow cytometry tubes, 250 nM Biotin-labeled TDO5 or TDO5-C was incubated with 3×105 Ramos or CCRF-CEM cells at 4°C in 200 µL of binding buffer for 30 minutes. The cells were washed twice with 1 mL of washing buffer, centrifuged at 1300 rpm for 3 min, and then resuspended in 200 µL of washing buffer. One µL (1:400) Streptavidin-conjugated PE dye was incubated with the cells for another 20 minutes and washed twice using washing buffer. The cells were analyzed on a FACScan Flow Cytometer by counting 30,000 events. The PE-labeled unselected ssDNA library was used as a negative control.
Confocal Imaging of Cells Bound with Aptamer
For confocal imaging, the Ramos and CCEF-CEM cells were incubated with 50 pmol of TAMRA-labeled TDO5-C in 100 µL of binding buffer containing 20% FBS on ice for 30 min. The cells were washed twice with 1 mL of washing buffer, centrifuged at 1300 rpm for 3 min, and then resuspended in 100 µL of washing buffer. Twenty microliters of cell suspension bound with TAMRA-labeled TDO5-C were dropped on a thin glass slide placed above a 60× objective on the confocal microscope. Imaging of the cells was performed on an Olympus FV500-IX81 confocal microscope. A 5-mW, 543-nm He-Ne laser was the excitation source for TAMRA throughout the experiments. The objective used for imaging was a PLAPO60XO3PH 60× oil-immersion objective with a numerical aperture of 1.40 (Olympus).
Cytotoxicity Study
The cytotoxicity study was performed using the Cell Titer 96 Aqueous One Solution cell proliferation assay (MTS) for Ramos and CCRF-CEM cell lines in a 96-well cell culture plate at 500k/well, 100 µL. Five groups of cell samples were set up as follows: group 1, cells only; group 2, cells+ 3.3 µL 30 µM A1, 1.3 µL 77 µM A2 and 2 µL 50 µM Ce6-modified R12; group 3, cells incubated with the same amount of A1, A2 and R12, together with 1 µL 10 µM free C; group 4, cells incubated with 1 µL 100 µM preannealed A12 and 2 µL 50 µM Ce6-modified R12. For group 5, the cells were incubated with TDO5-C conjugates for 30 min at 4°C, followed by centrifugation at 1300 rpm for 3 min to remove the unbound DNA. Then probes in the same amount as that of group 2 were added. All groups of cells were suspended in cell medium (No FBS) and then irradiated with white light on ice for 3 hr. After irradiation, the cells were incubated in a CO2 incubator for 36 hr. Finally, a 6×-concentrated MTS solution (120 µL/well) in RPMI 1640 medium solution was added to each well and incubated at 37 °C for 2 h. The absorbance value at 490 nm was determined by a VersaMax microplate reader (Molecular Devices, Inc., Sunnyvale, CA).
Supplementary Material
Acknowledgment
We thank Dr. K. R. Williams for manuscript review. This work is supported by grants awarded by the National Key Scientific Program of China (2011CB911000), the Foundation for Innovative Research Groups of NSFC (Grant 21221003), China National Instrumentation Program 2011YQ03012412 and by the National Institutes of Health (GM066137, GM079359 and CA133086).
Footnotes
Conflict of Interest: The authors declare no competing financial interest.
Supporting Information available: Materials and methods, DNA sequences, gel electrophoresis to demonstrate the catalysis effect of DNA circuit, and fluorescence measurement for the performance of the circuit. This material is available free of charge via the Internet at http://pubs.acs.org.”
References
- 1.Dougherty TJ, Gomer CJ, Henderson BW, Jori G, Kessel D, Korbelik M, Moan J, Peng Q. Photodynamic Therapy. J. Natl. Cancer Inst. 1998;90:889–905. doi: 10.1093/jnci/90.12.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bugaj AM. Targeted Photodynamic Therapy - A Promising Strategy of Tumor Treatment. Photoch. Photobio. Sci. 2011;10:1097–1109. doi: 10.1039/c0pp00147c. [DOI] [PubMed] [Google Scholar]
- 3.Mallikaratchy P, Tang Z, Tan W. Cell Specific Aptamer-Photosensitizer Conjugates as a Molecular Tool in Photodynamic Therapy. ChemMedChem. 2008;3:425–428. doi: 10.1002/cmdc.200700260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang X, Huang J, Wang K, Li W, Cui L, Li X. Angiogenin-Mediated Photosensitizer-Aptamer Conjugate for Photodynamic Therapy. ChemMedChem. 2011;6:1778–1780. doi: 10.1002/cmdc.201100226. [DOI] [PubMed] [Google Scholar]
- 5.Wang J, Zhu G, You M, Song E, Shukoor MI, Zhang K, Altman MB, Chen Y, Zhu Z, Huang CZ, et al. Assembly of Aptamer Switch Probes and Photosensitizer on Gold Nanorods for Targeted Photothermal and Photodynamic Cancer Therapy. ACS Nano. 2012;6:5070–5077. doi: 10.1021/nn300694v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng G, Chen J, Stefflova K, Jarvi M, Li H, Wilson BC. Photodynamic Molecular Beacon as an Activatable Photosensitizer Based on Protease-Controlled Singlet Oxygen Quenching and Activation. Proc. Natl. Acad. Sci. U.S.A. 2007;104:8989–8994. doi: 10.1073/pnas.0611142104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonnell SO, Hall MJ, Allen LT, Byrne A, Gallagher WM, O'Shea DF. Supramolecular Photonic Therapeutic Agents. J. Am. Chem. Soc. 2005;127:16360–16361. doi: 10.1021/ja0553497. [DOI] [PubMed] [Google Scholar]
- 8.Tang Z, Zhu Z, Mallikaratchy P, Yang R, Sefah K, Tan W. Aptamer-Target Binding Triggered Molecular Mediation of Singlet Oxygen Generation. Chem. Asian J. 2010;5:783–786. doi: 10.1002/asia.200900545. [DOI] [PubMed] [Google Scholar]
- 9.Zhu Z, Tang Z, Phillips JA, Yang R, Wang H, Tan W. Regulation of Singlet Oxygen Generation Using Single-Walled Carbon Nanotubes. J. Am. Chem. Soc. 2008;130:10856–10857. doi: 10.1021/ja802913f. [DOI] [PubMed] [Google Scholar]
- 10.Clo E, Snyder JW, Voigt NV, Ogilby PR, Gothelf KV. DNA-Programmed Control of Photosensitized Singlet Oxygen Production. J. Am. Chem. Soc. 2006;128:4200–4201. doi: 10.1021/ja058713a. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Stefflova K, Niedre MJ, Wilson BC, Chance B, Glickson JD, Zheng G. Protease-Triggered Photosensitizing Beacon Based on Singlet Oxygen Quenching and Activation. J. Am. Chem. Soc. 2004;126:11450–11451. doi: 10.1021/ja047392k. [DOI] [PubMed] [Google Scholar]
- 12.Dirks RM, Pierce NA. Triggered Amplification by Hybridization Chain Reaction. Proc. Natl. Acad. Sci. U.S.A. 2004;101:15275–15278. doi: 10.1073/pnas.0407024101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang J, Wu Y, Chen Y, Zhu Z, Yang X, Yang CJ, Wang K, Tan W. Pyrene-Excimer Probes Based on the Hybridization Chain Reaction for the Detection of Nucleic Acids in Complex Biological Fluids. Angew. Chem., Int. Ed. 2011;50:401–404. doi: 10.1002/anie.201005375. [DOI] [PubMed] [Google Scholar]
- 14.Zhang DY, Turberfield AJ, Yurke B, Winfree E. Engineering Entropy-Driven Reactions and Networks Catalyzed by DNA. Science. 2007;318:1121–1125. doi: 10.1126/science.1148532. [DOI] [PubMed] [Google Scholar]
- 15.Yin P, Choi HMT, Calvert CR, Pierce NA. Programming Biomolecular Self-Assembly Pathways. Nature. 2008;451:318–324. doi: 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- 16.Li B, Ellington AD, Chen X. Rational, Modular Adaptation of Enzyme-Free DNA Circuits to Multiple Detection Methods. Nucleic Acids Res. 2011;39:e110. doi: 10.1093/nar/gkr504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellington AD, Szostak JW. In Vitro Selection of RNA Molecules That Bind Specific Ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 18.Tuerk C, Gold L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 19.Robertson DL, Joyce GF. Selection in Vitro of an RNA Enzyme That Specifically Cleaves Single-Stranded DNA. Nature. 1990;344:467–468. doi: 10.1038/344467a0. [DOI] [PubMed] [Google Scholar]
- 20.Huizenga DE, Szostak JW. A DNA Aptamer That Binds Adenosine and ATP. Biochemistry. 1995;34:656–665. doi: 10.1021/bi00002a033. [DOI] [PubMed] [Google Scholar]
- 21.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of Single-Stranded DNA Molecules That Bind and Inhibit Human Thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 22.Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. Aptamers Evolved from Live Cells as Effective Molecular Probes for Cancer Study. Proc. Natl. Acad. Sci. U.S.A. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang Z, Shangguan D, Wang K, Shi H, Sefah K, Mallikratchy P, Chen HW, Li Y, Tan W. Selection of Aptamers for Molecular Recognition and Characterization of Cancer Cells. Anal. Chem. 2007;79:4900–4907. doi: 10.1021/ac070189y. [DOI] [PubMed] [Google Scholar]
- 24.Sefah K, Tang ZW, Shangguan DH, Chen H, Lopez-Colon D, Li Y, Parekh P, Martin J, Meng L, Phillips JA, et al. Molecular Recognition of Acute Myeloid Leukemia Using Aptamers. Leukemia. 2009;23:235–244. doi: 10.1038/leu.2008.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shangguan D, Meng L, Cao ZC, Xiao Z, Fang X, Li Y, Cardona D, Witek RP, Liu C, Tan W. Identification of Liver Cancer-Specific Aptamers Using Whole Live Cells. Anal. Chem. 2008;80:721–728. doi: 10.1021/ac701962v. [DOI] [PubMed] [Google Scholar]
- 26.Wang L, Yang CJ, Medley CD, Benner SA, Tan W. Locked Nucleic Acid Molecular Beacons. J. Am. Chem. Soc. 2005;127:15664–15665. doi: 10.1021/ja052498g. [DOI] [PubMed] [Google Scholar]
- 27.Tang Z, Mallikaratchy P, Yang R, Kim Y, Zhu Z, Wang H, Tan W. Aptamer Switch Probe Based on Intramolecular Displacement. J. Am. Chem. Soc. 2008;130:11268–11269. doi: 10.1021/ja804119s. [DOI] [PubMed] [Google Scholar]
- 28.Kim VN. Small RNAs: Classification, Biogenesis, and Function. Mol. Cells. 2005;19:1–15. [PubMed] [Google Scholar]
- 29.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y. Multi-Input RNAi-Based Logic Circuit for Identification of Specific Cancer Cells. Science. 2011;333:1307–1311. doi: 10.1126/science.1205527. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.