Abstract
Combinations of phosphorylation, acetylation, methylation, ubiquitylation, and sumoylation of histones comprise what is referred to as the “histone code”. These marks influence processes from transcription to DNA replication, where gaining access to DNA organized in chromatin is necessary. Much emphasis has been placed on the role of histone ubiquitylation and sumoylation during the process of transcription. Histone H2B is monoubiquitylated at lysine 123 in budding yeast and influences gene activation. All four of the core histones are sumoylated on their amino terminal tails in this organism, and this serves to negatively regulate gene expression. Because antibodies specific for ubiquitylated or sumoylated yeast histones are not commercially available, and these marks are highly sensitive to proteolysis in native cell extracts, special genetic and molecular tools have been developed to monitor these dynamic and often rare modifications in vivo. Here, we describe some of these tools, with emphasis on how they can be used for transcriptional studies.
Keywords: Ubiquitylation, Sumoylation, Histones, Yeast, Transcription
1. Introduction
The first ubiquitylated protein to be characterized in eukaryotes was H2A in 1975 [1]. Since then, this modification has been found on all four of the core histones, the histone variants H2A.Z and H2A.X, and the linker histone H1 in a wide range of organisms. Given the diverse roles of histone ubiquitylation, ranging from transcription to the DNA damage response, it is surprising that in budding yeast the only ubiquitylated histone identified to date is H2B. Here, its primary role is in gene activation. In contrast, all four core histones are sumoylated in this organism, and as first reported for sumoylated H4 in mammals, this mark is involved in gene repression [2]. The importance of these marks has generated intensive research in recent years, and in this article we present methods to isolate, identify, and characterize histone ubiquitylation and sumoylation in the budding yeast, Saccharomyces cerevisiae.
1.1 Monoubiquitylation of histone H2B
Monoubiquitylation of histone H2B (H2Bub1) has been associated with both transcriptional activation and gene silencing, and it is also required for the trans-histone methylation of histone H3 on lysine 4 (H3K4) by COMPASS [3–6] and lysine 79 (H3K79) by Dot1 [7, 8], modifications closely associated with the regulation of gene expression. In S. cerevisiae, H2B monoubiquitylation is mediated by the E2 conjugating enzyme, Rad6, in conjunction with the E3 ubiquitin ligase, Bre1 [8–10]. The reverse deubiquitylation process is carried out by two distinct ubiquitin specific proteases. Deubiquitylation by Ubp8, a component of the SAGA complex (Spt-Ada-Gcn5-Acetyltransferase), controls the transient H2B ubiquitylation associated with gene activation [11, 12]. In contrast, Ubp10 preferentially removes the mark at regions of heterochromatin such as telomeres and is important for gene silencing [13, 14]. In S. cerevisiae, the site of monoubiquitylation was found to be lysine 123 [10]. A conservative mutation changing this lysine to arginine (htb-K123R) prevents ubiquitylation but does not impart structural changes to the nucleosome core particle in vitro [10, 15]. However, chromatin becomes somewhat destabilized in vivo in an htb-K123R mutant [16, 17]. Strains with this mutation have been used by numerous laboratories to elucidate the role of H2Bub1 in a variety of cellular processes, most prominently transcription, where H2Bub1 has been shown to be dynamically regulated (Figure 1). During transcription elongation, the sequential ubiquitylation and deubiquitylation of histone H2B acts as a checkpoint for Ctk1-dependent H3 lysine 36 methylation [12] and regulates nucleosome reassembly in the wake of RNA polymerase II [17].
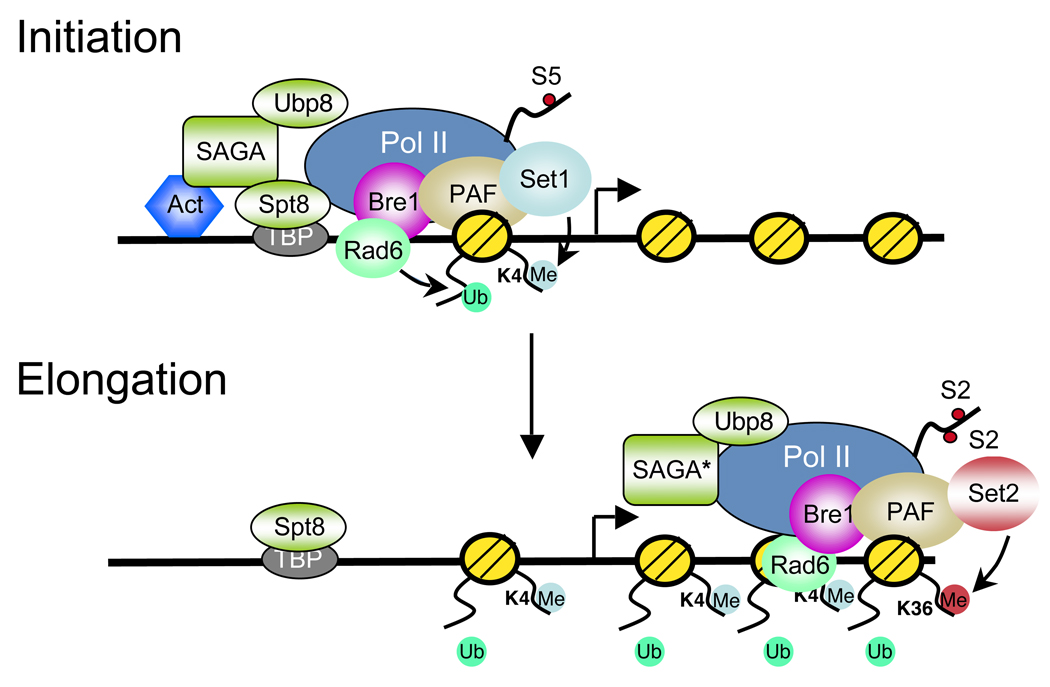
Figure 1. Dynamic ubiquitylation and deubiquitylation of H2B during transcription initiation and elongation.
Activators such as Gal4 recruit the Bre1/Rad6 and SAGA histone acetyltransferase/deubiquitinase complexes to gene promoters. RNA polymerase II (Pol II) CTD phosphorylation on serine 5 (S5) by Kin28 recruits the PAF complex, which in turn recruits Rad6/Bre1 and the Set1 histone methytransferase complex to Pol II. This association results in H2BK123ub1 and H3K4me3 formation at the promoter and 5’ coding region during the initiation phase of transcription. H2BK123ub1 is a prerequisite for Set1-mediated H3K4me3 in these regions. A SAGA subcomplex (*) that contains the Ubp8 H2B ubiquitin protease module also associates with Pol II and deubiquitylates H2BK123ub1. Both the Bre1/Rad6 and the SAGA-Ubp8 complexes travel with Pol II across the coding region and lead to a cycle of H2B ubiquitylation and deubiquitylation during transcription elongation. The dynamic regulation of H2B ubiquitylation has at least two functions during this phase of transcription. First, deubiquitylation of H2B by Ubp8 allows Ctk1 recruitment and Pol II CTD phosphorylation on serine 2(S2). This in turn results in the recruitment of the Set2 histone methyltransferase and the formation of H3K36me3 at the 3’ coding region. Second, H2B ubiquitylation plays a role in nucleosome dynamics by promoting nucleosome reassembly in the wake of elongating Pol II. Both functions are postulated to contribute to the fidelity of transcription elongation.
1.2 Histone sumoylation
In comparison to H2B ubiquitylation, less is known about histone sumoylation, which was characterised as the first histone modification to be associated with transcriptional repression in S. cerevisiae [18]. SUMO was found on all four of the core histones, and was suggested to exist in a dynamic interplay with histone acetylation and ubiquitylation [18]. Furthermore, a two fold increase in sumoylated histone H2B (H2B-SUMO) was found adjacent to telomeric repeats, in contrast to H2Bub1, which is depleted in these same heterochromatic regions [19]. SUMO (encoded by SMT3 in S. cerevisiae) is a conserved modifier protein of 101 amino acids that shares 3D structural similarity and a related pathway of activation with ubiquitin [20]. In yeast, the E1 is the heterodimer Aos1/Uba2 which activates SUMO [21]. The only known E2 conjugating enzyme in this organism is Ubc9, which is capable of ligating SUMO to its target. However, several E3s including Siz1, Siz2 and Mms21 confer specificity to the process [22–24]. Like histone ubiquitylation, sumoylation of histones is dynamically regulated, and the Smt4 isopeptidase specifically targets sumoylated proteins to remove the mark [25]. Sites of sumoylation on H2B were found to be K6/7 and K16/17, although mutation of the four lysines to alanine did not completely abolish sumoylation [18], suggesting that there are alternative sites of SUMO modification. In the same study, mass spectrometry indicated a sumoylation site at K126 of histone H2A, although an hta-K126R mutation did not cause a significant reduction in sumoylation levels compared to wild type. In agreement with studies on mammalian histones [2], deletion of the N-terminal tail of yeast histone H4 resulted in a considerable reduction in sumoylation, implicating the N tail of this histone as the primary target of this modification [18].
1.3 Detection of histone ubiquitylation and sumoylation
For researchers interested in studying the roles of histone ubiquitylation and sumoylation in yeast, several obstacles to the detection of these modifications have to be overcome. Until recently, there were no specific antibodies directed against these modified histones. Although a polyclonal antibody has recently been described for ubiquitylated H2B, this is not yet commercially available [26]. Attempts by the Berger laboratory to develop H2B-SUMO antibodies using branched peptides as antigens have also been unsuccessful [18]. Importantly, the modified histones are present as a very small fraction of total histones. H2Bub1 accounts for roughly 10% or less of total cellular H2B, while sumoylated histones are present at even lower levels [10, 18]. Finally, both histone modifications are very labile in native yeast extracts and highly susceptible to proteolysis.
In this article we will present tools that have been developed to circumvent these major obstacles by describing:
Strains used to bypass the requirement for specific antibodies against ubiquitylated H2B and sumoylated histones to allow detection of these low abundance histone modifications.
Methods for detecting the modifications on both global and local levels.
Specific examples from the literature that have been associated with the described methods, emphasizing the relationship of SUMO and ubiquitin modified histones to transcription.
2. Methods for the global detection of histone ubiquitylation and sumoylation
In this section, we have outlined genetic and immunological methods that can be applied to the enrichment and detection of ubiquitin or sumo-conjugated histones. These methods utilize special yeast strains that contain epitope tags on histones, ubiquitin, and SUMO and procedures to isolate the histone conjugates under denaturing conditions to prevent proteolysis of the marks.
2.1 Strains and plasmids
Table 1 lists some of the key strains that have been used to study H2Bub1 and histone sumoylation. S. cerevisiae has only two chromosomal copies of each of the core histone genes, which can be deleted and replaced with plasmid borne copies to maintain cell viability. In addition, several ‘shuffle strains’ exist for the study of H2A-H2B or H3-H4, having single copies of the wild type gene pairs on a URA3 plasmid. This plasmid can be replaced with one carrying an epitope tagged or mutant copy of the histone gene of interest by ‘plasmid shuffling’ using 5-FOA selection [27]. Typically, the histone genes have been tagged with Flag, HA, or Myc epitopes, which have been used for both western blot analysis and chromatin immunoprecipitation (ChIP). Research has shown that the Flag tag, in particular, does not affect the functions of H2B [26, 28]. It is also possible to tag yeast histone genes in the genome, although this method has not been routinely used [29]. While strains JR5-2A and Y131 were originally used to study H2Bub1, the discovery that the Y131 strain harbors a wild type copy of HTA2-HTB2 under the regulation of the GAL1/10 promoter on chromosome II makes FY406-derived strains essential for studies requiring prolonged (e.g. longer than 2 hours) galactose induction [26].
Table 1.
Strains used to study histone ubiquitylation and sumoylation
| Strain | Genotype | Source |
|---|---|---|
| YZS276 |
MATa, hta1-htb1∷LEU2, HTA2-GAL1/GAL10-HTB2, ura3-1, trp1-1, leu2-3,-112, his3-11, ade2-1, can1-100 <pZS145 HTA1-Flag-HTB1,CEN, HIS3> |
[5] |
| YZS277 |
MATa, hta1-htb1∷LEU2, HTA2-GAL1/GAL10-HTB2, ura3-1, trp1-1, leu2-3,-112, his3-11, ade2-1, can1-100 <pZS146 HTA1-Flag-htb1-K123R, CEN,HIS3> |
[5] |
| YSN545 | MATa (hta1-htb1)Δ∷LEU2, (hta2-htb2) Δ∷TRP1, his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128Δ <pZS145 HTA1-Flag-HTB1-HIS3> |
[26] |
| YSN763 | MATa (hta1-htb1)Δ∷LEU2, (hta2-htb2) Δ∷TRP1, his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128Δ <pZS145 HTA1-Flag- htb1K123A-HIS3> |
[26] |
| YSF200 | MATa his3 1 leu2 0 met15 0 ura3 hhf2-hht2∷NAT hta1- htb1∷HPH hht1-hht2∷KAN hta2-htb2∷NAT <pRS315- HHT2-HHF2-HTA1-Flag-HTB1> |
[26] |
| YSF201 | MATa his3 1 leu2 0 met15 0 ura3 hhf2-hht2∷NAT hta1- htb1∷HPH hht1-hht2∷KAN hta2-htb2∷NAT <pRS315- HHT2-HHF2-HTA1-Flag-htb1K123R> |
[26] |
| CFK920 |
MATa, hta1-htb1∷LEU2, hta2-htb2, ura3-1, trp1-1, leu2-3,- 112, his3-11, ade2-1, can1-100, GAPDH-HA-UBI4∷URA3 <pZS145 HTA1-Flag-HTB1, CEN, HIS3> |
[17] |
| CFK921 |
MATa, hta1-htb1∷LEU2, hta2-htb2, ura3-1, trp1-1, leu2-3,-112 his3-11, ade2-1, can1-100, GAPDH-HA-UBI4∷URA3 <pZS146 HTA1-Flag-htb1-K123R, CEN, HIS3> |
[17] |
|
Flag H2A strain |
MATa (hta1-htb1)Δ∷LEU2, (hta2-htb2)Δ∷TRP1, his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128Δ <pRS315 (Flag-HTA1- HTB1)> |
[18] |
|
Flag H3 strain |
MATa his3Δ200 leu2 Δ 1 ura3-52 trp1Δ63 lys2-128Δ (hht1- hhf1) Δ∷LEU2 (hht2-hhf2) Δ∷HIS3< pRM204 (FlagHHT1- HHF1)> |
[51] |
|
Flag H4 strain |
MATa his3Δ200 leu2 Δ 1 ura3-52 trp1Δ63 lys2-128Δ (hht1- hhf1) Δ∷LEU2 (hht2-hhf2) Δ∷HIS3 <pRS426 (HHT1-Flag- HHF1)> |
[18] |
YZS276 and YZS277 are based on the Y131 shuffle strain, which is in the W303 background [10]. YSN545 and YSN763 were generated following plasmid shuffling from FY406, which is in the S288c background [52]. YSF200 and YSF201 are based on the quadruple histone deletion strain, JHY205, also in the S288c background [53]. CFK920 and CFK921 are used for double ChIP analysis and were made by transforming YZS276 and YZS277 with an integrating URA3 plasmid containing HA tagged ubiquitin under control of the constitutive GAPDH promoter. The Flag tagged H2A, H3 and H4 strains were made by plasmid shuffling from the FY1716 strain, also in the S288c background [51].
To increase detection of histone ubiquitylation and sumoylation, ubiquitin and Smt3 can be tagged with small epitopes in strains that also carry epitope tagged histones. HA and Flag epitopes, as well as a 6xHis tag, have been used to tag these modifier proteins [12, 17, 18, 30, 31]. Placing the tagged ubiquitin or SMT3 gene under control of constitutive (GAPDH) or inducible (GAL, CUP1) promoters will also increase the expression of the modifier proteins. Additional strains can be engineered to further enhance detection of ubiquitylated or sumoylated histones. For example, deletion of the ubiquitin protease genes UBP8 and/or UBP10 can result in up to 50% of cellular H2B becoming ubiquitylated [19, 32].
2.2 Detecting H2B ubiquitylation in trichloroacetic acid (TCA) extracts
This method was first developed by the Schatz laboratory for the isolation and detection of cytochrome c and was later modified for the analysis of ubiquitin conjugated histones [10]. A major advantage of this procedure is that it can be used for immunoprecipitation of epitope tagged histones from a denatured extract [18, 33]. In addition, using standard spectrophotometric techniques, an accurate estimation of protein concentrations in these lysates can be obtained. This ensures equal and consistent loading of extracts for analysis on SDS-polyacrylamide gels. The method described below uses strains that carry a Flag-tagged HTB gene.
Collect 10–20 OD units of mid-log phase cells (OD600 between 0.4 and 0.8) by centrifugation in a table-top centrifuge at 1800 × g for 3 minutes.
Decant the medium and suspend the pellet in 1 ml of 20% TCA. Transfer to a 1.5 ml eppendorf tube and immediately freeze in a dry ice bath. Suspensions can be stored at −80 °C.
Thaw the cell suspension on ice and spin at maximum speed in a microfuge (13,000 × g for 1 minute).
Discard the supernatant and resuspend the pellet in 250 ml of fresh 20% TCA.
Add acid-washed glass beads (Sigma G9143, 212–300 mm diameter) to the suspension such that there is approximately 3–4 mm of space between the bead volume and the suspension surface. Disrupt cells at 4 °C by vortexing for 15 minutes at the highest speed.
Using a sequencing pipette tip, remove the crude extract and place into a fresh eppendorf tube on ice. Wash the remaining beads with 500 ml of 5% TCA and combine with the crude extract.
Spin the lysate at maximum speed in a microfuge for 2 minutes at 4 °C. Using a sequencing gel pipette tip, discard the supernatant, leaving only the pellet, which contains precipitated proteins and cellular debris.
Suspend the pellet in 300 ml of a modified 1x Laemmli buffer (75 mM Tris-Cl (unbuffered), 1% SDS, 10% glycerol, 0.01% bromophenol blue, 350 mM 2-mercaptoethanol). When larger cell pellets are processed (>20 OD units), it is not unusual for the suspension to turn yellow due to residual TCA lowering the pH. If this occurs, 1.5 M Tris-Cl (unbuffered) is titrated in 5 ml aliquots until the color turns dark blue (usually 10–20 ml total). It should also be noted that reduced levels of 2-mercaptoethanol relative to standard recipes for Laemmli buffer are important if the extracts are to be used for subsequent immunoprecipitation.
Boil the neutralized suspension for 5 minutes and clarify by centrifugation in a microfuge at maximum speed for 5 minutes. Transfer the clarified lysate to a fresh eppendorf tube.
Analyze the sample for total protein content via a Bradford assay [34] and load 15–30 mg onto an SDS polyacrylamide gel. As histones are small proteins, typically 15% PAGE gels are used.
Following electrophoresis, transfer proteins to an Immobilon-P membrane (Millipore cat#IPVH00010) in Tris-glycine buffer containing 20% methanol.
Block the membrane in TBS-Tween-20 (50 mM Tris pH 8.0, 138 mM NaCl, 2.7 mM KCl, 0.05% Tween-20) with 5% milk for 30–60 minutes.
Incubate the membrane with anti-Flag M2 monoclonal antibody (Sigma F3165) at a 1:10000 dilution in TBS-Tween-20 plus 5% milk overnight at 4 °C with constant rotation.
Wash the membrane 3 times with TBS-Tween-20, rocking for 10 minutes each time.
Incubate the membrane with anti-mouse HRP conjugated secondary antibody (GE Healthcare NA931V) diluted 1:6000 in TBS for 1 hour at room temperature.
Wash the membrane 3 times with TBS, rocking each time for 10 minutes, and develop using a BioRad enhanced chemiluminescence kit (cat# 170–5040) per manufacturer’s instructions.
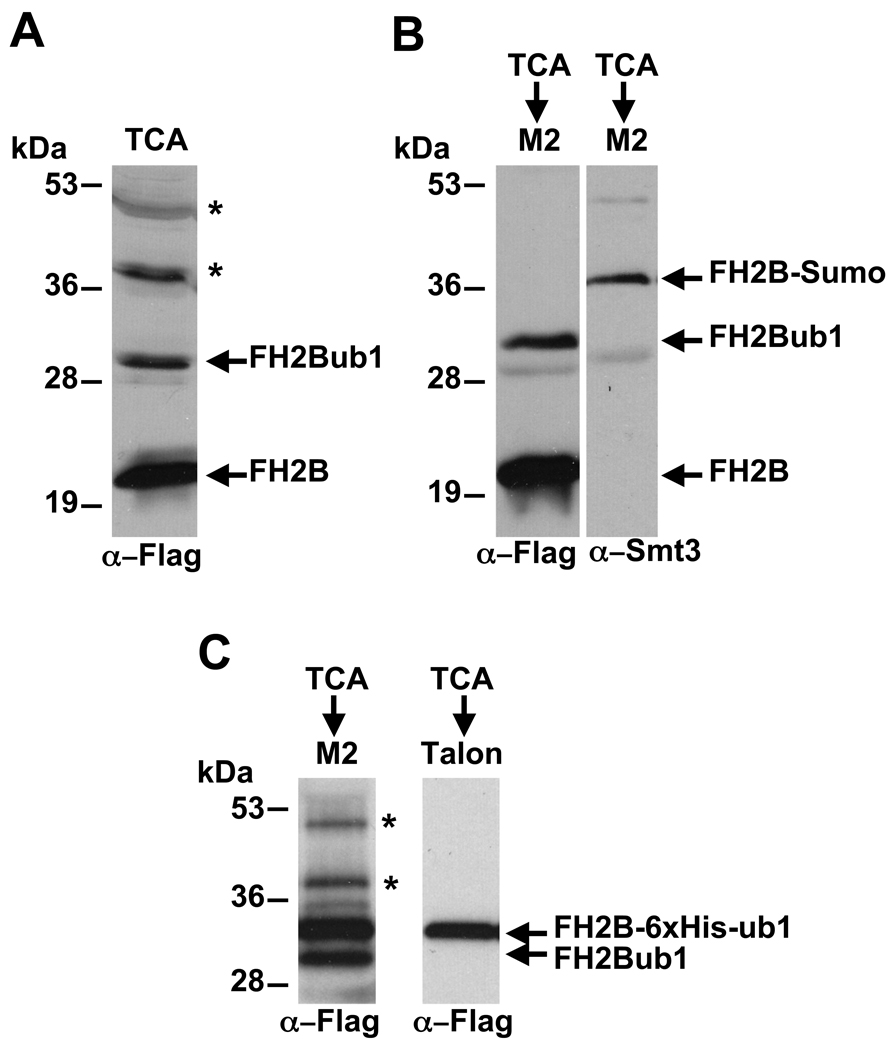
Figure 2A shows an anti-Flag Western blot of TCA extracts derived from the YZS276 strain (Table 1) in which the endogenous HTA1-HTB1 and HTA2-HTB2 loci were deleted and complemented with a plasmid expressing Flag-tagged H2B and untagged H2A. Both the ubiquitylated and unmodified Flag-H2B species can be readily detected by this method, with the ubiquitin-conjugated species exhibiting slower mobility than unmodified H2B.
Figure 2. Detection and enrichment of modified Flag-H2B from TCA extracts.
(A) 30 mg of total protein from a TCA extract of cells expressing Flag-tagged H2B were electrophoresed on a 15 % SDS polyacrylamide gel and transferred to an Immobilon-P membrane. Unmodified and ubiquitylated H2B were detected by Western blot analysis using anti-Flag antibodies. (B) 150 mg of the TCA extract in (A) were enriched on M2 agarose beads, and a portion of the eluate was electrophoresed on a 15 % SDS polyacrylamide gel and subjected to Western blot analysis. The left panel was probed with anti-Flag antibody and the right panel was blotted with antibodies raised against recombinant GST-Smt3 (a gift from Pam Meluh). (C) TCA extracts derived from a strain expressing Flag-H2B and containing a 6xHis-ubiquitin gene driven by a CUP1 promoter (in addition to the endogenous ubiquitin genes) were purified on either M2 agarose beads (left panel) or a Talon metal affinity resin (right panel). Western blot analysis using anti-Flag antibodies was performed to detect the modified Flag-H2B species. * Non-specific bands
2.3 Alternate methods for generating extracts under denaturing conditions
Besides TCA lysates, other methods to produce extracts under denaturing conditions have been described. One method uses urea and SDS as denaturants (SUMEB buffer: 1% SDS, 8 M urea, 10 mM MOPS pH 6.8, 10 mM EDTA, 0.01% bromophenol blue) and it provides a reliable means to quickly generate extracts for Western blot analysis of ubiquitin modified Flag-H2B [13, 32]. SUMEB lysates obtained from equivalent numbers of cells are typically analyzed by this method [32, 33]. Perhaps the simplest and quickest method for preparing denatured cell lysates entails boiling cell pellets resuspended in Laemmli buffer [35, 36]. However, in our hands we have found that these lysates are somewhat less reliable for the reproducible detection of H2Bub1. Finally, another method for generating denatured cell extracts was developed by the Tansey lab. This method involves bead beating cell pellets in a buffer containing 6M guanidine-HCl as the denaturant and has been used to identify polyubiquitylated forms of H2B in yeast [30].
2.4 Immunoprecipitation of ubiquitylated H2B and sumoylated histones from TCA extracts
While Western blot analysis of TCA extracts is sufficient for the detection of H2Bub1, more sensitive methods are required for detection of the much less abundant sumoylated histones [18]. A major advantage of the TCA method of lysis is that these extracts are suitable for immunoprecipitation of epitope tagged histones for Western blot analysis [10, 18, 33]. Here, we describe a method for the enrichment of ubiquitylated or sumoylated Flag-tagged H2B from TCA lysates using an M2 agarose matrix. Using an excess of M2 agarose beads ensures the complete extraction of the Flag-tagged histone from the lysate, thus ensuring a quantitative measurement of the fraction of modified H2B relative to the total pool of modified and unmodified H2B.
Bring 150 mg of a TCA lysate prepared as described in Section 2.2 to 500 ml with FA lysis buffer (50 mM HEPES-KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% TritonX-100, 0.1% sodium deoxycholate) supplemented with a 1x protease inhibitor cocktail (Sigma P2714) in an eppendorf tube. Set on ice for 20 minutes to allow proteins in the extract to renature.
Pre-wash an M2 agarose slurry (Sigma A2220) three times in 1.0 ml FA buffer +1.0 mg/ml BSA, and equilibrate in FA buffer for 60 minutes at 4 °C.
Add 100 ml of the bead suspension to the renatured lysate and rock overnight at 4 °C.
Centrifuge at 1500 × g for 1 minute in a microfuge and discard the supernatant using a sequencing gel pipette tip.
Wash beads three times with 1.0 ml FA buffer containing 500 mM NaCl.
Resuspend beads in 100 ml of the modified 1x Laemmli buffer described in section 2.1 and boil the suspension for 5 minutes.
Collect the eluate in a fresh eppendorf tube using a sequencing gel pipette tip. Load 10–20 ml on a 15 % SDS-polyacrylamide gel for Western blot analysis or store at −20 °C.
Figure 2B shows an example of how this method can be used to identify both H2Bub1 and H2B-SUMO conjugates. TCA extracts derived from the YZS276 strain (Table 1), which expresses a Flag-tagged histone H2B, were enriched on M2 agarose. Following SDS-PAGE, the membrane was blotted with anti-Flag or anti-Smt3 (SUMO) antibodies. In the left panel, both the unmodified and ubiquitylated forms of H2B can be observed. Note that the relative amounts of ubiquitylated H2B and unmodified H2B are similar with or without enrichment on M2 agarose (compare Fig. 2A to Fig. 2B, left panel), indicating that the affinity purification was accomplished in a quantitative manner. Because there is very little H2B-SUMO in yeast, a band corresponding to H2B-SUMO, which should migrate at ~38 kDa, typically cannot be detected with anti-Flag antibodies (Fig.1B, left panel). However, by blotting with anti-Smt3 antibodies, the sumoylated form of H2B is readily observed (Fig. 1B, right panel).
As a variation on this method, strains in which ubiquitin or Smt3 have been epitope tagged can also be employed to facilitate detection and provide an alternative means for enrichment of the modified histones [12, 30, 33]. Figure 2C demonstrates how affinity purification of 6xHis tagged ubiquitin can be used to enrich for H2Bub1 from TCA extracts. In the left panel of Figure 2C, TCA extracts containing Flag-H2B were subject to affinity purification on M2 agarose as described above. In addition to Flag-H2B, the yeast strain used in this experiment (YZS276) harbors a plasmid (pUB221 [31]) that carries a 6xHis-9Myc tagged ubiquitin gene under the control of the copper-inducible CUP1 promoter. In this strain, the endogenous ubiquitin genes are present and the tagged ubiquitin gene can be turned on by addition of copper sulfate to the medium. In the example shown in the right panel of Figure 2C, cells were grown to an OD600 of ~0.2 in synthetic minimal medium and then copper sulfate was added to a final concentration of 1 mM. The culture was incubated for another hour to allow for expression of the tagged ubiquitin before harvesting cells for preparation of TCA lysates. These lysates can be used for enrichment of ubiquitylated Flag-H2B on either M2 agarose or a metal affinity resin, such as Ni2+ agarose or Talon (Clontech, Mountain View CA).
Western blot analysis of an M2 agarose purified fraction with anti-Flag antibodies reveals two modified forms of Flag-H2B. One of these modified forms migrates at ~30kDa, corresponding to Flag-H2B marked with untagged ubiquitin, and the other band migrates at ~33kDa, corresponding to Flag-H2B marked with 6xHis-9Myc tagged ubiquitin (Fig. 2C, left panel).
For enrichment of ubiquitylated Flag-H2B on a Talon resin, 200 mg of the TCA lysate were brought to 1.2 ml with FA buffer (see section 3.1) supplemented with 10 mM MgCl2. The inclusion of additional Mg2+ is important to maintain the binding capacity of the matrix because the FA buffer contains EDTA, which could chelate the divalent metal ions in the resin. The diluted lysate is bound to 100 ml of the Talon resin for 2 hours, washed 3 times with 1 ml of FA buffer plus 10 mM MgCl2, and eluted by boiling in 100 ml of 1X Laemmli buffer. Western blot analysis of the eluate with anti-Flag antibodies (alternatively, anti-6xHis or anti-9Myc antibodies could also be used) allows for the specific detection of Flag-H2B modified with 6xHis-9Myc ubiquitin (Fig. 2C, right panel). Because the 6xHis-9Myc ubiquitin construct is under the control of an inducible promoter, this procedure could be used to monitor new H2B ubiquitylation as a biological response to specific cellular stimuli (e.g. after heat shock or DNA damage).
A similar approach was taken to identify sumoylated H2A.Z in yeast [37]. In this study, a strain was used that expressed an HA-tagged version of H2A.Z and a 6xHis tagged SUMO construct. Nickel agarose beads were used to enrich for all species of sumoylated proteins from those extracts, and sumoylated H2A.Z was specifically identified by Western blot analysis with anti-HA antibodies. The same 6xhis-Smt3 construct was also employed to enrich for the sumoylated forms of H2A and H2B for mass spectrometry analysis [18].
3. Double chromatin immunoprecipitation for detection of H2B ubiquitylation and histone sumoylation at specific chromosomal loci
Chromatin immunoprecipitation (ChIP) is a technique that was originally designed for the detection of histones associated with specific DNA sequences in vivo [38–40]. It has become a powerful tool for the analysis of many protein-DNA interactions and has provided a widely used method to monitor these interactions during processes such as transcription, DNA repair, and replication [17, 41–43]. Advancements in the amplification and detection of immunoprecipitated DNAs have allowed quantitative measurements of these interactions with a high degree of sensitivity.
In the field of transcription, ChIP has been used to analyze histone modification patterns in response to gene activation and repression, and has yielded great insight into the roles of specific modifications in gene regulation [17, 18]. As outlined in the accompanying article by Oren et al., a monoclonal antibody against ubiquitylated human H2B was recently developed and used to identify the genome-wide distribution of this mark [44, 45]. However, this antibody does not detect yeast H2Bub1. A polyclonal antibody against yeast H2Bub1 has been described by Schulze et al., but this antibody is not commercially available [26, 46]. Here, we describe an alternative method to identify and quantify the interaction of ubiquitylated H2B with its DNA targets. Briefly, this procedure, termed double-ChIP (ChDIP), utilizes sequential immunoprecipitation to separate ubiquitylated H2B from its unmodified form. The first step involves the enrichment of Flag-H2B and its associated DNA using an M2 agarose purification step in a strain that carries both Flag-H2B and HA-tagged ubiquitin (e.g., strain CFK920, Table 1). The second step involves an anti-HA immunoprecipitation from the eluted M2 agarose fraction to enrich for HA-ubiquitylated Flag-H2B. As a negative control, these experiments are also performed in a Flag-H2B strain in which lysine 123 has been mutated to an arginine (e.g., strain CYK921, Table 1). We have found that H2Bub1 represents the major chromatin-associated ubiquitylated protein using this method, with only trace amounts of other ubiquitylated proteins present in the final HA eluate (C-F Kao and M.A, Osley, unpublished observations). Thus, ChDIP provides a way to obtain reliable information on the localization and levels of H2Bub1 at specific genomic locations. In addition to presenting the basic methodology for performing ChDIP, the normalization of data derived by quantitative PCR will be discussed.
3.1. Fixation of cells and extract preparation
Due to the intimate contacts of histones with DNA, shorter fixation times are required to crosslink histones to their DNA targets [47]. The following protocol describes procedures for the generation of chromatin from extracts derived by bead beating and sonication, although alternative methods involving spheroplasting and micrococcal nuclease (MNase) digestion have also been employed [48]. In instances where a high degree of resolution is required, chromatin prepared by MNase digestion may be desirable. However, in most cases, sonication, which fragments chromatin to an average length of ~500–700bp (3–4 nucleosomes), is sufficient.
Collect 40 OD600 units of mid-log phase cells (strains CFK920 and CFK921, Table 1) in an Erlenmeyer flask and rotate at room temperature for 15 minutes with 1% formaldehyde (final concentration).
Add glycine to a final concentration of 125 mM and incubate for an additional 5 minutes.
Pellet cells at 1800 × g in a tabletop centrifuge and wash with ice-cold 1x TBS.
Centrifuge the cells again, quick freeze the pellet in an ethanol-dry ice bath, and store the pellets at −80 °C.
Resuspend the cell pellet in 1 ml FA Buffer (50 mM HEPES pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% TritonX-100, 0.1% sodium deoxycholate) plus a 1x protease inhibitor cocktail.
Distribute the cell suspension equally into two 1.5 ml eppendorf tubes and add acid washed glass beads to 3–4 mm below the sample surface (approx. 0.5 g).
Vortex 15 minutes at 4° C at highest speed and collect the crude extract in two fresh 1.5 ml eppendorf tubes. Wash the beads with 250 ml FA buffer and combine with the crude extract.
Sonicate 6 times for 10 seconds (with 1 minute rest periods on ice) using a Branson Sonifier set at 30% continuous output. (Note: conditions should be optimized for each sonicator by extracting the DNA and analyzing fragment sizes following electrophoresis on a 1 % agarose gel. A smear of DNA with an average size of 500–700 bp should be observed.)
Spin the sonicated extract in a microfuge at 4°C for 5 minutes to separate the insoluble material. Combine the supernatants from both tubes into a 2.0 ml eppendorf tube (~1.5 ml total volume).
Measure the total protein concentration via standard methods. Typical recovery is 4–6 mg of protein.
3.2. M2 agarose enrichment and elution under native conditions
The major advantage of using Flag-tagged histones is that they can be purified by elution from M2 agarose under native conditions with a competing peptide. This maintains the protein-DNA crosslinks and makes it possible for a sequential purification step (HA immunoprecipitation) to enrich for a pool of HA-ubiquitin modified Flag-H2B.
Bring 3 mg of the sonicated extract to 1.5 ml with FA buffer plus a 1x protease inhibitor cocktail in a 2 ml conical eppendorf tube.
Add 200 ml M2 agarose slurry (Sigma A2220; pre-washed 3 times in 5 volumes of 125 mM glycine, and equilibrated in FA buffer for 60 minutes at 4 °C). Incubate overnight at 4 °C with constant rotation.
Spin at 1500 × g in a microfuge for 1 minute and discard the supernatant. Wash the beads 3 times with 1 ml FA Buffer. After the final wash, remove any residual buffer with a sequencing gel pipette tip.
Resuspend the beads in 475 ml FA buffer plus 25 ml of 3x Flag peptide (Sigma F4799; 4 mg/ml stock concentration) and rock overnight at 4 °C to elute Flag-H2B.
Collect the eluate in a fresh 1.5 ml eppendorf tube using a sequencing gel pipette tip. Set aside 50 ml for a Flag-H2B input control and store at −80 °C. This material will be used for data normalization as discussed in section 3.5.
3.3 Secondary purification of HA-ubiquitylated histones from the Flag-H2B pool
At this point in the procedure, the pool of Flag-H2B that has been eluted from the M2 agarose includes both modified and unmodified forms of Flag-H2B. In order to isolate the ubiquitylated fraction of Flag-H2B, a secondary purification step is performed. In the ongoing example, we describe the purification of the ubiquitylated pool from strains that express HA-tagged ubiquitin. Variations to the secondary purification step will be discussed in section 3.4.
Add 3–5 ml of anti-HA antibody (Roche 12CA5, 3 mg/ml; Roche 3F10, 100 mg/ml) to the remaining Flag peptide eluate (~450 ml) and rock overnight at 4 °C.
Add 100 ml of Protein-A agarose beads (washed 3 times in 5 volumes of FA Buffer plus 1 mg/ml BSA and equilibrated in FA buffer without BSA for 60 minutes) and rock for 4–6 hours at 4°C.
Spin the mixture at 1500 × g in a microfuge for 1 minute and discard the supernatant. Wash the beads with 1 ml FA buffer, followed by an additional wash with FA buffer supplemented with 500 mM NaCl. A third wash in TL buffer (10 mM Tris-Cl pH 8.0, 250 mM LiCl, 0.5 % NP-40, 0.5 % sodium deoxycholate, 1 mM EDTA) is followed by a final wash in 1x TE. Remove the residual TE using a sequencing gel pipette tip.
Elute HA-ubiquitin tagged Flag-H2B from the Protein-A beads with 250 ml EB buffer (50 mM Tris-Cl pH 8.0, 1 % SDS) by heating the sample to 65 °C for 20 minutes and vortexing briefly while still warm. At the same time, thaw the Flag-H2B input sample that was set aside after the 3x Flag peptide elution (3.2 step 5) and add 250 ml EB buffer. Heat the Flag-H2B input sample at 65 °C for 20 minutes as well.
Transfer both the HA-ubiquitin and Flag-H2B samples to 0.5 ml PCR tubes and add 50 ml pronase (Sigma P6911, 20 mg/ml stock in ddH2O) and 1.5 ml 1 M CaCl2 to each sample. Using a thermocycler, incubate the samples for 2 hours at 42 °C, followed by 12–16 hours at 65°C. This step de-proteinizes the samples and reverses the formaldehyde-induced crosslinks.
Transfer the samples to a 2 ml eppendorf tube containing 1.5 ml PBI buffer from a Qiagen PCR Purification Kit (cat# 28106) and 20 µl 3 M NaOAC pH 5.2. Proceed with the purification of the DNA as outlined in the manufacturer’s instructions. DNA samples can be stored at −20 °C. However, multiple freeze/thaw cycles are not recommended, and dilutions for quantitative PCR analysis should be stored at 4 °C.
3.4. Variations on the standard ChDIP procedure
An alternative approach has been to use antibodies raised against ubiquitin-protein conjugates for the second purification step. These antibodies are commercially available (Enzo Life Sciences cat#PW8810 clone FK2) [16] and they eliminate the need for a strain in which ubiquitin has been epitope tagged. It should also be noted that in addition to HA tagged ubiquitin, strains harboring other epitope tags on ubiquitin could be employed to enrich for the ubiquitylated pool using the appropriate commercially available antibodies to these epitopes. This latter approach has been successfully used to enrich for and localize sumoylated histones. For example, strains expressing a 6xHis tagged Smt3 have been used in ChDIP experiments to isolate sumoylated histones from a Flag-H2B pool using a nickel-agarose purification procedure [18]. In that study, a strain in which the putative sumoylation sites on Flag-tagged H2B were mutated to alanine residues was included to control for background. Similarly, antibodies against SUMO (Rockland, cat# 200–401–428) [16] or antibodies raised against GST-Smt3 (obtained from Pam Meluh) have also been used in a second IP of eluted Flag-H2B chromatin to enrich for sumoylated H2B in ChDIP studies.
3.5. Real time quantitative PCR (RT-qPCR) and normalization of ChDIP data
The most sensitive method for analyzing ChDIP data is real time quantitative PCR (RT-qPCR). This method utilizes fluorescent DNA dyes to monitor DNA amplification by PCR using primer pairs corresponding to specific genomic regions of interest. Several constitutively expressed and inducible genes have been commonly analyzed by RT-qPCR to identify changes in H2Bub1 levels during transcription, and the primer pairs used to amplify these loci have been published [41, 49]. Among the most commonly studied constitutively expressed genes are PMA1, ACT, and PYK1, which have high levels of H2Bub1 over their promoter and coding regions. The GAL1 gene has been extensively used to monitor H2Bub1 levels before, during, and after transcriptional activation with galactose, and during subsequent repression with glucose [17, 41]. Additionally, because H2Bub1 levels are low at regions of yeast heterochromatin, primer pairs corresponding to telomere adjacent regions or to the silent mating type loci are frequently included as negative or normalization controls [49].
Typically, normalization of ChIP data requires the division of the immunoprecipitation (IP) signal by the total input DNA signal for a given primer pair. This ratio accounts for variations in the amount of starting material between experimental samples. The IP/input ratio is also determined at a control region (e.g., TELVI), where no change is expected during the course of the experiment. Dividing the IP/input ratio at the experimental locus by the IP/input ratio at a control locus is a common way to normalize ChIP data. Other examples of regions with stable H2Bub1 have been described [41] and include HMR, intragenic regions and inducible genes under repressive conditions (e.g., GAL1). The investigator should take care to establish a suitable control region dependent upon the specific experimental conditions.
For the ChDIP procedure, however, a different approach is taken to normalize data. In this case, the PCR signal from the second immunoprecipitation (e.g., the HA-ubiquitin signal) is divided by the PCR signal derived from the M2 agarose eluted chromatin (i.e. the Flag-H2B signal). The HA/Flag ratio thus describes the relative proportion of Flag-H2Bub1 present in the Flag-H2B eluate, which contains both modified and unmodified H2B. Because the htb-K123R mutant cannot be ubiquitylated, measuring the HA/Flag ratio in this strain accounts for background levels under each experimental condition.
4. Biochemical Assays
4.1 Isolation of H2Bub1 for deubiquitylating assays
Using epitope tagged H2B and ubiquitin, it is possible to isolate sufficient H2Bub1 from denatured extracts to use as a substrate for in vitro biochemical assays, such as deubiquitylation assays [11, 12, 50]. The following method was used to isolate H2Bub1 for deubiquitylating assays with purified Ubp8 and Ubp10 ubiquitin proteases [12].
Grow strains that contain Flag-tagged H2B and HA-tagged ubiquitin (Table 1) to mid-log phase [12].
Prepare TCA extracts as described in section 2.1 and isolate Flag-H2B using M2 agarose beads, followed by elution with 3xFlag peptide (section 3.2). The eluate will contain both Flag-H2B and Flag-H2Bub1 (in which ubiquitin is unmodified or tagged with HA). This pooled substrate can be used in an in vitro deubiquitylating assay, such as one employed to identify the Ubp8 subunit of SAGA as an H2Bub1-specific protease [12].
After determining the protein concentration of the eluate, combine 250–500 ng of the substrate with purified Ubp8, Ubp10, or another putative deubiquitylase, in DUB buffer (10 mM Tris-Cl pH8, 1 mM DTT, 1 µM PMSF, and 1 µg/ml of aprotinin and pepstatin) for 60 minutes at 30 °C.
Stop the reaction by freezing in liquid nitrogen.
Boil the samples in Laemmli buffer for 5 minutes and perform SDS-PAGE, followed by Western blot analysis with anti-Flag and anti-HA antibodies (section 2.1). The inclusion of an HA ubiquitylated Flag-H2B substrate in this assay increases sensitivity, as the HA species disappears during the course of the deubiquitylating reaction, while the amount of the unmodified Flag-H2B species remains unchanged [12].
Enrichment of the ubiquitylated form of H2B for deubiquitylating assays is also possible using sequential Flag/HA IPs as described for ChDIP (section 3.3). Using this approach, Daniel and co-workers performed a deubiquitylation assay with purified SAGA while HA ubiquitylated-Flag-H2B was immobilised on an HA affinity matrix [11].
5. Conclusions
Because of the large array of genetic and molecular tools available in budding yeast, its value as a model for studying transcription and other DNA-mediated processes has been immeasurable. Numerous studies have used antibody-based approaches to understand the relationship of specific histone modifications to these processes. Detection of histone ubiquitylation and sumoylation in this organism has presented special challenges due to the low cellular abundance of these modified histones, the presence of highly active proteases that target these species in yeast extracts, and the absence of commercially available antibodies against these modified histones. In this article, we have reviewed the available genetic and molecular tools that can be used to reliably detect and quantify both ubiquitylated and sumoylated histones on global and local levels. These methods rely on the marking of both histones and ubiquitin or SUMO with small epitope tags that can be subsequently employed in a variety of standard immunological techniques. Epitope tagging has become routine in most yeast labs, and the equipment necessary to make extracts, perform immunoprecipitations, affinity purifications, Western blotting, and RT-qPCR analysis are commonplace.
In the case of gene transcription, understanding the relationship between histone ubiquitylation and sumoylation is an important but poorly understood issue, as one mark (ubiquitylation) correlates with active transcription, while the other (sumoylation) correlates with repressed transcription. Reduction of H2B sumoylation leads to increased histone acetylation and H2B ubiquitylation [18]. Conversely, substitution of the ubiquitylation site on H2B results in higher levels of H2B sumoylation. Exactly how histone sumoylation interferes with activating histone modifications such as acetylation and ubiquitylation is unknown, but one possibility is that there exists a direct competition between the modifications for some transcription factors. Such ideas can be tested using the techniques described in this article.
6. Acknowledgements
We gratefully acknowledge Pam Meluh (Johns Hopkins University, School of Medicine) for the gift of the GST-Smt3 antibody. Research in the authors’ laboratories is funded by grants from the NIH (GM40118; M.A.O; GM55360, S.L.B.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
7. References
- 1.Goldknopf IL, Taylor CW, Baum RM, Yeoman LC, Olson MO, Prestayko AW, Busch H. Isolation and characterization of protein A24, a "histone-like" non-histone chromosomal protein. J Biol Chem. 1975;250:7182–7187. [PubMed] [Google Scholar]
- 2.Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci U S A. 2003;100:13225–13230. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- 4.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 5.Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 6.Wood A, Schneider J, Shilatifard A. Cross-talking histones: implications for the regulation of gene expression and DNA repair. Biochem Cell Biol. 2005;83:460–467. doi: 10.1139/o05-116. [DOI] [PubMed] [Google Scholar]
- 7.Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, Allis CD, Strahl BD. Gene silencing: trans-histone regulatory pathway in chromatin. Nature. 2002;418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- 8.Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, Johnston M, Shilatifard A. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell. 2003;11:267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- 9.Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell. 2003;11:261–266. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- 10.Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- 11.Daniel JA, Torok MS, Sun ZW, Schieltz D, Allis CD, Yates JR, 3rd, Grant PA. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J Biol Chem. 2004;279:1867–1871. doi: 10.1074/jbc.C300494200. [DOI] [PubMed] [Google Scholar]
- 12.Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardner RG, Nelson ZW, Gottschling DE. Ubp10/Dot4p regulates the persistence of ubiquitinated histone H2B: distinct roles in telomeric silencing and general chromatin. Mol Cell Biol. 2005;25:6123–6139. doi: 10.1128/MCB.25.14.6123-6139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kahana A, Gottschling DE. DOT4 links silencing and cell growth in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:6608–6620. doi: 10.1128/mcb.19.10.6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies N, Lindsey GG. Histone H2B (and H2A) ubiquitination allows normal histone octamer and core particle reconstitution. Biochim Biophys Acta. 1994;1218:187–193. doi: 10.1016/0167-4781(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 16.Chandrasekharan MB, Huang F, Sun ZW. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc Natl Acad Sci U S A. 2009;106:16686–16691. doi: 10.1073/pnas.0907862106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 18.Nathan D, Ingvarsdottir K, Sterner DE, Bylebyl GR, Dokmanovic M, Dorsey JA, Whelan KA, Krsmanovic M, Lane WS, Meluh PB, Johnson ES, Berger SL. Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications. Genes Dev. 2006;20:966–976. doi: 10.1101/gad.1404206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emre NC, Ingvarsdottir K, Wyce A, Wood A, Krogan NJ, Henry KW, Li K, Marmorstein R, Greenblatt JF, Shilatifard A, Berger SL. Maintenance of low histone ubiquitylation by Ubp10 correlates with telomere-proximal Sir2 association and gene silencing. Mol Cell. 2005;17:585–594. doi: 10.1016/j.molcel.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 20.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 21.Johnson ES, Schwienhorst I, Dohmen RJ, Blobel G. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. Embo J. 1997;16:5509–5519. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson ES, Gupta AA. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 2001;106:735–744. doi: 10.1016/s0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi Y, Toh-e A, Kikuchi Y. A novel factor required for the SUMO1/Smt3 conjugation of yeast septins. Gene. 2001;275:223–231. doi: 10.1016/s0378-1119(01)00662-x. [DOI] [PubMed] [Google Scholar]
- 24.Zhao X, Blobel G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci U S A. 2005;102:4777–4782. doi: 10.1073/pnas.0500537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li SJ, Hochstrasser M. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol Cell Biol. 2000;20:2367–2377. doi: 10.1128/mcb.20.7.2367-2377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakanishi S, Lee JS, Gardner KE, Gardner JM, Takahashi YH, Chandrasekharan MB, Sun ZW, Osley MA, Strahl BD, Jaspersen SL, Shilatifard A. Histone H2BK123 monoubiquitination is the critical determinant for H3K4 and H3K79 trimethylation by COMPASS and Dot1. J Cell Biol. 2009;186:371–377. doi: 10.1083/jcb.200906005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 28.Davie JR, Murphy LC. Level of ubiquitinated histone H2B in chromatin is coupled to ongoing transcription. Biochemistry. 1990;29:4752–4757. doi: 10.1021/bi00472a002. [DOI] [PubMed] [Google Scholar]
- 29.Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, Knop M. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 30.Geng F, Tansey WP. Polyubiquitylation of histone H2B. Mol Biol Cell. 2008;19:3616–3624. doi: 10.1091/mbc.E08-01-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yaglom J, Linskens MH, Sadis S, Rubin DM, Futcher B, Finley D. p34Cdc28-mediated control of Cln3 cyclin degradation. Mol Cell Biol. 1995;15:731–741. doi: 10.1128/mcb.15.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gardner R, Cronin S, Leader B, Rine J, Hampton R. Sequence determinants for regulated degradation of yeast 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol Biol Cell. 1998;9:2611–2626. doi: 10.1091/mbc.9.9.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kao CF, Osley MA. In vivo assays to study histone ubiquitylation. Methods. 2003;31:59–66. doi: 10.1016/s1046-2023(03)00088-4. [DOI] [PubMed] [Google Scholar]
- 34.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 35.Horvath A, Riezman H. Rapid protein extraction from Saccharomyces cerevisiae. Yeast. 1994;10:1305–1310. doi: 10.1002/yea.320101007. [DOI] [PubMed] [Google Scholar]
- 36.Swerdlow PS, Schuster T, Finley D. A conserved sequence in histone H2A which is a ubiquitination site in higher eucaryotes is not required for growth in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:4905–4911. doi: 10.1128/mcb.10.9.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalocsay M, Hiller NJ, Jentsch S. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol Cell. 2009;33:335–343. doi: 10.1016/j.molcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 38.Gilmour DS, Lis JT. Detecting protein-DNA interactions in vivo: distribution of RNA polymerase on specific bacterial genes. Proc Natl Acad Sci U S A. 1984;81:4275–4279. doi: 10.1073/pnas.81.14.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilmour DS, Lis JT. In vivo interactions of RNA polymerase II with genes of Drosophila melanogaster. Mol Cell Biol. 1985;5:2009–2018. doi: 10.1128/mcb.5.8.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilmour DS, Lis JT. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol Cell Biol. 1986;6:3984–3989. doi: 10.1128/mcb.6.11.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kao CF, Hillyer C, Tsukuda T, Henry K, Berger S, Osley MA. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev. 2004;18:184–195. doi: 10.1101/gad.1149604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Papamichos-Chronakis M, Peterson CL. The Ino80 chromatin-remodeling enzyme regulates replisome function and stability. Nat Struct Mol Biol. 2008;15:338–345. doi: 10.1038/nsmb.1413. [DOI] [PubMed] [Google Scholar]
- 43.Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol. 2008;10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- 45.Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, Raver-Shapira N, Minsky N, Pirngruber J, Tarcic G, Hublarova P, Moyal L, Gana-Weisz M, Shiloh Y, Yarden Y, Johnsen SA, Vojtesek B, Berger SL, Oren M. The histone H2B–specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev. 2008;22:2664–2676. doi: 10.1101/gad.1703008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schulze JM, Jackson J, Nakanishi S, Gardner JM, Hentrich T, Haug J, Johnston M, Jaspersen SL, Kobor MS, Shilatifard A. Linking cell cycle to histone modifications: SBF and H2B monoubiquitination machinery and cell-cycle regulation of H3K79 dimethylation. Mol Cell. 2009;35:626–641. doi: 10.1016/j.molcel.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jackson V. Formaldehyde cross-linking for studying nucleosomal dynamics. Methods. 1999;17:125–139. doi: 10.1006/meth.1998.0724. [DOI] [PubMed] [Google Scholar]
- 48.Tsukuda T, Lo YC, Krishna S, Sterk R, Osley MA, Nickoloff JA. INO80-dependent chromatin remodeling regulates early and late stages of mitotic homologous recombination. DNA Repair (Amst) 2009;8:360–369. doi: 10.1016/j.dnarep.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 49.Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA, Strahl BD. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol Cell Biol. 2005;25:637–651. doi: 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee KK, Swanson SK, Florens L, Washburn MP, Workman JL. Yeast Sgf73/Ataxin-7 serves to anchor the deubiquitination module into both SAGA and Slik(SALSA) HAT complexes. Epigenetics Chromatin. 2009;2:2. doi: 10.1186/1756-8935-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mann RK, Grunstein M. Histone H3 N-terminal mutations allow hyperactivation of the yeast GAL1 gene in vivo. Embo J. 1992;11:3297–3306. doi: 10.1002/j.1460-2075.1992.tb05408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hirschhorn JN, Bortvin AL, Ricupero-Hovasse SL, Winston F. A new class of histone H2A mutations in Saccharomyces cerevisiae causes specific transcriptional defects in vivo. Mol Cell Biol. 1995;15:1999–2009. doi: 10.1128/mcb.15.4.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ahn SH, Cheung WL, Hsu JY, Diaz RL, Smith MM, Allis CD. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell. 2005;120:25–36. doi: 10.1016/j.cell.2004.11.016. [DOI] [PubMed] [Google Scholar]