Abstract
Upon activation, the epidermal growth factor (EGF) receptor becomes phosphorylated and triggers a vast signaling network that has profound effects on cell growth. The EGF receptor is observed to assemble into clusters after ligand binding and tyrosine kinase autophosphorylation, but the role of these assemblies in the receptor signaling pathway remains unclear. To address this question, we measured the phosphorylation of EGFR when the EGF ligand was anchored onto laterally mobile and immobile surfaces. We found that cells generated clusters of ligand-receptor complex on mobile EGF surfaces, and displayed a lower ratio of phosphorylated EGFR to EGF when compared to immobilized EGF that is unable to cluster. This result was verified by tuning the lateral assembly of ligand-receptor complexes on the surface of living cells using patterned supported lipid bilayers. Nanoscale metal lines fabricated into the supported membrane constrained lipid diffusion and EGF receptor assembly into micron and sub-micron scale corrals. Single cell analysis indicated that clustering impacts EGF receptor activation, and larger clusters (> 1 µm2) of ligand-receptor complex generated lower EGF receptor phosphorylation per ligand than smaller assemblies (< 1 µm2) in HCC1143 cells that were engaged to ligand-functionalized surfaces. We investigated the mechanism of EGFR clustering by treating cells with compounds that disrupt the cytoskeleton (Latrunculin-B), clathrin-mediated endocytosis (Pitstop2), and inhibit EGFR activation (Gefitinib). These results help elucidate the nature of large-scale EGFR clustering, thus underscoring the general significance of receptor spatial organization in tuning function.
Introduction
Cellular communication is vital for the survival of multicellular organisms, and dictates cellular processes that range from tissue patterning and organization to mounting an immune response to specific threats. Much of the information exchanged between cells is in the form of chemical signals that are received and interpreted by thousands of receptors found in the cell membrane. While the field has generally been focused on chemical inducers that regulate specific pathways, recent evidence has shown that the spatial organization of cell surface receptors on length scales spanning the molecular to the size of the cell itself can also play a role in cellular signal regulation.1–4 Some of the earliest evidence suggesting that oligomerization of membrane receptors plays a role in signaling comes from studies of the FCεRI receptor, where oligomers showed increasing levels of activation in comparison to monomers and dimers.5 Further experiments making use of synthetic multivalent ligands have likewise revealed signaling outcomes that are unique to multivalent ligands.6, 7
Many membrane receptors, including the toll-like,8 EGF,9–17 ErbB family,17, 18 T-cell,2, 19, 20 Fas (CD95),21–23 and Ephrin1 have been found to assemble into higher-order structures comprised of tens to thousands of receptors whose signaling levels are correlated to cluster formation. In the immunological synapse, it has been observed that spatial patterning of antigens and their cognate T-cell receptors dictates the intensity of T-cell activation.2, 19, 24 Recent studies have also shown that levels of metalloprotease recruitment to the EphA2 receptor is related to receptor clustering.1, 25 Additionally, progressive clustering of Fas (CD95) induced by ligand binding has been shown to stabilize the individually weak interactions of the death-induced signaling complex.23 These results suggest that signal transduction is not exclusively the result of ligand-receptor binding, and is rather an ensemble process that transcends chemical recognition to include supramolecular organization and spatial patterning within the fluid membranes of cells. In light of this, a more detailed understanding of how ligand organization influences cell signaling pathways is needed in order to achieve system-level control of cell fate and function.
The epidermal growth factor receptor (EGFR) is one of the most well studied receptors in cell biology, and is a member of the ErbB family of receptor tyrosine kinases that also includes ErbB2 (HER2), ErbB3, and ErbB4. It has long been a receptor of interest due to its involvement in many types of cancer, and was one of the first mitogenic receptors to be characterized.26 In the conventional model of EGFR activation, the receptor binds its ligand (EGF) and undergoes dimerization and autophosphorylation, thus triggering a signaling cascade that proceeds to the transcriptional level.18, 27 Homo- and hetero-dimerization are hallmarks of ErbB family signaling, and these dynamic receptor associations have broad biochemical and biomedical significance.18
Several lines of biophysical studies confirm the formation of EGFR clusters across different cell line models. For example, near-field scanning optical fluorescence microscopy (a super-resolution imaging technique) provided high-resolution images of ligand-bound EGFR clusters on the surface of fixed HeLa cells. The data showed that cells formed a range of cluster sizes with an average diameter of 150 nm ± 80 nm after 10 min of ligand incubation.10 Another validation comes from an investigation using transmission electron microscopy (TEM) of gold nanoparticle-antibody conjugates specific for EGFR which showed the assembly of nanometer scale clusters on the surface of SKBR3 cells.28 Image correlation spectroscopy (ICS) and its derivative techniques have also contributed to the quantification and characterization of EGFR clustering. Number and brightness analysis (a type of ICS) revealed the formation of EGFR oligomers in A431 cells after 30 min of EGF treatment. Importantly, these oligomers contained three-fold higher levels of phosphorylated molecules of EGFR than bound EGF molecules.29 In another study, imaging of surface plasmon coupling between gold nanoparticles attached to EGFR revealed the formation of large EGFR assemblies on the surface of A431 cells but cluster size was not quantified.17 Förster resonance energy transfer-fluorescence lifetime imaging microscopy (FRET-FLIM) imaging in combination with ICS also showed that ligand bound and activated EGFR exists primarily as a higher-order oligomer, and that clusters of activated EGFR contain at minimum four times the amount of receptor as inactive clusters in HEK293 cells.13 When combined with flow cytometry, FRET-FLIM data showed that after ligand treatment, EGFR homocluster size increased from 4 to 10 molecules per cluster on the surface of A431 cells.14 Finally, single molecule tracking experiments showed a slowed rate of EGFR diffusion within 20 s of ligand binding, which suggested that EGFR forms large oligomers.9 Taken together, these experiments indicate that upon ligand binding EGFR assembles into clusters that may contain tens to tens of thousands of molecules and diffuse as a single unit in the plasma membrane.
One of the major challenges in the field pertains to developing a quantitative relationship between EGFR clustering and receptor activation. To address this goal, we use the recently developed nanopatterned supported lipid bilayer technique along with surface immobilization methods to control EGFR clustering levels in living cells and then quantify the effect of EGFR organization on its phosphorylation levels.1, 30 We find that large-scale clustering of EGFR dampens its phosphorylation intensity in HCC1143 breast cancer cells, and that kinase and cytoskeleton activity, and endocytosis machinery contribute to this clustering behavior.
Results and Discussion
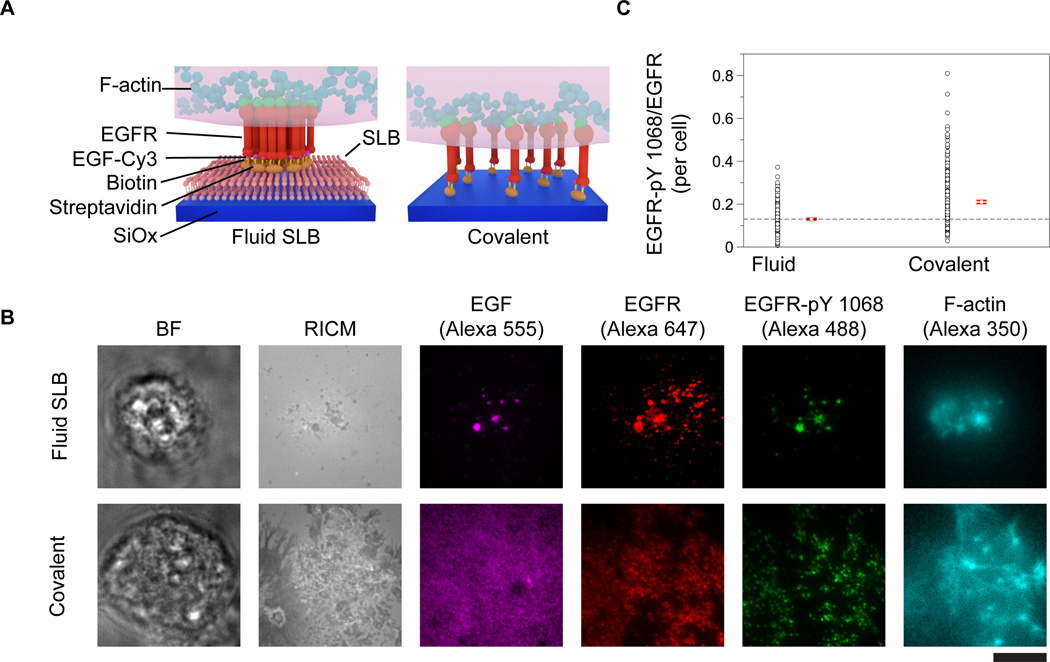
To investigate the role of EGFR cluster formation on activation levels, it is necessary to use methods that allow one to externally control receptor clustering within the cell membrane and to measure its corresponding level of activation within individual cells. To initially explore this question, we tethered the EGF ligand onto two different types of surfaces (Fig. 1A). The EGF ligand was anchored either to a laterally mobile fluid supported lipid bilayer, or to a covalently functionalized glass slide that prevents lateral diffusion of ligand (Fig. 1A). The motivation for using these surfaces was to contrast the activation levels of ligand-bound receptors that self-assemble into molecular clusters compared to receptor complexes that are spatially constrained.
Figure 1. EGFR stimulation using mobile and immobile surface-tethered ligands.
(A) Schematic illustrations of the three EGF-functionalized surfaces employed in this study. Supported lipid bilayer surfaces were formed by exposing lipid vesicles to etched glass surfaces, while the covalent substrates were fabricated using silane coupling as described in the methods section. In both of these surfaces, a biotinylated EGF ligand was anchored to streptavidin-functionalized surfaces. (B) HCC1143 cells were plated onto the EGF-functionalized surfaces and incubated for 1 h at 37° C, 5% CO2. After fixing and staining, cells were imaged, revealing the formation of large ligand (EGF, Alexa-555) clusters on the fluid surfaces that colocalized with EGFR-pY 1068 (Alexa 488), EGFR (Alexa 647), and F-actin (Alexa 350). Cells plated on covalently functionalized substrates displayed activated (phosphorylated) EGFR, however cell-induced clustering of the ligand was not observed. Scale bar is 10 µm. (C) Plot showing the EGFR-pY 1068/EGFR ratio for each type of surface. The red dot and lines represent the mean and standard error of the ratio for individual cells, respectively. n = 257 and 332 cells and the standard error = 0.004, and 0.007 a.u. for the fluid and covalent surfaces, respectively.
Fluid supported lipid bilayers were formed on the surface of clean glass slides by exposing the surface to 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) vesicles that spontaneously rupture and fuse onto the glass in the presence of aqueous buffer.2, 16, 20, 30 The supported lipid bilayers were doped with biotin-functionalized lipids (0.01%- 0.1%) to tether the EGF through biotin-streptavidin conjugation. The lipid membrane and surface-anchored EGF were laterally fluid as confirmed by fluorescence recovery after photobleaching experiments (Supplementary Fig. 1). The surface density of EGF was found to be 280 ± 20 EGF molecules per µm2 for the membrane surfaces doped with 0.1% biotin-functionalized lipid, and this value was determined by using quantitative fluorescence imaging using TRITC-DHPE doped supported lipid bilayers as a reference (see methods for details).31 The density of tethered EGF on the 0.1% biotin-doped fluid membrane matches the density of receptor expressed on the surface of breast cancer cell lines, such as BT474 (~900 EGFR molecules/µm2 assuming a 15 µm diameter cell), that overexpress EGFR, within one order of magnitude.32
In order to immobilize the EGF ligand onto a surface and to prevent its lateral mobility, we directly anchored the EGF ligand to glass surfaces covalently functionalized with biotin and bound to streptavidin (Fig. 1A). Substrates covalently functionalized with biotin groups were prepared by etching glass coverslips in piranha (3:1 H2SO4 (glacial):H2O2 (30%)) and then functionalizing with aminopropyltriethoxy silane. The terminal amine of the silane was then coupled to an N-hydroxysuccinimidyl ester-biotin linker and subsequently incubated with streptavidin. These surfaces presented the EGF ligand at a density of 250 ± 20 molecules/µm2 and this density matched the density of a 0.075% biotin-doped lipid bilayer (230 ± 20 EGF molecules/ µm2). Therefore, these two types of surfaces were used to stimulate cells since they display similar EGF surface densities and use the same conjugation chemistry, but the difference is that the ligand is not capable of lateral movement with the covalently modified glass slides.
In a typical experiment, human breast cancer cells that overexpress the EGFR (HCC1143) were plated onto each surface and allowed to incubate for 1 h at 5% CO2, 37°C. Cells were then fixed and stained via immunohistochemistry for the EGFR, phosphorylated tyrosine residue 1068 of the intracellular domain of EGFR (EGFR pY 1068), and also stained for F-actin using phalloidin-Alexa 350. We found that cells engaged the EGF-functionalized fluid supported lipid bilayer surface and formed large-scale assemblies of receptor that were colocalized with EGF, EGFR-pY 1068 and F-actin (Fig. 1B). In contrast, cells that were plated on the non-mobile EGF ligand substrates were strongly adhered to the surface through EGF-EGFR binding interactions, but did not translocate ligand-receptor complexes (Fig. 1B). Nonetheless, ligand-bound receptors, as indicated by the EGF and EGFR immunostains, were highly phosphorylated and colocalized with F-actin in both cases (Fig. 1B). Note that although the fluorescence immunostaining images of EGFR on the fluid and non-fluid surfaces suggests the formation of dense receptor aggregates, only the fluid membrane surfaces allow for lateral translocation and molecular assembly of ligand-receptor complexes. Note that the lack of complete colocalization between the covalently tethered EGF and receptor is due to incomplete ligand-receptor binding. In contrast, the fluid EGF is free to diffuse and concentrate at the locations of EGFR cluster assemblies (Fig 1B).
To compare EGFR clustering induced by membrane-tethered ligand to that induced by soluble EGF, HCC1143 cells were incubated on cyclic Arg-Gly-Asp-Phe-Lys (cRGDfK, an integrin-binding peptide that facilitates cell adhesion) functionalized supported membranes for 1h and then treated with 1.65 nM EGF-Alexa 647 for 5 min. Soluble ligand-stimulation generated clusters of EGFR on the surface of HCC1143 cells, in agreement with literature precedent.10, 29 We found that the mean diameter of soluble EGF-induced clusters was generally smaller than that observed during stimulation with fluid membrane-tethered EGF. While this experiment does show that clusters form when EGFR-expressing cells are treated with soluble EGF, any further comparison is not possible due to the inherent differences between tethered and soluble ligand.33, 34 (Supplementary Fig. 2).
To compare receptor activation levels across the immobilized and laterally fluid EGF surfaces, we normalized the EGFR-pY 1068 level by the EGFR signal intensity in order to determine the ratio of activated receptor to total receptor within individual cells plated on each surface type. We did not use the EGF intensity under each cell due to the fact that not all ligand was bound to receptor on the non-mobile surfaces. Image analysis indicated differences between the average EGFR-pY 1068/EGFR ratio for cells on each type of substrate (Supplementary Fig. 3). The covalent surfaces displayed a 62% (0.21 ± 0.007) larger EGFR-pY 1068/EGFR ratio, respectively, compared to the fluid supported lipid bilayer surface ratio (0.13 ± 0.004). This suggests that the assembly of ligand-receptor complexes within clusters on the fluid membrane surfaces dampens receptor activation at this experimental time point (t = 60 min) (Fig. 1C).
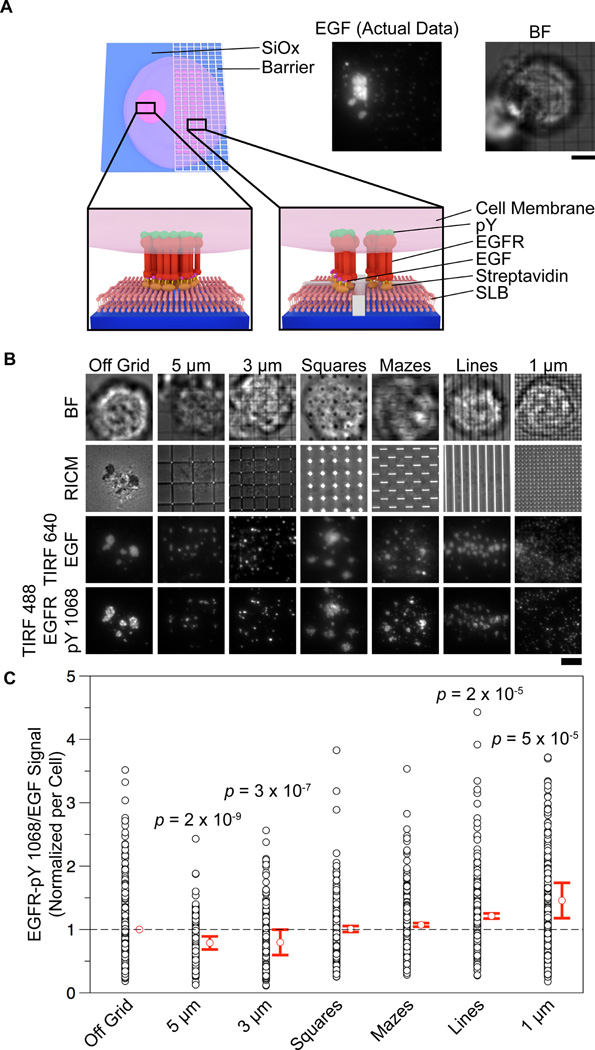
To quantify how clustering influences receptor activation, we manipulated receptor cluster size by patterning metal lines into the supported lipid bilayer. In this approach, nanopatterned chromium lines are fabricated on a glass slide and act as physical diffusion barriers to confine lipid diffusion within discrete corrals (schematic: Fig. 2A, Supplementary Fig. 4).24 The diffusion barriers were fabricated using e-beam lithography (see methods for fabrication details). The typical width of each line was ~100 nm, and the height was commiserate with that of a streptavidin conjugated lipid bilayer (~ 10 nm) (Supplementary Fig. 4 A-B). The patterned diffusion barriers only affect cells through the specific ligand-receptor interaction rather than surface topography, thus diffusion constraints are exerted only on ligand-bound EGF receptors. This technique allows the direct manipulation of EGFR receptor clustering while also permitting quantification of receptor activation in living cells. Metal patterning of surfaces with supported lipid bilayers has previously been used to elucidate the role of spatial organization in T-cell receptor activation, 2, 24 EphA2 receptor activation,1, 25 and has also been used to track single protein diffusion in a position dependent manner via a nano-antenna patterning approach.35
Figure 2. Lipid diffusion barriers limit EGFR clustering and allow quantification of the relationship between cluster size and activation level.
(A) Scheme illustrating a cell that interacts with a patterned and non-patterned region of a supported lipid membrane juxtaposed with a fluorescence image of the EGF organization under a cell on the border of a 3 µm nanopatterned grid functionalized supported lipid bilayer. Scale bar is 5 µm. (B) Representative images of cells in the BF, RICM, TIRF 640 (EGF), and TIRF 488 (EGFR-pY 1068) channels interacting with nanopatterns (or non-patterned areas) on a supported lipid bilayer functionalized with EGF. Barriers directly controlled cluster size and pattern according to their respective dimensions and shapes. (See Supplementary Fig. 5 for cluster size histograms). Image contrast levels have been adjusted to illustrate cluster size for each representative image. Scale bar is 5 µm. (C) Vertical scatter plot showing the relationship between feature size and EGFR-pY 1068/EGF ratio. Ratios were taken from the mean intensities of the TIRF 488 (EGFR-pY 1068) and TIRF 647 (EGF) channels (see methods and Supplementary Figure 3 for data processing details). The p values listed for the 5 µm, 3 µm, 1 µm, and line control features correspond to a Student’s t-test between the given feature and the off-grid control. n = 401, 206, 210, 418, 245, 221, and 243 cells for the off-grid, 5 µm, 3 µm, 1 µm, square control, maze control, and 2 µm lines, respectively.
In these experiments, each substrate had an array of six types of nanopatterns: 5 µm, 3 µm, and 1 µm pitch grids, 3 µm pitch squares, 2 µm pitch mazes, and 2 µm pitch lines (Supplementary Fig. 4 C). The total size of each patterned area within the array was 400 by 400 µm2. The grid nanopatterns were designed to allow for molecular reorganization and assembly, while preventing large-scale clustering across the chromium barriers. The square and maze features were included as additional controls for potential artifacts due to chromium metal. For all nanopatterned experiments, supported lipid bilayers were formed on chromium barrier surfaces and functionalized with Alexa 647-EGF as described above.
HCC1143 cells were seeded onto nanopatterned EGF- Alexa 647 supported lipid bilayer surfaces and incubated for 1 h at 5% CO2, 37°C. Cells were subsequently fixed and permeabilized and then immunostained for EGFR-pY 1068. The cells that were engaged to each of the six different types of nanopatterns, along with the non-patterned regions, were imaged and analyzed. Representative images and live cell imaging showed that the patterns restricted EGF-EGFR clustering (Fig. 2B, Supplementary Movie 1), and altered the average cluster size. To quantify the average cluster size for cells that engaged each type of pattern, we applied a threshold to each EGFR image and used an automated script to detect particles and generate histograms of their sizes. The 1 µm, 3 µm, 5 µm pitch grid, and line patterns restricted the average cluster area to ~0.5 µm2, ~0.6 µm2, ~0.8 µm2, and ~1 µm2, respectively (Fig. 2B, Supplementary Fig. 5).
In order to determine if cluster size influenced activation levels, the normalized EGFR-pY-1068/EGF fluorescence ratio was measured for individual cells on each type of pattern, as well as cells outside of the patterned region (Fig. 2C). This measurement was performed without any thresholding of the raw fluorescence intensities, assuring that the ratio was a faithful measure of the amount of receptor activated per ligand on the surface. This experiment was repeated four times, and each run was combined into a single data set (see online methods and Supplementary Fig. 3 for analysis details, and Supplementary Fig. 6 for raw EGF and pY values). The mean EGFR-pY 1068/EGF value decreases slightly (~10%) when cells engaged the 5 µm or 3 µm grid in comparison to the non-patterned region. However, cells engaged to 1 µm grid patterns exhibited a significant (46%) increase in EGFR-pY1068/EGF ratio. The control patterns that allow free diffusion of the lipids (mazes and squares) showed minimal deviation from the off-pattern cell mean, while the line pattern showed a ~20% increase in ratio. This result suggests that large scale clustering (> 1 µm grid pattern) dampens receptor activation levels (Fig. 2C). In order to further assess the validity of these results, population statistics were performed on each pattern subtype. The p value (according to Student’s t test) between the off grid control and the 1 µm grid was 5 × 10−5, indicating that the increase in ratio due to limiting the cluster size of the EGFR receptor is significant (Fig. 2C).
The smallest pitch nanopatterns (1 µm spaced grid array) were found to reduce the effective EGF ligand density by ~50% due to blocking access to the glass surface, and thus reducing the lipid membrane total area. To verify that the observed increase in EGFR-pY 1068/EGF ratio was not due to reduced EGF surface density, we measured the ratio across a series of substrates that presented decreasing concentrations of EGF (less than ~280 ligand molecules/µm2). Supported lipid bilayer surfaces with reduced amounts of EGF ligand were generated by doping the 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) lipid vesicles with decreasing concentrations of biotinylated lipid (0.1%, 0.075%, and 0.045%) through mixing with DOPC/Biotin-DPPE vesicles (99.9%, .1%), resulting in substrates of varying EGF densities. Importantly, the lipid membranes doped with 0.035% and 0.075% biotin presented EGF densities that were ~63% and ~26% smaller than the EGF density of the control surface, respectively. These densities were chosen because they better matched the range of EGF densities observed in the nanopattern experiments. When we measured the EGFR-pY 1068/EGF ratio for these cells, we observed no correlation between EGF concentration for each cell and EGFR-pY 1068/EGF ratio (Fig. 3). Therefore, the observed increase in phosphorylation per receptor on the nanopatterned substrates (1 µm grid, Fig. 2C) is not due to reduced EGF concentration. We also performed controls to determine if the experimental time point had an effect on the EGFR-pY 1068/EGF ratio. HCC1143 cells were plated onto fluorescent EGF functionalized supported lipid bilayers and fixed after 30, 45, and 60 min incubation times. The samples were then stained for EGFR-pY 1068 and imaged. Ratio analysis did not find a significant difference in EGFR-pY 1068 to EGF ratio between the tested time points (Supplementary Fig. 7). This is in agreement with literature precedent measuring the time dependence of receptor activation by surface tethered EGF ligand.33
Figure 3. Ligand concentration does not alter the EGFR-pY 1068/EGF ratio within experimentally relevant EGF densities.
(A) Representative images of cells plated on EGF functionalized supported lipid bilayers featuring EGF densities that were ~40% (110 ± 20 molecules/µm2) and ~70% (200 ± 20 molecules/µm2) of the EGF density on the 0.1% biotin membrane (280 ± 20 molecules/µm2). EGF intensity values are averages of the mean EGF signal intensities per cell for each population of cells. Scale bar is 10 µm. (B) Scatter plot of the EGFR-pY 1068/EGF ratio versus mean EGF intensity for approximately 1700 cells on the surfaces described in (A).
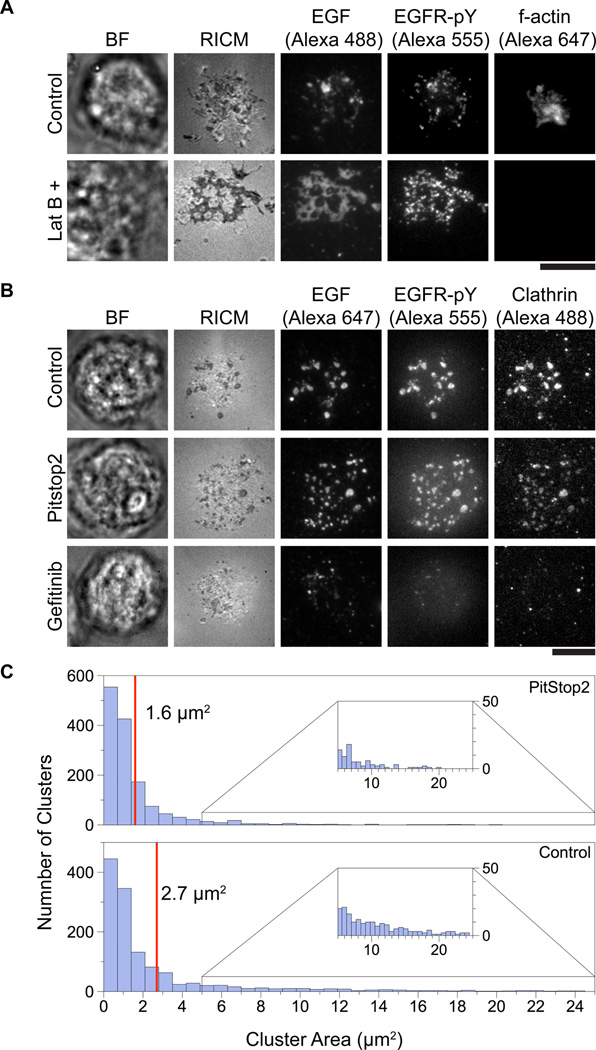
Having shown that EGFR clustering impacts its phosphorylation level we next wanted to investigate the mechanism of EGFR assembly into larger clusters. Live cell time-lapse TIRF microscopy imaging indicated that there were two stages of EGFR clustering (Supplementary Movies 2 and 3). The initial stage consisted of spontaneous formation of small sub-micron receptor clusters that formed across the cell-supported membrane contact area. After approximately 15 min, clusters coalesced into larger (sub-micron to micron sized) clusters that generally translocated to the center of the cell-supported membrane junction. The directed transport of EGFR assemblies suggested that the cytoskeleton was involved in receptor translocation. Clustering of other receptors has been shown to depend on actin polymerization,36 and EGF-EGFR complexes are subject to retrograde flow of actin.37, 38 In light of this, we tested the role of F-actin EGFR clustering. HCC1143 cells were pre-treated with 25 µM Latrunculin B (a drug that prevents actin polymerization by binding actin monomers, LatB) for 20 min or DMSO control and incubated on EGF-functionalized supported lipid bilayers for ~30 min. The cells were then fixed and stained for F-actin to confirm the effect of the drug at these concentrations. Surprisingly, cells treated with LatB exhibited large irregular EGFR clusters that typically covered the entire cell-supported membrane contact area (Fig. 4A). In contrast, control cells treated with DMSO exhibited typical clusters that were considerably smaller as previously shown (Fig. 1B, 2B, Fig. 4A). This suggests that F-actin association with EGFR oligomers initially prevents coalescence into large clusters.
Figure 4. Inhibition of F-actin, EGFR kinase activity, and clathrin terminal domain associations alters EGFR clustering.
(A) HCC1143 cells were serum starved overnight and treated with either Latrunculin B (25 µM) or vehicle DMSO for 20 min. Cells were then plated on supported lipid bilayers functionalized with EGF-Alexa 488 and incubated for 1 h at 37° C, 5% CO2. After incubation, cells were stained for EGFR-pY 1068 and F-actin. Latrunculin B treatment resulted in the near complete abrogation of F-actin and the formation of extremely large EGFR clusters as compared to the control. Scale bar is 10 µm.
(B) Cells were treated with either Gefitinib (10 µM, 20 min), PitStop2 (30 µM, 20 min), or vehicle DMSO and then incubated on supported lipid bilayers functionalized with EGF for 1 h at 37° C, 5% CO2 after which they were fixed and permeabilized (see methods for details). The cells were then stained for clathrin and EGFR-pY 1068. Control cells exhibited typical clusters, see Fig. 1B and Fig. 2B. Cells treated with PitStop2 showed decreased cluster size, while clustering was abolished in cells affected by Gefitinib. Additionally, clusters colocalized with clathrin-eGFP. Scale bar is 10 µm. (C) Histogram comparing cluster sizes in control and PitStop2 samples. PitStop2 decreased the cluster size by ~1.1 µm2 on average as compared to control. n = 204 control cells and 138 PitStop2 treated cells.
To further probe the dynamics of this F-actin association, HCC1143 cells were transfected with an actin-eGFP plasmid and plated on EGF functionalized supported lipid bilayers. Dual-channel time-lapse TIRF microscopy of these samples revealed F-actin and EGF co-movement (Supplementary Movie 3). This suggests coupling between F-actin and EGFR, which has been reported previously using single particle tracking and agrees with the initial phalloidin stain results.37
The surface concentration of EGFR is primarily regulated by clathrin-mediated endocytosis and recycling, a process by which the cell exerts a force on the receptor during internalization.16, 39 To investigate the role of this process in EGFR clustering and translocation, _cells were transfected with a clathrin light chain-eGFP construct and incubated on EGF-functionalized supported lipid bilayers (see methods for experimental details). Live cell TIRF imaging of EGF and CLC-eGFP revealed assemblies that colocalized, suggesting a role for clathrin in clustering (Supplementary Fig. 8). Clathrin binds to a host of adapter proteins through its terminal domain motif, or TD, and these proteins play a vital role in regulating the process of endocytosis.40 In light of this, we used PitStop2, a small molecule which inhibits clathrin TD associations (but does not prevent clathrin assembly), to interrogate whether clathrin terminal domain associated proteins contribute to EGFR clustering.41 When cells were treated with PitStop2, EGFR-EGF clusters were still observed, however, the average cluster size was reduced to ~1.6 µm2 (as compared to ~2.7 µm2 for the control) and the clusters were more evenly distributed across the cell membrane (Fig. 4B-C). Furthermore, cells in which the EGFR kinase domain was inhibited by Gefitinib showed the formation of small receptor clusters but drastically reduced formation of larger-scale assemblies (Fig. 4B). Taken together, these data suggest that receptor activation (but not ligand binding), and clathrin terminal domain-association are critical for the formation of large-scale EGFR clusters.
One possible mechanism to explain the correlation between cluster size and EGFR receptor phosphorylation relates to non-ligand activation of the receptor. For example, it has been observed that kinase-deficient mutants of EGFR can be activated by association with kinase-active receptor,42 and that inactive, un-liganded EGFR can be activated through lateral interaction with activated EGFR.12 Taken with the fact that the EGFR has the capability of dynamic self-association and disassociation, it is possible that the observed increase in activity (larger EGFR-pY per ligand) for small clusters is due to a cross-activation effect whereby ligand-bound phosphorylated receptor activates un-liganded receptor at its periphery in a diffusion dependent manner. As small clusters have a higher circumference to area ratio and lower densities, it may suggest that they are capable of activating a larger number of receptors than larger clusters using the same number of ligands.
Conclusion
Herein we demonstrated that spatial organization of the EGFR modulates receptor activation levels, and conversely we showed that EGFR clustering depends on receptor phosphorylation and interaction with the cytoskeleton as well as involvement of clathrin-mediated internalization. Through the use of nanopatterned diffusion barriers and supported lipid bilayers, EGFR clustering was altered without modifying native proteins. This approach revealed that EGFR cluster size modulates receptor phosphorylation, as clusters smaller than 1 µm2 are more efficiently phosphorylated than larger clusters.
These findings may be useful in the area of biomaterials, where surface-tethered growth factors are commonly employed, or for use in drug-delivery strategies that prevent receptor aggregation. It is important to note that while EGF is generally a soluble ligand, it has been shown that tethering EGF does not prevent receptor activation,33, 34, 43, 44 and supported lipid bilayer surfaces have previously been used for EGF surface display.15 Importantly, tethering of EGF prevents internalization of the receptor, resulting in sustained activation that differs from the activation spikes seen with soluble EGF.33 For this reason, surface tethered EGF-polymers have been used to improve the survival of mesenchymal stem cells (MSCs) and promote attachment, opening up the possibility for functionalized polymer scaffolds that could be used to repair tissue damage.34 Additionally, EGFR has natural juxtacrine ligands such as the EGF homologues heparin-binding EGF-like growth factor and transforming growth factor alpha (TGF-α).45 Thus, differences in the spatial arrangement of juxtacrine ligands may play a role in signal regulation. Moreover, these findings help elucidate how the cytoskeleton and the endocytotic machinery can influence the signaling outputs of membrane tethered growth factors. Therefore, investigating surface-tethered ligand activation of EGFR is broadly relevant despite some limitations in specifically applying the results of this study to the typical soluble ligand stimulation pathway.
Importantly, while these results represent a step towards understanding how receptor clustering affects the EGFR signaling cascade, more thorough investigations are hindered by the difficulty of combining conventional biochemical techniques such as qRT-PCR and Western blotting with nanoscale cell receptor manipulation methods.46 Future studies will need to overcome these limitations to further probe the effects of EGFR clustering and determine the extent of its regulatory influence over the global EGFR signaling network.
Materials and Methods
Data Processing
Data processing was performed with the imageJ (NIH) software package using the LOCI bioformats plugin for proprietary image handling.
Determination of EGFR-pY 1068/EGF (EGFR) Ratios
In order to generate the EGFR-pY 1068/EGF (EGFR) ratios, we performed a series of operations that are illustrated in Supplementary Fig. 3. First, the raw fluorescence images from each channel were background subtracted. Next, each subtracted image was divided by a normalization image taken at identical settings in order to correct for the TIRF excitation intensity of each channel. Finally, a uniform cell-sized circular region of interest (ROI) was manually placed over regions of cell fluorescence and used to measure the intensity in each channel. Note that there was no thresholding applied to these images, thus the mean fluorescence under each cell was not altered from the original 16 bit image format. The ratio of the intensities was used to calculate the given signal ratio and averaged for many cells on each type of surface.
Cell Fixation Procedure
After incubating cells on the specific surface, the wells were rinsed with 5 mL of 4° C 1X sterile PBS to remove media from the sample. Next, each sample was rinsed with 5 mL of 4° C 4% paraformaldehyde (Sigma) in 1X PBS and incubated for 12 min at RT. After incubation, the sample was again rinsed with 5 mL of 1X PBS, and then permeabilized with 5 mL 0.1% Triton-X (Sigma) in 1X PBS (vol/vol %) and incubated for 5 min at RT. Finally, cells were washed with a 5 mL aliquot of 1X PBS and 5 mL 1% BSA (fraction V, heat shock isolation, Calbiochem) in 1X PBS. Cells were then blocked overnight at 4° C.
Antibody Staining
The anti-EGFR-pY-1068 primary rabbit IgG antibody (Cell Signaling Technologies, #3777S) was diluted at a 1:800 volume:volume ratio in 1% BSA (1X PBS) and added to each sample and incubated for 1h at RT. In experiments where the EGFR was also stained the anti-EGFR rat IgG primary antibody (Santa Cruz #71035) was incubated at the same dilution concurrently. Each sample was then rinsed with 5 mL of 1% BSA in 1X PBS. Next, a 1:1000 dilution of anti-rabbit Alexa 488 (Invitrogen) and anti-rat Alexa 647 (when the EGFR primary was used, Invitrogen) were added to each sample and incubated for 30 min. The samples were then rinsed once more with 5 mL of 1% BSA in 1X PBS and then imaged. In experiments where clathrin was imaged, samples were stained with 1:800 Covance antibody # MMS-423P (clathrin light chain, mouse IgG) and an appropriate anti-mouse secondary following the same procedure as described above.
Cell Culture
All cell lines were maintained at 37° C and 5% CO2. HCC1143 cells were cultured in RPMI-1640 media with the addition of 10% fetal bovine serum (FBS, Mediatech), 100 IU ml−1 penicillin G (Mediatech), 100 μg ml−1 streptomycin (Mediatech) and 2.5 mM L-Glutamine (Mediatech) following the ATCC formulation guidelines.
Cell Transfection
HCC1143 cells were seeded on a 24 well plate in antibiotic-free media at a density of ~30,000 cells per well overnight at 37° C, 5% CO2. The cells were then transfected with the construct (either CLC-eGFP or F-actin-eGFP) using Lipofectamine 2000 (Invitrogen) following standard transfection protocols. These cells were then serum starved overnight and used for experiments as indicated within 24 h of the transfection.
Fluorescence Microscopy
Live cells were imaged in serum free RPMI 1640 (Mediatech) media formulated as described in the cell culture section at 37 °C, and fixed cells were imaged in 1% BSA in 1X PBS at RT. During live-cell imaging, physiological temperatures were maintained with a warming apparatus consisting of a sample warmer and an objective warmer (Warner Instruments part numbers 641674D and 640375). The microscope used was an Eclipse Ti driven by the Nikon Elements software package (Nikon). The microscope features an Evolve EM CCD (Photometrics), an Intensilight epifluorescence source (Nikon), a CFI Apo 100x (NA = 1.49) objective (Nikon), and a TIRF launcher with two laser lines: 488 nm (10 mW) and 640 nm (20 mW). This microscope also includes the Nikon Perfect Focus System, an interferometry-based focus lock that allowed the capture of multipoint and time-lapse images without loss of focus. The microscope is equipped with the following Chroma filter cubes: TIRF 488, TIRF 640, Cy5, TRITC, FITC, DAPI and RICM.
Preparation of Small Unilamellar Lipid Vesicles
After being mixed in the correct proportions in chloroform, lipids (Avanti Polar Lipids) were dried with a rotary evaporator and placed under a stream of N2 to ensure complete evaporation of the solvent. These lipid samples were then resuspended in Nanopure (~18.2 mΩ) and subjected to three freeze/thaw cycles by alternating immersions in an acetone and dry ice bath and a warm water bath (45 °C). To obtain small unilamellar vesicles (SUV’s), lipids were forced through a high-pressure extruder with a 100 nm nanopore membrane (Whatman).
Supported Lipid Bilayer Preparation
Supported lipid bilayer surfaces were prepared via the vesicle fusion method as previously described in the literature.16, 30 Preparation varied depending on the nature of the vessel used:
A. Glass-Bottomed 96 Well Plate
Wells were rinsed with 5 mL H2O (18.2 mΩ) and then incubated for 1–2 hours in 1 M NaOH, which was followed by a 5 mL wash with H2O. All water was then removed from wells and 100 µL of a 1:3 mixture of the desired lipid vesicles and 1X sterile PBS was added to each well and incubated for 5 min at RT. Each well was then rinsed with 5 mL of H2O and 5 mL of 1X PBS. The wells were then blocked for 30 min at RT with 40 µL of 1 mg/mL BSA (fraction V, cold alcohol isolation, EMD Chemicals), rinsed with 10 mL 1X PBS, and functionalized as described according to the individual experiment. When performing experiments with covalently functionalized glass substrates, it was necessary to adjust the ligand density of the fluid surfaces to the same level. In order to accomplish this, the biotin lipid doping level in the fluid supported lipid bilayers was adjusted by mixing DOPC vesicles with the DOPC/DPPE-Biotin vesicles in a molar ratio of 99.925% to 0.075%. This ratio generated a surface EGF density of 230 ± 20 molecules/µm2, which closely matches the density of the covalently functionalized glass surfaces.
B. Attofluor Cell Chamber
No 2. Microscope cover glasses (35 mm, VWR) were sonicated in water for 10 min and then etched for 10 min in piranha (a 3:1 mixture of sulfuric acid and 30% hydrogen peroxide, respectively. Please use caution as piranha is extremely corrosive and may explode if it comes into contact with organics). After etching, cover glasses were rinsed well with H2O and smacked onto a 30 µL droplet of 1:1 lipid vesicle:1X PBS mixture. Cover glasses were then removed from the droplet in a bath of water, and assembled into the attofluor cell chambers (Invitrogen). Chambers were then rinsed with 50 mL H2O and 50 mL 1X sterile PBS. All samples were then blocked with 100 µL of 1 mg/mL BSA for 30 min at room temperature, then rinsed again with 50 mL 1X PBS and functionalized according to the experiment performed.
Supported Lipid Bilayer Density Calibration
The EGF density on the fluid supported lipid bilayers was measured using quantitative fluorescence microscopy as described in the literature.31 Briefly, a fluorescence standard curve was determined using fluid supported lipid bilayers (in a 96 well plate) containing various concentrations of TRITC-DHPE lipids. Next, the fluorescence ratio (F factor) between TRITC (the standard dye) and Alexa 555 (the ligand label) was determined by comparing the fluorescence of a range of concentrations of either unilamellar lipid vesicles or Alea 555-EGF-biotin in 1X PBS (fluorescence intensity was determined using the 100X objective focused ~100 µm above the well surface). The F factor was found to be 0.62 ± 0.16. After obtaining the standard curve and the F factor, any image of Cy3-EGF acquired with the same settings as the standard curve can be converted into molecules of EGF molecules/µm2 through background subtraction and subsequent division by the F factor and the slope of the standard curve.
Synthesis of Biotin-EGF-Dye Conjugates
EGF (R&D Biosystems) was labeled with either Alexa 647, 488, or Cy3 succinimidyl esters (Invitrogen) and NHS-biotin (Pierce) in a single-pot reaction as previously described.16 Briefly, 100 µL of EGF (1 mg/mL) was combined with 10 µL of 1 M sodium bicarbonate and a ten-fold molar excess of dye and NHS-biotin. The reaction was incubated for 1 h after which it was purified with Bio-Gel P4 resin (Bio-Rad). This reaction typically yielded protein with a labeling ratio between 1 and 2.
Fabrication of Chromium Diffusion Barriers
Clean glass coverslips were spin-coated at 2500 RPM with electron beam resist, NANO™PMMA 495 A4 (MicroChem Co., Newton, MA). A chromium dissipation layer with a thickness of 10 nm was deposited onto the samples via electron beam evaporation. Samples were exposed in a JEOL JBX9300FS electron beam lithography tool operating at 100 kV and 2 nA with a dose of 1200 µC/cm2. Following removal of the Cr dissipation layer in Cr etchant, samples were developed for 1 minute in 1:3 Methyl isobutyl ketone (MIBK): isopropyl alcohol (IPA), rinsed with IPA and dried with N2. Samples were subjected to brief (6 s) oxygen plasma treatments to remove any organic residue in the developed areas. Electron beam evaporation was used to deposit a thin film of Cr as a barrier layer. Sonication of the samples in acetone for approximately 2 minutes removed the patterned PMMA leaving Cr barriers in the areas previously exposed and developed during EBL processing.
Supplementary Material
Acknowledgements
We would like to thank the Oak Ridge National Lab Center for Nanophase Materials Sciences (CNMS 2009-269) for fabricating the chromium diffusion barrier substrates, and Dr. Rita Nahta at the Winship Cancer Center for the gift of cell lines used herein. K.S. would like to acknowledge support from the National Institutes of Health (NIH) through R01-GM097399-01, seed funding through the NIH NHLBI Program Excellence in Nanotechnology (HHSN268201000043C), and seed funding through the Emory University Research Committee (URC) 00016401.
References
- 1.Salaita K, Nair PM, Petit RS, Neve RM, Das D, Gray JW, Groves JT. Science. 2010;327:1380–1385. doi: 10.1126/science.1181729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manz BN, Jackson BL, Petit RS, Dustin ML, Groves J. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:9089. doi: 10.1073/pnas.1018771108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orth RN, Wu M, Holowka DA, Craighead HG, Baird BA. Langmuir. 2003;19:1599–1605. [Google Scholar]
- 4.Wu M, Holowka D, Craighead HG, Baird B. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:13798–13803. doi: 10.1073/pnas.0403835101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fewtrell C, Metzger H. Journal of Immunology. 1980;125:701–710. [PubMed] [Google Scholar]
- 6.Kiessling LL, Gestwicki JE, Strong LE. Angewandte Chemie International Edition. 2006;45:2348–2368. doi: 10.1002/anie.200502794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cochran JR, Cameron TO, Stern LJ. Immunity. 2000;12:241–250. doi: 10.1016/s1074-7613(00)80177-6. [DOI] [PubMed] [Google Scholar]
- 8.Triantafilou M, Gamper FGJ, Haston RM, Mouratis MA, Morath S, Hartung T, Triantafilou K. Journal of Biological Chemistry. 2006;281:31002–31011. doi: 10.1074/jbc.M602794200. [DOI] [PubMed] [Google Scholar]
- 9.Chung I, Akita R, Vandlen R, Toomre D, Schlessinger J, Mellman I. Nature. 2010;464:783–787. doi: 10.1038/nature08827. [DOI] [PubMed] [Google Scholar]
- 10.Abulrob A, Lu Z, Baumann E, Vobornik D, Taylor R, Stanimirovic D, Johnston L. Journal of Biological Chemistry. 2010;285:3145. doi: 10.1074/jbc.M109.073338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hofman EG, Bader AN, Voortman J, Van Den Heuvel DJ, Sigismund S, Verkleij AJ, Gerritsen HC, Van Bergen En Henegouwen PMP. Journal of Biological Chemistry. 2010;285:39481–39489. doi: 10.1074/jbc.M110.164731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ichinose J, Murata M, Yanagida T, Sako Y. Biochemical and Biophysical Research Communications. 2004;324:1143–1149. doi: 10.1016/j.bbrc.2004.09.173. [DOI] [PubMed] [Google Scholar]
- 13.Clayton A, Orchard S, Nice E, Posner R, Burgess A. Growth Factors. 2008;26:316–324. doi: 10.1080/08977190802442187. [DOI] [PubMed] [Google Scholar]
- 14.Szabó Á, Horváth G, Szöllosi J, Nagy P. Biophysical Journal. 2008;95:2086–2096. doi: 10.1529/biophysj.108.133371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nam J-M, Nair PM, Neve RM, Gray JW, Groves JT. ChemBioChem. 2006;7:436–440. doi: 10.1002/cbic.200500479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stabley DR, Jurchenko C, Marshall SS, Salaita KS. Nat Meth. 2012;9:64–67. doi: 10.1038/nmeth.1747. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, Yu X, Boriskina SV, Reinhard BrM. Nano Letters. 2012 doi: 10.1021/nl3012227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yarden Y, Sliwkowski M. Nature Reviews Molecular Cell Biology. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 19.Qi S, Groves J, Chakraborty A. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:6548. doi: 10.1073/pnas.111536798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mossman KD, Campi G, Groves JT, Dustin ML. Science. 2005;310:1191–1193. doi: 10.1126/science.1119238. [DOI] [PubMed] [Google Scholar]
- 21.Grassmé H, Jekle A, Riehle A, Schwarz H, Berger J, Sandhoff K, Kolesnick R, Gulbins E. Journal of Biological Chemistry. 2001;276:20589–20596. doi: 10.1074/jbc.M101207200. [DOI] [PubMed] [Google Scholar]
- 22.Grassmé H, Cremesti A, Kolesnick R, Gulbins E. Oncogene. 2003;22:5457–5470. doi: 10.1038/sj.onc.1206540. [DOI] [PubMed] [Google Scholar]
- 23.Scott FL, Stec B, Pop C, Dobaczewska MK, Lee JJ, Monosov E, Robinson H, Salvesen GS, Schwarzenbacher R, Riedl SJ. Nature. 2009;457:1019–1022. doi: 10.1038/nature07606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jay T G. Current Opinion in Chemical Biology. 2006;10:544–550. doi: 10.1016/j.cbpa.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 25.Xu Q, Lin W-C, Petit RS, Groves JT. Biophysical Journal. 2011;101:2731–2739. doi: 10.1016/j.bpj.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen S, Carpenter G, King L. Journal of Biological Chemistry. 1980;255:4834–4842. [PubMed] [Google Scholar]
- 27.Lax I, Mitra AK, Ravera C, Hurwitz DR, Rubinstein M, Ullrich A, Stroud RM, Schlessinger J. Journal of Biological Chemistry. 1991;266:13828–13833. [PubMed] [Google Scholar]
- 28.Yang S, Raymond-Stintz MA, Ying W, Zhang J, Lidke DS, Steinberg SL, Williams L, Oliver JM, Wilson BS. Journal of Cell Science. 2007;120:2763–2773. doi: 10.1242/jcs.007658. [DOI] [PubMed] [Google Scholar]
- 29.Nagy P, Claus J, Jovin T, Arndt-Jovin D. Proceedings of the National Academy of Sciences. 2010;107:16524. doi: 10.1073/pnas.1002642107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narui Y, Salaita KS. Chemical Science. 2012;3:794–799. [Google Scholar]
- 31.Galush WJ, Nye JA, Groves JT. Biophysical Journal. 2008;95:2512–2519. doi: 10.1529/biophysj.108.131540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu M, Scollard D, Chan C, Chen P, Vallis K, Reilly RM. Nuclear Medicine and Biology. 2007;34:887–896. doi: 10.1016/j.nucmedbio.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 33.Platt M, Roman A, Wells A, Lauffenburger D, Griffith L. Journal of Cellular Physiology. 2009;221:306–317. doi: 10.1002/jcp.21854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan VH, Au A, Tamama K, Littrell R, Richardson LB, Wright JW, Wells A, Griffith LG. Stem Cells. 2007;25:1241–1251. doi: 10.1634/stemcells.2006-0320. [DOI] [PubMed] [Google Scholar]
- 35.Lohmüller T, Iversen L, Schmidt M, Rhodes C, Tu HL, Lin WC, Groves JT. Nano Letters. 2012;12:1717–1721. doi: 10.1021/nl300294b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiang EN, Dong R, Ober CK, Baird BA. Langmuir. 2011 doi: 10.1021/la200093e. 110510130404039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lidke DS, Nagy P, Heintzmann R, Arndt-Jovin DJ, Post JN, Grecco HE, Jares-Erijman EA, Jovin TM. Nature Biotechnology. 2004;22:198–203. doi: 10.1038/nbt929. [DOI] [PubMed] [Google Scholar]
- 38.Lidke DS, Lidke KA, Rieger B, Jovin TM, Arndt-Jovin DJ. J. Cell Biol. 2005;170:619–626. doi: 10.1083/jcb.200503140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vieira A, Lamaze C, Schmid S. Science. 1996;274:2086. doi: 10.1126/science.274.5295.2086. [DOI] [PubMed] [Google Scholar]
- 40.Lemmon SK, Traub LM. Traffic. 2012;13:511–519. doi: 10.1111/j.1600-0854.2011.01321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.von Kleist L, Stahlschmidt W, Bulut H, Gromova K, Puchkov D, Robertson MJ, MacGregor KA, Tomilin N, Pechstein A, Chau N, Chircop M, Sakoff J, von Kries JP, Saenger W, Kräusslich H-G, Shupliakov O, Robinson PJ, McCluskey A, Haucke V. Cell. 2011;146:471–484. doi: 10.1016/j.cell.2011.06.025. [DOI] [PubMed] [Google Scholar]
- 42.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 43.Marcantonio N, Boehm C, Rozic R, Au A, Wells A, Muschler G, Griffith L. Biomaterials. 2009;30:4629–4638. doi: 10.1016/j.biomaterials.2009.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuhl PR, Griffith-Cima LG. Nature Medicine. 1996;2:1022–1027. doi: 10.1038/nm0996-1022. [DOI] [PubMed] [Google Scholar]
- 45.Singh AB, Harris RC. Cellular Signalling. 2005;17:1183–1193. doi: 10.1016/j.cellsig.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 46.Nair PM, Salaita K, Petit RS, Groves JT. Nature protocols. 2011;6:523–539. doi: 10.1038/nprot.2011.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.