Abstract
Background
Mucoepidermoid carcinoma (MEC) can be classified into low-, intermediate-, and high-grade tumors based on its histological features. MEC is mainly composed of three cell types (squamous or epidermoid, mucous and intermediate cells), which correlates with the histological grade and reflects its clinical behavior. Most cancers exhibit reduced methylation of repetitive sequences such as Long INterspersed Element-1 (LINE-1) and Alu elements. However, to date very little information is available on the LINE-1 and Alu methylation status in MEC. The aim of this study was to investigate LINE-1 and Alu element methylation in MEC and compare if key differences in the methylation status exist between the three different cell types, and adjacent normal salivary gland cells, to see if this may reflect the histological grade.
Methods
LINE-1 and Alu element methylation of 24 MEC, and 14 normal salivary gland tissues were compared using Combine Bisulfite Restriction Analysis (COBRA). Furthermore, the three different cell types from MEC samples were isolated for enrichment by laser capture microdissection (LCM), essentially to see if COBRA was likely to increase the predictive value of LINE-1 and Alu element methylation.
Results
LINE-1 and Alu element methylation levels were significantly different (p<0.001) between the cell types, and showed a stepwise decrease from the adjacent normal salivary gland to the intermediate, mucous and squamous cells. The reduced methylation levels of LINE-1 were correlated with a poorer histological grade. In addition, MEC tissue showed a significantly lower level of LINE-1 and Alu element methylation overall compared to normal salivary gland tissue (p<0.001).
Conclusions
Our findings suggest that LINE-1 methylation differed among histological grade mucoepidermoid carcinoma. Hence, this epigenetic event may hold value for MEC diagnosis and prognostic prediction.
Keywords: Mucoepidermoid carcinoma (MEC), Methylation, Long INterspersed Element-1s (LINE-1s), Alu element, Laser capture microdissection
Background
Mucoepidermoid carcinoma (MEC) is a malignant neoplasm of salivary glands that occurs in both adults and children [1-4]. MEC typically occurs in 40–60-year-old patients, and with a median age of approximately 45 years. There is a 3:2 male:female gender preference for MECs with the exception of the tongue and retromolar area, which are more common in females [1,5]. Fifty-three percent of MECs are found in the major salivary glands, especially the parotid glands, while the palate and the buccal mucosa are the most common intraoral sites [1].
Histologically, MECs are primarily composed of three morphological cell types, which include squamous or epidermoid, mucous and intermediate cells, and these can take the form of a solid nest or cystic structure. According to the WHO classification system, MECs are classified as low-, intermediate- or high-grade based on five histological features: the presence of a cystic component, neural invasion, necrosis, mitotic activity and anaplasia [1]. However, many systems have been proposed for grading this tumor type, but none have been universally accepted [1,6-10]. Sadly, the outcome of MEC patients is influenced by the clinical stage and histological grade [11], whereby patients with high-grade, the rate of recurrence and metastasis is increased and thus compromising survival [1,12,13]. Furthermore, it has been suggested that the histological grade of MEC can considerably impact the treatment outcome of affected patients.
To date, only a few genetic studies have proposed mechanisms for the etiology of MEC. Some MECs have been reported to have a t(11:19)(q21:p13) translocation, and abnormality [14-16] that is also shared by acute leukemias [1,17-19]. Furthermore, a study reported that 18% of MECs analyzed demonstrated mutations in H-ras gene at codon 12 and/or 13 (and none at codon 61), but however these were essentially detected in high-grade cases [1,20].
One of the most common epigenetic changes found in cancer is the genome-wide decrease in methylation (genome-wide hypomethylation) [21-23]. Long INterspersed Element-1s (LINE-1s) are retrotransposons with highly repetitive, interspersed sequences which are distributed randomly throughout the genome, and constituting 17% of the total human genome [24,25]. Furthermore, Alu represents the most abundant Short INterspersed Element (SINE) repetitive sequence, representing 11% of total human genome [26]. Hypomethylation of LINE-1s, which occurs in many malignancies [21,27-31], generally results in chromosomal aberrations [32-35], hypermethylation, mutations of key tumor suppressor genes [36,37], and changes in oncogene transcription [38] resulting in the altered expression of cancer-related genes [39]. In addition, LINE-1 hypomethylation levels may hold value as a prognostic marker for epithelial solid cancers, for example cervical [30], hepatocellular [31] and ovarian [29]. Similarly, Alu hypomethylation have also been reported for many types of cancers, such as colorectal [27], gastric [28], and hepatocellular [40]. Thus, both LINE-1 and Alu element hypomethylation may play a notable role in different histological feature of cancer.
Most methylation studies report only quantitative information about the methylation level. Recently, we reported that the methylation patterns of LINE-1s could provide more crucial information regarding carcinogenesis. For instance, the percentage of hypomethylation loci (%uCuC) had a value that could significantly distinguish between normal peripheral blood mononuclear cells (PBMCs) and PBMCs from patients with cancers of the oral cavity, liver, colon, lung and the nasopharynx [41,42]. In this regard, no study has been carried out to analyze LINE-1 and Alu element methylation in human MEC. Thus, the goal of this study was to investigate levels and patterns of LINE-1 and Alu element methylation in MEC and also in the three cell types that are affected by this malignancy. The relationship of methylation status and histological grade in MEC was also assessed to obtain a better understanding of the clinical behavior of the tumor. Here, we demonstrate the methylation level of LINE-1 was different among the three histological grades of mucoepidermoid carcinoma.
Methods
Samples and LCM
The research protocol together with the experimental design underwent approval by the Institutional Review Board of the Faculty of Medicine, Chulalongkorn University (IRB006/53). Paraffin-embedded tissues from 24 salivary glands from MEC patients (diagnosed by histology) and 14 normal salivary glands from unrelated patients were obtained from the Department of Pathology, Faculty of Medicine, Chulalongkorn University. The limited clinical data available for each MEC patient was obtained from records, and this is shown in Table 1. The MEC group consisted of 14 women and 10 men (mean age ± SD = 39.62 ± 12.37 years).
Table 1.
Demographic data of MEC patients
| Sample | Sex | Age | Grade | Site | Cell type | |||
|---|---|---|---|---|---|---|---|---|
| |
|
|
|
|
N |
I |
M |
S |
|
MEC1 |
M |
60 |
Low |
Palate |
|
|
√ |
√ |
|
MEC2 |
F |
30 |
Low |
Palate |
√ |
|
√ |
|
|
MEC3 |
M |
35 |
Low |
Palate |
|
√ |
√ |
|
|
MEC4 |
M |
47 |
High |
Palate |
√ |
|
|
√ |
|
MEC5 |
F |
38 |
Low |
Palate |
√ |
|
|
|
|
MEC6 |
M |
31 |
Low |
Palate |
√ |
√ |
√ |
|
|
MEC7 |
F |
32 |
Low |
Palate |
|
|
√ |
|
|
MEC8 |
F |
53 |
High |
Anterior mandible |
|
|
|
√ |
|
MEC9 |
M |
41 |
Low |
Palate |
√ |
|
√ |
|
|
MEC10 |
F |
43 |
Low |
Palate |
|
|
√ |
√ |
|
MEC11 |
M |
33 |
Low |
Palate |
|
|
√ |
|
|
MEC12 |
F |
55 |
Intermediate |
Palate |
√ |
|
|
√ |
|
MEC13 |
M |
54 |
Low |
Palate |
|
|
√ |
√ |
|
MEC14 |
F |
34 |
Intermediate |
Palate |
|
|
|
√ |
|
MEC15 |
M |
35 |
Low |
Palate |
|
|
√ |
|
|
MEC16 |
F |
16 |
Intermediate |
Palate |
√ |
|
√ |
√ |
|
MEC17 |
F |
21 |
Intermediate |
Palate |
|
|
√ |
√ |
|
MEC18 |
F |
45 |
Intermediate |
Palate |
|
√ |
|
√ |
|
MEC19 |
F |
51 |
High |
Parotid gland |
√ |
|
|
√ |
|
MEC20 |
M |
31 |
Intermediate |
Parotid gland |
√ |
|
√ |
|
|
MEC21 |
F |
53 |
Intermediate |
Parotid gland |
√ |
|
√ |
√ |
|
MEC22 |
F |
17 |
Low |
Palate |
√ |
|
|
|
|
MEC23 |
F |
41 |
Intermediate |
Palate |
√ |
√ |
√ |
√ |
| MEC24 | M | 55 | Intermediate | Palate | √ | |||
MEC: Mucoepidermoid carcinoma, M: Male, F: Female, Low: Low-grade MEC, Intermediate: Intermediate-grade MEC,
High: High-grade MEC, N: Adjacent normal salivary gland cell, I: Intermediate cell, M: Mucous cell, S: Squamous cell.
These specimens were cut into 3-μm-thick sections and mounted onto histological glass slides. After deparaffinization, and hydration, the sections underwent standard hematoxylin and eosin (H&E) staining. After, each slide underwent, histopathological evaluation by three independent pathologists (SK, KD and NK), and those cases correctly identified as MECs were histologically graded according to the WHO diagnostic criteria [1]. The MEC samples assessed yielded low (n=12), intermediate (n=9) and high-grade (n=3) samples based on the 5 histological features (the presence of a cystic component, neural invasion, necrosis, mitotic activity and anaplasia) [1,6,43,44]. For the control group, normal salivary gland tissues were obtained (n=14) from patients undergoing radical neck dissections. All of the normal salivary glands were confirmed by histological analysis to be free of tumor cells.
MEC tissues underwent laser capture microdissection (LCM) using the method described in our previous study [23]. Using our expertise in LCM, we isolated pure cell population of different MEC subtype, as well as normal salivary gland cells adjacent to the lesion. From 24 MEC samples, cell subtypes isolated included squamous (n=13), intermediate (n=4), mucous (n=16), and adjacent normal salivary gland (n=12). Approximately 1,500 cells were isolated from each specimen and used for DNA extraction to yield sufficient amount and quality for PCR analysis (Table 1).
DNA extraction
DNA was extracted from laser-captured microdissected tissue by proteinase K digestion and a standard phenol-chloroform extraction protocol [45]. For whole MEC tissue anaysis, the paraffin-embedded tissues were cut into 4-μm-thick sections, and DNA was extracted using a DNA extraction kit (QIAamp® DNA FFPE Tissue, Qiagen, Valencia, CA, USA), and the method described previously [46]. The quality of DNA was assessed by NANO Drop 2000C, spectrophotometer with ratio of 1.8-2.0.
Combine Bisulfite Restriction Analysis (COBRA) of LINE-1 and Alu element
All DNA samples were treated with sodium bisulfite essentially following guidelines provided (EZ DNA Methylation-Gold™ Kit, Zymo research corp, Orange, CA, USA). For COBRALINE-1, the bisulfate-treated DNA was subjected to 40 PCR cycles with LINE-1-F (5’-CCGTAAGGGGTTAGGGAGTTTTT-3’) and LINE-1-R (5’-RTAAAACCCTCCRAACCAAATATAAA-3’) primers at an annealing temperature of 50°C. For COBRAAlu, the bisulfite-treated DNA was subjected to 40 cycles of PCR with two primers, Alu-F (5’-GGCGCGGTGGTTTACGTTTGTAA-3’) and Alu-R (5’-TTAATAAAAACGAAAT TTCACCATATTAACCAAAC-3’) at an annealing temperature of 53°C. After PCR amplification, the LINE-1 amplicons (160 bp) were digested with TaqI and TasI in NEB buffer 3 (New England Biolabs, Ontario, Canada), while the Alu amplicons (117 bp) were digested with TaqI in TaqI buffer (MBI Fermentas, Burlington, Canada). Both digestion reactions were incubated at 65°C overnight. The LINE-1 and Alu element digested products were then electrophoresed on an 8% non-denaturing polyacrylamide gel and stained with the SYBR green nucleic acid gel stain (Gelstar, Lonza, Rockland, ME, USA). Distilled water was used as negative control. All experiments were performed in duplicate.
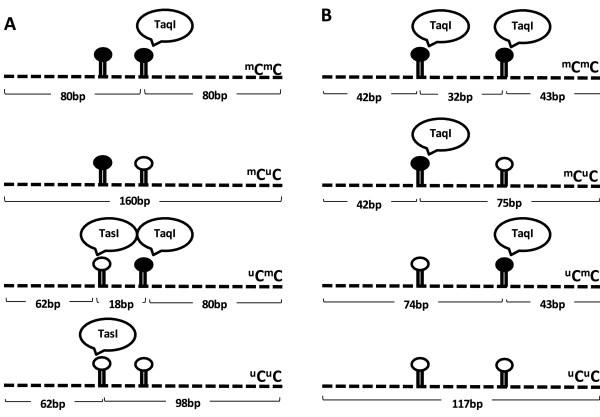
LINE-1 methylation analysis
The intensities of the COBRALINE-1 fragments on the polyacrylamide gel were quantified and analyzed using a Phosphoimager and the ImageQuant Software (Molecular-Dynamics, GE Healthcare, Slough, UK). COBRALINE-1 generated 4 products depending on the methylation state of the 2 CpG dinucleotides, as follows: partial methylation (mCuC, 160 bp), hypomethylation (uCuC, 98 bp), 1 methylated CpG (mC, 80 bp) and 1 unmethylated CpG (uC, 62 bp) (Figure 1A). LINE-1 methylation levels and patterns were calculated to determine the precise percentage of methylated CpG dinucleotides. The percentage was calculated as follows. First, the intensity of each band was divided by the length (bp) of the double-stranded DNA: %160/160 = A, %98/94 = B, %80/78 = C and %62/62 = D. Next, the frequency of each methylation pattern was calculated: percentage of mC = 100×(C+A)/(C+A+A+B+D), percentage of mCuC = 100×(A)/(((C-D+B)/2)+A+D), percentage of uCmC = 100×(D-B)/(C-D+B)/2)+A+D, percentage of hypomethylated loci (uCuC) = 100×B/(((C-D+B)/2)+A+D) and percentage of hypermethylated loci (mCmC) = 100×((C-D+B)/2)/(((C-D+B)/2)+D+A). DNA samples isolated from HeLa, Jurkat and Daudi cell lines were used as positive controls in each experiment and for interassay variation normalization [21].
Figure 1.
LINE-1 and Alu methylation patterns. The dark circles represent methylated cytosine, while the hollow circles represent unmethylated cytosine. There are four possible methylation patterns for the LINE-1 and Alu amplicons, including hypermethylated loci (mCmC), hypomethylated loci (uCuC), and 2 partially methylated loci (mCuC and uCmC). In each model, TaqI specifically identified methylated cytosine, while TasI specifically identified unmethylated cytosine. (A) The different methylation patterns of LINE-1 resulted in four differently sized digested products of 160 bp, 98 bp, 80 bp and 62 bp. (B) The different methylation patterns of the Alu element resulted in four differently sized digested products of 117 bp, 74/75 bp, 42/43 bp and 32 bp.
Alu element methylation analysis
The ImageQuant Software (Molecular-Dynamics) was used to quantify the intensities of COBRAAlu fragments on the polyacrylamide gel. COBRAAlu generated 3 bands based on the methylation status: hypomethylation (uCuC, 117 bp), partial methylation (mCuC and uCmC, 74 and 75 bp, respectively) and methylated loci (mC, 42 and 43 bp) (Figure 1B). Alu element methylation levels and patterns were calculated to determine the precise frequency of each pattern. The calculation was performed as the follows. First, the intensity of each band was divided by the length (bp) of the double-stranded DNA: %117/117 = A, %74 and 75/74.5 = B, %42 and 43/43.5 = D, and D-B = C (C= hypermethylated loci, mCmC). Next, the frequency of each Alu element methylation pattern was calculated as follows: percentage of methylated loci (mC) = 100×(2C+2B)/(2A+2B+2C) = 100×(2D)/(2A+2D), percentage of hypermethylated loci (mCmC) = 100× C/(A+B+C), percentage of partially methylated loci (uCmC+mCuC) = 100×B/(A+B+C) and percentage of hypomethylated loci (uCuC) = 100×A/(A+B+C). DNA samples from HeLa, Jurkat and Daudi cell lines were used as positive controls in every experiment and to standardize interassay variation [21].
Statistical analysis
Analysis of variance (ANOVA) was used to compare methylation patterns of LINE-1 and Alu elements among squamous, mucous, intermediate and adjacent normal salivary gland cells present in MEC lesions, as well as a paired t-test to analyze among cell subtypes in paired samples. An independent sample t-test was performed to determine differences between LINE-1 and Alu element methylation patterns in total MEC tissue and normal tissue of the salivary gland. A receiver operating characteristic (ROC) analysis was performed to verify the ability of COBRALINE-1 and COBRAAlu to differentiate MEC lesions from normal salivary gland tissue. An area under the ROC curve (AUC) value of 1.0 indicated perfect accuracy, while an AUC value of 0.5 indicated an inability to distinguish between samples. The cut-off values were selected to determine the diagnostic value of this approach. All calculations were performed using the SPSS software for Windows, version 17.0 (SPSS Inc., Chicago, IL) and the MedCalc statistical software. The results were considered statistically significant when the p-value was less than 0.05.
Results
LINE-1 methylation in microdissected MEC tissue
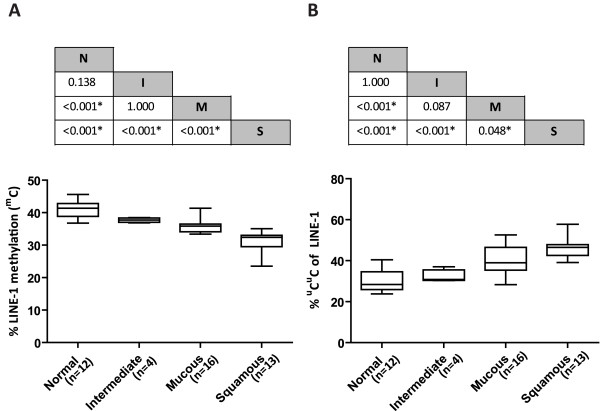
The frequency of each LINE-1 methylation pattern is shown in Table 2. The total LINE-1 methylation level (mC) decreased from the adjacent normal salivary gland cells (N) to the intermediate cells (I), mucous cells (M) and squamous cells (S). The results showed significant differences between S:M, S:I, S:N and M:N (p<0.001). However, there was no significant difference between M:I and N:I (p=1.000 and 0.138, respectively) (Figure 2A).
Table 2.
Frequency of LINE-1 methylation patterns in MEC cell subtypes, whole MEC tissues and normal salivary glands
| LINE-1 patterns | %mC | %mCmC | %mCuC | %uCmC | %uCuC |
|---|---|---|---|---|---|
| Adjacent normal salivary gland cell (N) |
41.13±2.51 |
12.49±4.61 |
26.21±4.62 |
31.06±7.35 |
30.22±5.08 |
| Intermediate cell (I) |
37.69±0.69 |
7.63±3.15 |
27.17±0.50 |
32.94±5.92 |
32.24±3.20 |
| Mucous cell (M) |
35.84±2.24 |
11.98±7.93 |
22.90±6.43 |
24.80±9.75 |
40.30±6.92 |
| Squamous cell (S) |
31.27±3.07 |
8.74±5.20 |
24.17±4.00 |
20.89±8.10 |
46.18±4.75 |
| Normal salivary gland (NG) |
41.79±1.90 |
21.03±2.31 |
28.13±2.95 |
13.38±3.26 |
37.44±2.86 |
| Whole MEC tissue (MEC) | 35.69±2.23 | 11.52±4.71 | 26.64±3.20 | 21.69±6.96 | 40.13±3.71 |
Figure 2.
Comparison of the frequency of total LINE-1 methylation (mC) and uCuC of LINE-1s among MEC cell subtypes. (A) The frequency of mC of LINE-1s among MEC cell subtypes showed a stepwise decrease from normal cells (N) to intermediate cells (I), mucous cells (M) and squamous cells (S). The p‐value between each group is shown in the table above the graph. (B) The frequency of uCuC of LINE-1s among cell types showed a stepwise increase from normal cells to intermediate cells, mucous cells and squamous cells. The p‐value between each group is shown in the table above the graph.
Additionally, the frequency of unmethylated (uCuC) LINE-1s increased from N to M, I and S. Significant differences were found between S:M (p=0.048), S:I, S:N and M:N (p<0.001). However, no significant difference was found between M:I and N:I (p=0.087 and 1.000, respectively) (Figure 2B). A significant difference in the uCmC level of LINE-1s in N, M, I and S was found only between S:N (p=0.027). There was no significant difference between S:I (p=0.099), M:I (p=0.551), M:N (p=0.353), S:M and N:I (p=1.000).
The paired comparisons among cell types displayed strongly significant difference (p<0.001) of mC between N:M, N:S and M:S. Furthermore, the uCuC is also different when compared between N:M (p=0.013), N:S (p<0.001) and I:M (p=0.006). The detailed data of paired comparisons are shown in Additional file 1: Table S1.
Alu element methylation in microdissected MEC tissue
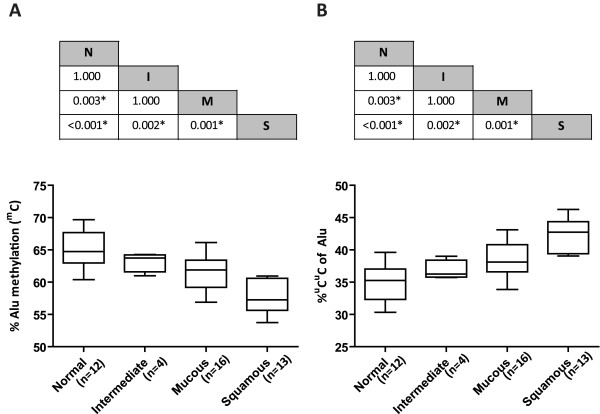
The frequency of each Alu element methylation pattern is shown in Table 3. Similar to LINE-1, total Alu element methylation (mC) decreased from N to M, I and S. The results showed significant differences between S:M (p=0.001), S:I (p=0.002), S:N (p<0.001) and M:N (p=0.003). However, there was no significant difference between M:I and N:I (p=1.000) (Figure 3A).
Table 3.
Frequency of Alu element methylation patterns in MEC cell subtypes, whole MEC tissues and normal salivary glands
| Alu patterns | %mC | %mCmC | %mCuC+uCmC | %uCuC |
|---|---|---|---|---|
| Adjacent normal salivary gland cell (N) |
65.10±2.80 |
23.36±6.42 |
41.74±4.43 |
34.89±2.80 |
| Intermediate cell (I) |
63.18±1.51 |
23.66±10.76 |
39.52±9.32 |
36.81±1.51 |
| Mucous cell (M) |
61.48±2.46 |
21.02±6.83 |
40.45±7.09 |
38.51±2.46 |
| Squamous cell (S) |
57.51±2.46 |
23.74±5.25 |
33.77±4.39 |
42.48±2.46 |
| Normal salivary gland (NG) |
64.52±4.66 |
18.53±10.16 |
45.99±8.97 |
35.47±4.66 |
| Whole MEC tissue (MEC) | 57.49±5.35 | 22.21±5.13 | 35.27±5.02 | 42.51±5.35 |
Figure 3.
Comparison of the frequency of total Alu element methylation (mC) and uCuC of Alu elements among MEC cell subtypes. (A) Alu element methylation among cell types showed a stepwise decrease from normal cells to intermediate cells, mucous cells and squamous cell. The p‐value between each group is shown in the table above the graph. (B) The frequency of uCuC of Alu elements among cell types showed a stepwise increase from normal cells to intermediate cells, mucous cells and squamous cells. The p‐value between each group is shown in the table above the graph.
On the contrary, the frequency of uCuC of Alu elements increased from N to M, I and S, respectively. A significant difference was found between S:M (p=0.001), S:I (p=0.002), S:N (p<0.001) and M:N (p=0.003). No significant difference was found between M:I and N:I (p=1.000) (Figure 3B).
A significant difference in the percentage of mCuC + uCmC of Alu elements was found between S:M (p=0.028) and S:N (p=0.011). However, there was no significant difference between S:I (p=0.061), M:N, M:I and N:I (p=1.000). Moreover, the frequency of mCmC of Alu elements showed no significant difference between groups of microdissected cells.
For the paired comparisons, significant differences in both mC and uCuC of Alu elements were observed as followed: N:M (p=0.032), N:S (p<0.001) and I:M (p=0.008). The detailed data of paired comparisons are shown in Additional file 1: Table S1.
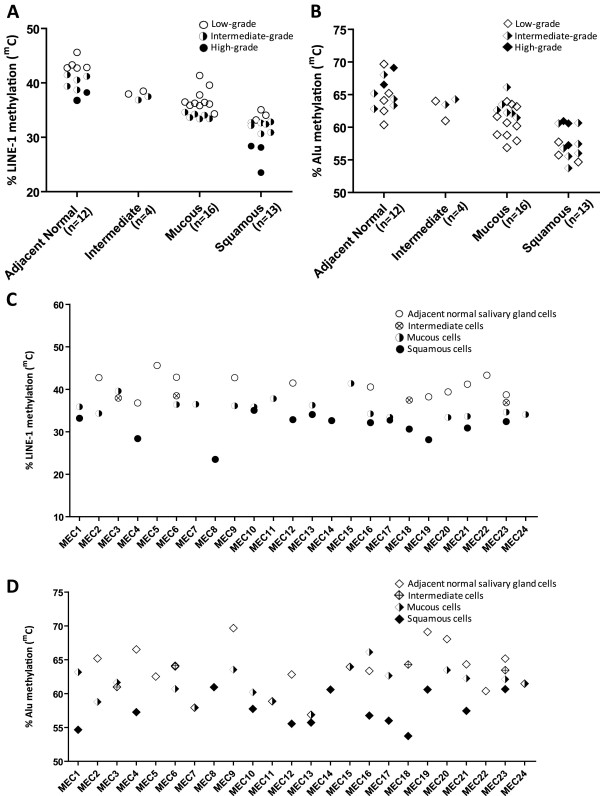
LINE-1 and Alu element methylation in MECs of various histological grades
Total LINE-1 methylation levels (mC) of microdissected cells in each cell type decreased from low-grade to intermediate-grade and high-grade MEC, p<0.001 (Figure 4A). However, total Alu element methylation (mC) in microdissected cells was not related to the histological grade of the MEC (Figure 4B). Interestingly, when we compared the total LINE-1 and Alu element methylation levels of microdissected cells in each specimen, more than 80% of the cases showed decreasing levels of LINE-1 and Alu element methylation from N to I, M and S. (Figure 4C, D). These results demonstrate that genomic hypomethylation, and specifically LINE-1 methylation, essentially correlates with poorer histological grade of MECs.
Figure 4.
LINE-1 and Alu element methylation levels among MEC cell subtypes. (A) LINE-1 methylation in MEC cell subtypes correlated with the histological grade of the MEC. (B) Alu element methylation level in MEC cell subtypes did not correlate with the histological grade of the MEC. (C) LINE-1 methylation level of each microdissected MEC specimen. (D) Alu element methylation level of each microdissected MEC specimen.
LINE-1 and Alu element methylation in whole MEC tissue
We next asked whether these methods could be used to detect and correctly classify MECs. To address this question, we analyzed LINE-1 and Alu element methylation in whole MEC tissues, and compared with normal salivary gland material.
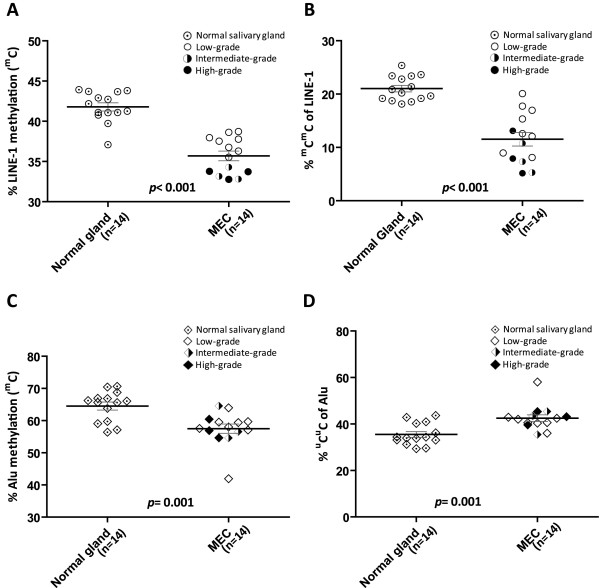
The frequency of each LINE-1 methylation pattern in whole MEC tissue is shown in Table 2. The frequency of mC and mCmC LINE-1s were significantly lower in MEC tissue than in normal salivary gland tissue (p<0.001) (Table 2, Figure 5A and B). Moreover, the frequency of mC of LINE-1s in the low-grade MECs was higher than in intermediate-grade (p<0.001) and high-grade MECs (p<0.001), respectively (Figure 5A and B).
Figure 5.
Comparison of total LINE-1 and Alu element methylation between normal salivary gland tissue and whole MEC tissue. (A, B) The frequency of mC and mCmC of LINE-1 methylation in whole MEC tissue was significantly lower than in normal salivary gland tissue (p<0.001). (C) The frequency of mC of Alu elements in whole MEC tissue was significantly lower than in normal salivary gland tissue (p=0.001). (D) The frequency of uCuC Alu elements in whole MEC tissue was significantly higher than in normal salivary gland tissue (p=0.001).
The frequency of each Alu element methylation pattern in whole MEC tissue is shown in Table 3. Similar to LINE-1, the total Alu element methylation level (mC) in MEC tissue was also significantly lower than in normal salivary gland tissue (p=0.001). In agreement with these results, the frequency of uCuC of Alu elements in MEC tissue was significantly higher than in normal salivary gland tissue (p=0.001) (Table 3, Figure 5C and D). However, Alu element methylation in whole MEC tissue was not related to the histological grade of the MEC (Figure 5C and D).
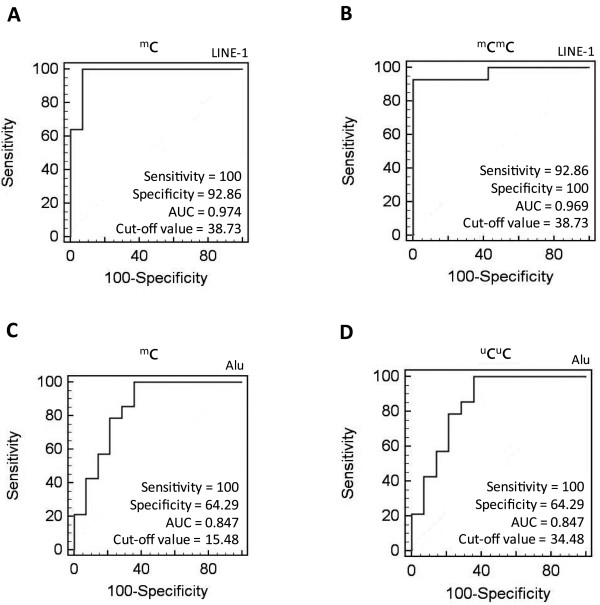
Receiver operating characteristic (ROC) analysis of LINE-1 and Alu element methylation
Next, we assessed the ability of these methods to discriminate between MEC tissue and normal salivary gland tissue using an ROC analysis. Among the various patterns of LINE-1 methylation, both the mC and the mCmC patterns yielded ROC values indicative of diagnostic reliability. For the mC pattern of LINE-1, the area under the ROC curve (AUC) value was 0.974, while the cut-off value, sensitivity and specificity were 38.73%, 100% and 92.86%, respectively (Figure 6A). The AUC value of the mCmC pattern of LINE-1 was 0.969, while the cut-off value, sensitivity and specificity were 38.73%, 92.86% and 100%, respectively (Figure 6B).
Figure 6.
ROC curve analysis of LINE-1 and Alu element methylation for MEC detection. (A) The total LINE-1 methylation level (mC). (B) The mCmC level of LINE-1 methylation. (C) The total Alu element methylation level (mC). (D) The uCuC level of Alu element methylation.
Among the various patterns of Alu element methylation, the mC and uCuC patterns demonstrated reasonable diagnostic values. Both the mC and the uCuC of Alu element methylation patterns had AUC values, sensitivity and specificity of 0.847, 100% and 64.29%, respectively. The cut-off values for the mC and uCuC of Alu element methylation patterns were 15.48% and 34.48%, respectively (Figure 6C and D). These results indicate that ROC analysis of LINE-1 methylation may have a stronger diagnostic value than analysis of Alu element methylation. This analysis is especially more effective when both the mC and mCmC patterns of LINE-1 methylation are assessed.
Discussion
To the best of our knowledge, this report represents the first epigenetic study of human MEC. We characterized the methylation status of the repetitive sequences in clinical samples of MEC, and our results clearly show that LINE-1 hypomethylation is in concordant with a poorer histological grade. The COBRA technique represents an excellent approach for detecting the methylation status [41,47], and using for example COBRALINE-1 and COBRAAlu, both are effective in detecting genome-wide methylation status of LINE-1s and Alu elements, respectively, in genomic DNA [48]. In our study, we used a modified method for COBRALINE-1 and COBRAAlu assessing the methylation status, as shown in Figure 1. These methods detected 2 CpG dinucleotide sites and can explain not only methylation level but also methylation patterns which was not reveal by pyrosequencing technique [41].
Although MEC is the most common salivary gland cancer, the overall incidence of occurrence in human is extremely low and thus rarely diagnosed. Therefore, one of the limitations of our study is the small number of MEC samples available for investigation. Theoretically, the parotid gland is the most common site of this tumor; however most of MEC samples used in this study were collected from the minor salivary glands of the palate. In this context, to maximize the number of samples available for analysis, we used laser capture microdissection (LCM), essentially as we have previously shown that this is a very sensitive method for isolating a minimal number cells of interest from whole tissue sections and performing molecular analysis on the extracted nucleic acid. In this study, we procured ~1,500 microdissected cells, which provided sufficient DNA to allow the detection of LINE-1 and Alu methylation levels and pattern. However, LCM did not allow the sufficient isolation of all three cell types from most of the MEC cases, but with some exceptions, for example cases MEC23 (Figure 4C and D), we could efficiently collected every cell population for analysis.
From our data we found that LINE-1 and Alu methylation levels were different among the three histological grades of MEC. We also observed that LINE-1 hypomethylation in adjacent normal salivary gland cells was dependent on the histological grade of the MEC (Figure 4A). This appearance may be explained by some signaling proteins released by cancer cells that can have an influence on normal surrounding tissue in the nearby vicinity [49]. Moreover, the level of LINE-1 methylation in intermediate cells was between that of adjacent normal salivary glands and mucous cells which seem to be correlated with the hypothesis proposed by Luna (2006). According to this author, the intermediate cells, which are derived from reserve cells of salivary duct unit, are believed to be the progenitor cells of the other three cell types of MEC (mucous cells, epidermoid cells and clear cells), and thus they may represent cells in halfway of differentiation between normal reserve cells and the other three cell types of MEC [50]. Since we could not measure the methylation level directly from the reserve cells as they are extremely hard to be identified by microscopic examination, we cannot conclude that the LINE-1 methylation level decrease along the pathway of cell differentiation from the reserve cells of salivary duct unit to the other three cell types of MEC as proposed by Luna (2006).
In conclusion, our findings provide preliminary information of methylation levels between different cell components in MEC, which may be related to histological grading and prognosis of the neoplasm. The knowledge may be applied as a diagnostic tool or a prognostic marker for these tumors in addition to histological grading.
Abbreviations
MEC: Mucoepidermoid carcinoma; LINE-1: Long INterspersed Element-1; LCM: Laser capture microdissection; COBRA: Combine Bisulfite Restriction Analysis.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
PS carried out all experiments, performed the statistical analysis and drafted the manuscript. SK and KD participated in the patient enrollment. AM, KS and NK conceived the study, participated in its design and coordination, and revised the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Paired comparison of LINE-1 and Alu methylation patterns among MEC cell subtypes.
Contributor Information
Porntipa Sirivanichsuntorn, Email: lippolon@hotmail.com.
Somboon Keelawat, Email: trcskl@gmail.com.
Kittipong Danuthai, Email: fibroma123@yahoo.com.
Apiwat Mutirangura, Email: apiwat.mutirangura@gmail.com.
Keskanya Subbalekha, Email: skeskanya@gmail.com.
Nakarin Kitkumthorn, Email: nakarinkit@gmail.com.
Acknowledgement
We would like to thank Dr. Vyomesh Patel (NIDCR/NIH) for critically review the manuscript, Mr. Dusit Bumalee for his help in preparing tissue samples; Mr. Prakasit Rattanatanyong and Mr. Surasak Yooyongsatit for their assistance during laboratory procedures. We also thank the Department of Microbiology, Faculty of Medicine, Chulalongkorn University for providing the laser capture microdissection system. This study was financially supported by The 90th Anniversary of Chulalongkorn University Fund and Ratchadapiseksomphot Endowment Fund, the Center of Excellence in Molecular Genetics of Cancer and Human Diseases, Department of Anatomy, Faculty of Medicine, Chulalongkorn University, Research Chair Grant 2011 from National Science and Technology Development Agency (NSTDA), Thailand, and TRF-MRG young scientific researcher grant No.MRG5380010 by Thailand Research Fund.
References
- Barnes LEJ, Reichart P, Sidransky D. World Health Organization Classification of Tumours. Pathology and Genetics of Head and Neck Tumours. Lyon: IARC press; 2005. [Google Scholar]
- Lopes MA, Kowalski LP, da Cunha Santos G, Paes De Almeida O. A clinicopathologic study of 196 intraoral minor salivary gland tumours. J Oral Pathol Med. 1999;28(6):264–267. doi: 10.1111/j.1600-0714.1999.tb02036.x. [DOI] [PubMed] [Google Scholar]
- Vargas PA, Gerhard R, Araujo Filho VJ, de Castro IV. Salivary gland tumors in a Brazilian population: a retrospective study of 124 cases. Rev Hosp Clin Fac Med Sao Paulo. 2002;57(6):271–276. doi: 10.1590/s0041-87812002000600005. [DOI] [PubMed] [Google Scholar]
- Waldron CA, El-Mofty SK, Gnepp DR. Tumors of the intraoral minor salivary glands: a demographic and histologic study of 426 cases. Oral Surg Oral Med Oral Pathol. 1988;66(3):323–333. doi: 10.1016/0030-4220(88)90240-X. [DOI] [PubMed] [Google Scholar]
- Ellis GL. Tumours of the salivary glands. 3. Washington: Armed Forced Institute of Pathology; 1996. [Google Scholar]
- Auclair PL, Goode RK, Ellis GL. Mucoepidermoid carcinoma of intraoral salivary glands. Evaluation and application of grading criteria in 143 cases. Cancer. 1992;69(8):2021–2030. doi: 10.1002/1097-0142(19920415)69:8<2021::AID-CNCR2820690803>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Brandwein MS, Ivanov K, Wallace DI, Hille JJ, Wang B, Fahmy A, Bodian C, Urken ML, Gnepp DR, Huvos A. Mucoepidermoid carcinoma: a clinicopathologic study of 80 patients with special reference to histological grading. Am J Surg Pathol. 2001;25(7):835–845. doi: 10.1097/00000478-200107000-00001. [DOI] [PubMed] [Google Scholar]
- Evans HL. Mucoepidermoid carcinoma of salivary glands: a study of 69 cases with special attention to histologic grading. Am J Clin Pathol. 1984;81(6):696–701. doi: 10.1093/ajcp/81.6.696. [DOI] [PubMed] [Google Scholar]
- Nascimento AG, Amaral LP, Prado LA, Kligerman J, Silveira TR. Mucoepidermoid carcinoma of salivary glands: a clinicopathologic study of 46 cases. Head Neck Surg. 1986;8(6):409–417. doi: 10.1002/hed.2890080604. [DOI] [PubMed] [Google Scholar]
- Spiro RH, Huvos AG, Berk R, Strong EW. Mucoepidermoid carcinoma of salivary gland origin. A clinicopathologic study of 367 cases. Am J Surg. 1978;136(4):461–468. doi: 10.1016/0002-9610(78)90262-3. [DOI] [PubMed] [Google Scholar]
- da Silveira EJ, Veras Barros SS, de Amorim RF, Queiroz LM, Freitas Rde A, de Souza LB. Cytokeratin profile in mucoepidermoid carcinoma is not related to its histological grading of malignancy. Exp Mol Pathol. 2006;81(1):72–76. doi: 10.1016/j.yexmp.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Skalova A, Lehtonen H, von Boguslawsky K, Leivo I. Prognostic significance of cell proliferation in mucoepidermoid carcinomas of the salivary gland: clinicopathological study using MIB 1 antibody in paraffin sections. Hum Pathol. 1994;25(9):929–935. doi: 10.1016/0046-8177(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Tipoe GL, White FH. Proliferative activity as detected by immunostaining with Ki-67 and proliferating cell nuclear antigen in benign and malignant epithelial lesions of the human parotid gland. Anal Quant Cytol Histol. 1999;21(4):336–342. [PubMed] [Google Scholar]
- Stenman G. Fusion oncogenes and tumor type specificity–insights from salivary gland tumors. Semin Cancer Biol. 2005;15(3):224–235. doi: 10.1016/j.semcancer.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Okumura Y, Miyabe S, Nakayama T, Fujiyoshi Y, Hattori H, Shimozato K, Inagaki H. Impact of CRTC1/3-MAML2 fusions on histological classification and prognosis of mucoepidermoid carcinoma. Histopathology. 2011;59(1):90–97. doi: 10.1111/j.1365-2559.2011.03890.x. [DOI] [PubMed] [Google Scholar]
- Nakayama T, Miyabe S, Okabe M, Sakuma H, Ijichi K, Hasegawa Y, Nagatsuka H, Shimozato K, Inagaki H. Clinicopathological significance of the CRTC3-MAML2 fusion transcript in mucoepidermoid carcinoma. Mod Pathol. 2009;22(12):1575–1581. doi: 10.1038/modpathol.2009.126. [DOI] [PubMed] [Google Scholar]
- El-Naggar AK, Lovell M, Killary AM, Clayman GL, Batsakis JG. A mucoepidermoid carcinoma of minor salivary gland with t(11;19)(q21;p13.1) as the only karyotypic abnormality. Cancer Genet Cytogenet. 1996;87(1):29–33. doi: 10.1016/0165-4608(95)00266-9. [DOI] [PubMed] [Google Scholar]
- Horsman DE, Berean K, Durham JS. Translocation (11;19)(q21;p13.1) in mucoepidermoid carcinoma of salivary gland. Cancer Genet Cytogenet. 1995;80(2):165–166. doi: 10.1016/0165-4608(94)00187-G. [DOI] [PubMed] [Google Scholar]
- Nordkvist A, Gustafsson H, Juberg-Ode M, Stenman G. Recurrent rearrangements of 11q14-22 in mucoepidermoid carcinoma. Cancer Genet Cytogenet. 1994;74(2):77–83. doi: 10.1016/0165-4608(94)90001-9. [DOI] [PubMed] [Google Scholar]
- Yoo J, Robinson RA. H-ras gene mutations in salivary gland mucoepidermoid carcinomas. Cancer. 2000;88(3):518–523. doi: 10.1002/(SICI)1097-0142(20000201)88:3<518::AID-CNCR4>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Chalitchagorn K, Shuangshoti S, Hourpai N, Kongruttanachok N, Tangkijvanich P, Thong-ngam D, Voravud N, Sriuranpong V, Mutirangura A. Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene. 2004;23(54):8841–8846. doi: 10.1038/sj.onc.1208137. [DOI] [PubMed] [Google Scholar]
- Piyathilake CJ, Frost AR, Bell WC, Oelschlager D, Weiss H, Johanning GL, Niveleau A, Heimburger DC, Grizzle WE. Altered global methylation of DNA: an epigenetic difference in susceptibility for lung cancer is associated with its progression. Hum Pathol. 2001;32(8):856–862. doi: 10.1053/hupa.2001.26471. [DOI] [PubMed] [Google Scholar]
- Kitkumthorn N, Mutirangura A. LINE-1 methylation difference between ameloblastoma and keratocystic odontogenic tumor. Oral Dis. 2010;16(3):286–291. doi: 10.1111/j.1601-0825.2009.01640.x. [DOI] [PubMed] [Google Scholar]
- Kazazian HH Jr, Moran JV. The impact of L1 retrotransposons on the human genome. Nat Genet. 1998;19(1):19–24. doi: 10.1038/ng0598-19. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH Jr. Biology of mammalian L1 retrotransposons. Annu Rev Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- Kwon HJ, Kim JH, Bae JM, Cho NY, Kim TY, Kang GH. DNA methylation changes in ex-adenoma carcinoma of the large intestine. Virchows Arch. 2010;457(4):433–441. doi: 10.1007/s00428-010-0958-9. [DOI] [PubMed] [Google Scholar]
- Park SY, Yoo EJ, Cho NY, Kim N, Kang GH. Comparison of CpG island hypermethylation and repetitive DNA hypomethylation in premalignant stages of gastric cancer, stratified for Helicobacter pylori infection. J Pathol. 2009;219(4):410–416. doi: 10.1002/path.2596. [DOI] [PubMed] [Google Scholar]
- Pattamadilok J, Huapai N, Rattanatanyong P, Vasurattana A, Triratanachat S, Tresukosol D, Mutirangura A. LINE-1 hypomethylation level as a potential prognostic factor for epithelial ovarian cancer. Int J Gynecol Cancer. 2008;18(4):711–717. doi: 10.1111/j.1525-1438.2007.01117.x. [DOI] [PubMed] [Google Scholar]
- Shuangshoti S, Hourpai N, Pumsuk U, Mutirangura A. Line-1 hypomethylation in multistage carcinogenesis of the uterine cervix. Asian Pac J Cancer Prev. 2007;8(2):307–309. [PubMed] [Google Scholar]
- Tangkijvanich P, Hourpai N, Rattanatanyong P, Wisedopas N, Mahachai V, Mutirangura A. Serum LINE-1 hypomethylation as a potential prognostic marker for hepatocellular carcinoma. Clin Chim Acta. 2007;379(1–2):127–133. doi: 10.1016/j.cca.2006.12.029. [DOI] [PubMed] [Google Scholar]
- Bollati V, Fabris S, Pegoraro V, Ronchetti D, Mosca L, Deliliers GL, Motta V, Bertazzi PA, Baccarelli A, Neri A. Differential repetitive DNA methylation in multiple myeloma molecular subgroups. Carcinogenesis. 2009;30(8):1330–1335. doi: 10.1093/carcin/bgp149. [DOI] [PubMed] [Google Scholar]
- Cho NY, Kim BH, Choi M, Yoo EJ, Moon KC, Cho YM, Kim D, Kang GH. Hypermethylation of CpG island loci and hypomethylation of LINE-1 and Alu repeats in prostate adenocarcinoma and their relationship to clinicopathological features. J Pathol. 2007;211(3):269–277. doi: 10.1002/path.2106. [DOI] [PubMed] [Google Scholar]
- Ogino S, Kawasaki T, Nosho K, Ohnishi M, Suemoto Y, Kirkner GJ, Fuchs CS. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int J Cancer. 2008;122(12):2767–2773. doi: 10.1002/ijc.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz WA, Elo JP, Florl AR, Pennanen S, Santourlidis S, Engers R, Buchardt M, Seifert HH, Visakorpi T. Genomewide DNA hypomethylation is associated with alterations on chromosome 8 in prostate carcinoma. Genes Chromosomes Cancer. 2002;35(1):58–65. doi: 10.1002/gcc.10092. [DOI] [PubMed] [Google Scholar]
- Choi IS, Estecio MR, Nagano Y, Kim Do H, White JA, Yao JC, Issa JP, Rashid A. Hypomethylation of LINE-1 and Alu in well-differentiated neuroendocrine tumors (pancreatic endocrine tumors and carcinoid tumors) Mod Pathol. 2007;20(7):802–810. doi: 10.1038/modpathol.3800825. [DOI] [PubMed] [Google Scholar]
- Kim MJ, White-Cross JA, Shen L, Issa JP, Rashid A. Hypomethylation of long interspersed nuclear element-1 in hepatocellular carcinomas. Mod Pathol. 2009;22(3):442–449. doi: 10.1038/modpathol.2008.203. [DOI] [PubMed] [Google Scholar]
- Wolff EM, Byun HM, Han HF, Sharma S, Nichols PW, Siegmund KD, Yang AS, Jones PA, Liang G. Hypomethylation of a LINE-1 promoter activates an alternate transcript of the MET oncogene in bladders with cancer. PLoS Genet. 2010;6(4):e1000917. doi: 10.1371/journal.pgen.1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woloszynska-Read A, Mhawech-Fauceglia P, Yu J, Odunsi K, Karpf AR. Intertumor and intratumor NY-ESO-1 expression heterogeneity is associated with promoter-specific and global DNA methylation status in ovarian cancer. Clin Cancer Res. 2008;14(11):3283–3290. doi: 10.1158/1078-0432.CCR-07-5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Kim BH, Cho NY, Yoo EJ, Choi M, Shin SH, Jang JJ, Suh KS, Kim YS, Kang GH. Prognostic implications of and relationship between CpG island hypermethylation and repetitive DNA hypomethylation in hepatocellular carcinoma. Clin Cancer Res. 2009;15(3):812–820. doi: 10.1158/1078-0432.CCR-08-0266. [DOI] [PubMed] [Google Scholar]
- Pobsook T, Subbalekha K, Sannikorn P, Mutirangura A. Improved measurement of LINE-1 sequence methylation for cancer detection. Clin Chim Acta. 2011;412(3–4):314–321. doi: 10.1016/j.cca.2010.10.030. [DOI] [PubMed] [Google Scholar]
- Kitkumthorn N, Mutirangura A. Long interspersed nuclear element-1 hypomethylation in cancer: biology and clinical applications. Clin Epigenet. 2011;2:315–330. doi: 10.1007/s13148-011-0032-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzo M, Andreola S, Sirizzotti G, Cantu G. Mucoepidermoid carcinoma of the salivary glands: clinicopathologic review of 108 patients treated at the National Cancer Institute of Milan. Ann Surg Oncol. 2002;9(7):688–695. doi: 10.1007/BF02574486. [DOI] [PubMed] [Google Scholar]
- Monoo K, Sageshima M, Ito E, Nishihira S, Ishikawa K. [Histopathological grading and clinical features of patients with mucoepidermoid carcinoma of the salivary glands] Nihon Jibiinkoka Gakkai Kaiho. 2003;106(3):192–198. doi: 10.3950/jibiinkoka.106.192. [DOI] [PubMed] [Google Scholar]
- Wangsri S, Subbalekha K, Kitkumthorn N, Mutirangura A. Patterns and Possible Roles of LINE-1 Methylation Changes in Smoke-Exposed Epithelia. PLoS One. 2012;7(9):e45292. doi: 10.1371/journal.pone.0045292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanatatsaneeji P, Kitkumthorn N, Dhammawipark C, Rabalert J, Patel V, Mutirangura A. Codon72 polymorphism in the p53 tumor suppressor gene in oral lichen planus lesions in a Thai population. Asian Pac J Cancer Prev. 2010;11(4):1137–1141. [PubMed] [Google Scholar]
- Kitkumthorn N, Tuangsintanakul T, Rattanatanyong P, Tiwawech D, Mutirangura A. LINE-1 methylation in the peripheral blood mononuclear cells of cancer patients. Clin Chim Acta. 2012;413(9-10):869–874. doi: 10.1016/j.cca.2012.01.024. [DOI] [PubMed] [Google Scholar]
- Kitkumthorn N, Keelawat S, Rattanatanyong P, Mutirangura A. LINE-1 and Alu Methylation Patterns in Lymph Node Metastasis of Head and Neck Cancers. Asian Pac J Cancer Prev. 2012;13(9):4469–4475. doi: 10.7314/APJCP.2012.13.9.4469. [DOI] [PubMed] [Google Scholar]
- Aboussekhra A. Role of cancer-associated fibroblasts in breast cancer development and prognosis. Int J Dev Biol. 2011;55(7–9):841–849. doi: 10.1387/ijdb.113362aa. [DOI] [PubMed] [Google Scholar]
- Luna MA. Salivary mucoepidermoid carcinoma: revisited. Adv Anat Pathol. 2006;13(6):293–307. doi: 10.1097/01.pap.0000213058.74509.d3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Paired comparison of LINE-1 and Alu methylation patterns among MEC cell subtypes.