Abstract
Humans have seven APOBEC3 DNA cytosine deaminases. The activity of these enzymes allows them to restrict a variety of retroviruses and retrotransposons, but may also cause pro-mutagenic genomic uracil lesions. During interphase the APOBEC3 proteins have different subcellular localizations: cell-wide, cytoplasmic or nuclear. This implies that only a subset of APOBEC3s have contact with nuclear DNA. However, during mitosis, the nuclear envelope breaks down and cytoplasmic proteins may enter what was formerly a privileged zone. To address the hypothesis that all APOBEC3 proteins have access to genomic DNA, we analyzed the localization of the APOBEC3 proteins during mitosis. We show that APOBEC3A, APOBEC3C and APOBEC3H are excluded from condensed chromosomes, but become cell-wide during telophase. However, APOBEC3B, APOBEC3D, APOBEC3F and APOBEC3G are excluded from chromatin throughout mitosis. After mitosis, APOBEC3B becomes nuclear, and APOBEC3D, APOBEC3F and APOBEC3G become cytoplasmic. Both structural motifs as well as size may be factors in regulating chromatin exclusion. Deaminase activity was not dependent on cell cycle phase. We also analyzed APOBEC3-induced cell cycle perturbations as a measure of each enzyme’s capacity to inflict genomic DNA damage. AID, APOBEC3A and APOBEC3B altered the cell cycle profile, and, unexpectedly, APOBEC3D also caused changes. We conclude that several APOBEC3 family members have access to the nuclear compartment and can impede the cell cycle, most likely through DNA deamination and the ensuing DNA damage response. Such genomic damage may contribute to carcinogenesis, as demonstrated by AID in B cell cancers and, recently, APOBEC3B in breast cancers.
Keywords: APOBEC3, cancer, cell cycle, DNA cytosine deamination, mitosis, subcellular localization, uracil
Introduction
The seven APOBEC3 proteins are part of a larger polynucleotide cytosine deaminase family that in humans also includes activation-induced cytosine deaminase (AID), APOBEC1, APOBEC2 and APOBEC4.1 All of the APOBEC3 proteins have the ability to convert single-stranded DNA cytosines to uracils, and this is the main mechanism by which several of these enzymes restrict the replication of HIV-1 and other retroviruses.1,2 Retroviral restiction occurs when relevant APOBEC3s are packaged into viral particles along with the viral RNA genome. During reverse transcription, the single-stranded viral cDNA becomes a target for cytosine deamination; replication across this cDNA template fixes uracils as point mutations in the retroviral genome and results in hypermutation and either inactivation or degradation.2 This mechanism is clear for APOBEC3G (A3G) and APOBEC3F (A3F) on HIV-1, as well as for other APOBEC3 enzymes on a broad number of other viral substrates.2-4 In addition, APOBEC3 proteins have a role in restricting retrotransposons (including LINE-1, Alu, IAP and MusD), primarily, although not solely, through a deamination process similar to retrovirus restriction.4-6
However, APOBEC3 enzymatic activity may also be problematic for genomic DNA. As an important precedent, AID-dependent uracil lesions in the antibody locus can be processed into chromosomal translocations, and additional AID-dependent uracil lesions in other genomic areas may also be pro-carcinogenic.7-9 For example, transgenic expression of AID causes T cell cancer in mice.7 Likewise, transgenic expression of APOBEC1 can cause hepatocellular carcinoma.10,11 Although a role for APOBEC3A (A3A) in cancer development is unclear, heterologous expression causes S-phase arrest, γH2Ax focus formation and mutational events.12,13 In addition, we recently demonstrated APOBEC3B (A3B) upregulation in a majority of breast cancers, with corresponding increases in A3B-dependent mutation signatures and overall mutation loads.14 The role of the other five APOBEC3 proteins in genomic deamination is less clear.
Subcellular regulation allows cells to compartmentalize proteins that could be genotoxic or cytotoxic. For example, caspase-activated deoxyribonuclease (CAD) is a DNase containing a nuclear localization signal that is complexed with an inhibitory protein in the cytoplasm.15 Cleavage of this inhibitor by caspase-3 allows CAD to enter the nucleus and degrade the genome as part of the natural apoptotic pathway. Likewise, the transcription factors STAT1 and NFκB are kept in the cytoplasm until activated, when they transport to the nucleus and bind promoters to enhance or suppress transcription.16,17 Proteins can be actively targeted to a region of the cell through localization sequences within the protein or in an interacting partner. They can also be excluded from the nuclear compartment passively, based on size and shape, with a limit of approximately 50 kDa and 6 nm diameter.18
To complete mitosis, a cell must split its replicated genome between two daughter cells. In mammals, mitosis is facilitated by breaking down the nuclear envelope to allow for spindle formation and physical segregation of the chromosomes. The nuclear envelope re-forms shortly after cytokinesis.19 Some proteins change localization during mitosis. For example, activated NFAT, which is nuclear during interphase, is excluded from DNA during mitosis and remains cytoplasmic until it is activated again.20 RUNX proteins, which form nuclear foci, dissolve and reform after mitosis, unlike histones, which remain bound to the DNA.21
During interphase, the APOBEC3 proteins vary in subcellular localization. A3A, APOBEC3C (A3C) and APOBEC3H (A3H) are the smallest deaminases, each having a single deaminase domain (~25 kDa). A3A and A3C have shown consistent cell-wide distributions, whereas A3H is more variable, but A3H haplotype II is both cytoplasmic and nucleolar.6,22-29 As a comparison, AID, which is a similarly sized, single domain deaminase, appears cytoplasmic at steady-state, but clearly shuttles between the nuclear and cytoplasmic compartment.30-33 APOBEC1 is also a shuttling protein with a predominantly cytoplasmic steady-state distribution in the absence of its interacting partner ACF.34-36 The double domain APOBEC3s (~50 kDa) are composed of two conserved deaminase domains and are either nuclear, A3B, or cytoplasmic, APOBEC3D, APOBEC3F and APOBEC3G (A3D, A3F, A3G) during interphase.30,37-44 Interestingly, DNA damage can cause cytoplasmic APOBEC3G to enter the nucleus,45 as well as affecting the shuttling of AID, shifting it from primarily cytoplasmic to more nuclear.46 It is not clear what the importance of subcellular localization is for the function of the APOBEC3 proteins, although a cytoplasmic distribution may be related to antiviral activity.39,47
Recently, we analyzed A3B and AID during mitosis.30 A3B is excluded from nuclear DNA during the entire process of mitosis, while AID associates with nuclear DNA during telophase. We thus became interested in the potential for other APOBEC3 enzymes to bind nuclear DNA during mitosis. The present study tests the hypothesis that nuclear envelope breakdown during mitosis allows all of the APOBEC3 proteins access to genomic DNA and provides comparative and mechanistic information on the mitotic regulation of the members of the APOBEC3 subfamily. We also use cell cycle profiling as a measure of each DNA deaminase’s capacity to damage nuclear DNA and trigger DNA damage responses.
Results
A3A, A3C and A3H have access to genomic DNA during interphase and telophase
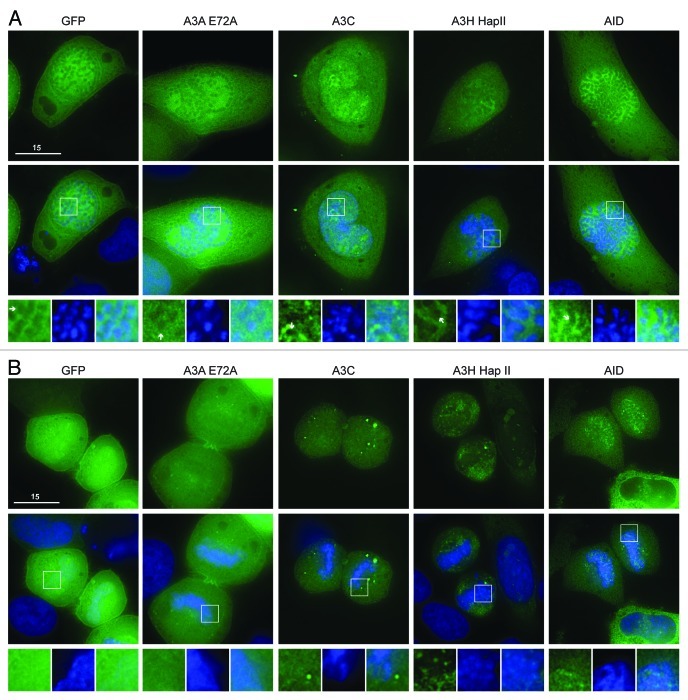
We first considered the single domain APOBEC3s, which are similar in size to AID and APOBEC1. The predicted molecular weight for A3A, A3C and A3H is 25 kDa (50 kDa with eGFP). The interphase localization of these single domain proteins has been described.6,22-29 For all our experiments, we used the stable A3H haplotype II, which is more cytoplasmic than A3A and A3C but can be seen in the nucleoli of HeLa cells29,48,49 (Fig. S1). An E72A catalytic mutant was used for A3A, because the wild-type enzyme killed almost all cells after 48 h of transient expression (concordant with prior reports13,14). As described,30 AID was excluded from chromatin during prophase, metaphase and anaphase, but co-localized specifically with DNA during telophase (Fig. 1A and B; Fig. S2). As expected, eGFP alone was excluded from the condensed DNA in prophase, metaphase and anaphase50 (Fig. 1A; Fig. S2), but resumed its cell-wide distribution once the chromosomes began to relax during telophase (Fig. 1B). Likewise, A3A-E72A and A3C were excluded from condensed chromosomes during prophase, metaphase and anaphase (Fig. 1A; Fig. S2). Once the cells reached telophase A3A-E72A and A3C became fully cell-wide (Fig. 1B). Thus, A3A-E72A and A3C are excluded from DNA during early mitosis in a manner similar to eGFP, implying that these proteins do not bind to DNA and/or may simply be excluded physically from highly condensed chromosomes [unlike AID or eGFP-tagged histone 2B (H2B), Fig. 1A; Vid. S2]. These data are supported by two-color live cell microscopy using A3A-E72A-mCherry and eGFP-H2B (Vids. S1 and S2). A3H-eGFP was also excluded from DNA during prophase, metaphase and anaphase (Fig. 1A; Fig. S2). However, during telophase, some A3H-eGFP remained excluded from DNA, but it could also be seen associated with punctate nuclear bodies, possibly part of re-assembling nucleoli (Fig. 1B). This continuing exclusion of A3H-Hap II during telophase is similar to the behavior of the double domain APOBEC3 proteins (below).
Figure 1. A3A-E72A, A3C and A3H are excluded from DNA as the chromosomes condense but become cell-wide during telophase. (A) Images of HeLa cells in prophase expressing the indicated APOBEC3-eGFP constructs (top) merged with Hoechst stain to visualize the nuclei (merge, middle). Boxed regions (bottom) are magnified below each image with APOBEC3-eGFP exclusion from DNA indicated (white arrows). (B) Images of HeLa cells in telophase expressing indicated APOBEC3-eGFP constructs (top), merged with nuclear stain (middle) and magnified (bottom). All images are representative of at least three mitotic cells. See Figure S2 for APOBEC3-eGFP localization during metaphase and anaphase and Videos S1 and S2 for time-lapse images of A3A-E72A-mCherry localization during mitosis.
A3B, A3D, A3F and A3G are excluded from the DNA during mitosis
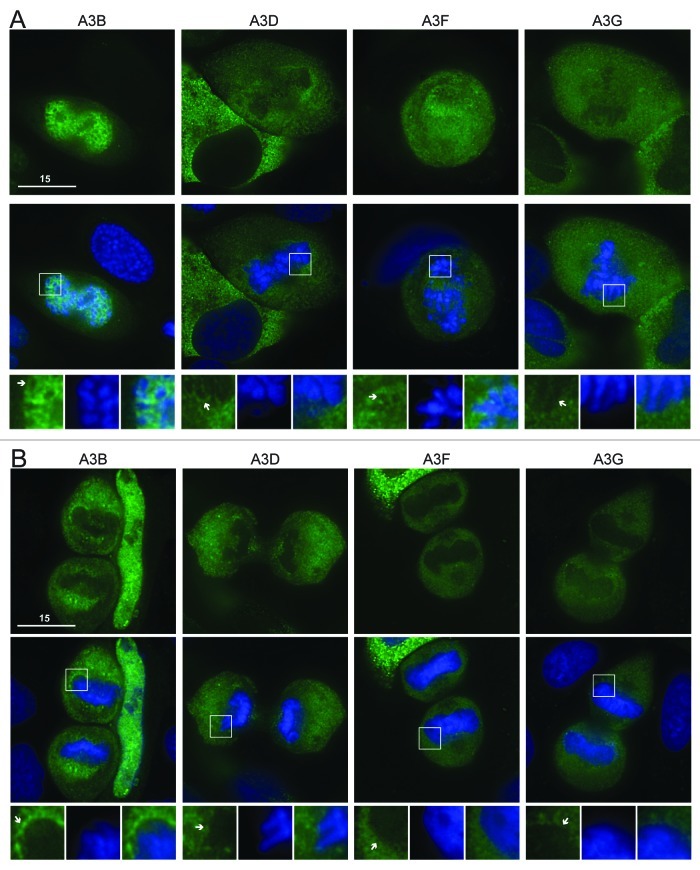
We next considered the double domain APOBEC3s. These proteins are twice the size of the single domain proteins (~50 kDa native or ~75 kDa with eGFP) and are generally composed of a C-terminal active deaminase domain and a less active or inactive N-terminal deaminase domain. A3B is nuclear during interphase, while A3D, A3F and A3G are cytoplasmic30,37-44,47 (Fig. S3). Despite A3B’s nuclear interphase localization, we had previously seen that it was excluded from mitotic chromosomes30 (Fig. 2B). Likewise, A3D, A3F and A3G were all excluded from condensed chromosomes during prophase, metaphase, anaphase and telophase (Fig. 2A and B; Fig. S4). This contrasts with AID, which associates with chromatin during telophase, and with A3A-E72A, A3C and eGFP alone, which begin to resume their cell-wide distributions during telophase (Fig. 1B). These data are supported by time-lapse microscopy using A3F-mCherry and eGFP-H2B (Vids. S3 and S4). These results indicate that that A3D, A3F and A3G have little opportunity for contact with genomic DNA during interphase or throughout mitosis.
Figure 2. A3B, A3D, A3F and A3G are excluded from DNA during cell division. (A) Images of HeLa cells in prophase expressing the indicated APOBEC3-eGFP constructs (top). Cells were stained with Hoechst dye to identify the nuclei (merge, middle). Boxed regions (bottom) are blown up below each image with APOBEC3 exclusion indicated (white arrows). (B) Images of HeLa cells in telophase expressing indicated APOBEC3-eGFP constructs (top), merged with nuclear stain (middle) and magnified (bottom). All images are representative of at least three mitotic cells. See Figure S4 for APOBEC3-eGFP localization during metaphase and anaphase and Videos S3 and S4 for time-lapse images of A3F-mCherry localization during mitosis.
Size-dependent and -independent effects on mitotic localization
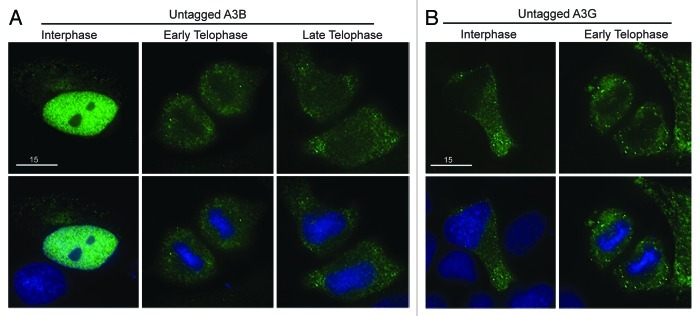
All the double domain proteins were excluded from genomic DNA. Since these tagged APOBEC3 proteins are approximately 75 kDa, they are considerably larger than the native double domain enzymes or the tagged 50 kDa single domain enzymes. This size difference could be an important biological means of control or an effect of the tag. To test this possibility, we analyzed all four double domain proteins with a smaller HA-tag. Similar to our previous data, we saw that A3B, A3D, A3F and A3G-HA were all excluded from mitotic DNA during telophase (Fig. S5). This triple HA-tag is less than 5 kDa. Since A3A-eGFP is cell-wide (51.3 kDa), A3B-HA (50.1 kDa) should not be impeded by its tag from its normal cellular distribution. To further rule out the effect of a tag on the localization of the double domain APOBEC3s, we used recently developed rabbit polyclonal sera to analyze the subcellular distribution of untagged A3B and A3G (Fig S6). After transient transfection, the interphase localization of untagged A3B was nuclear, and, as expected, it was excluded from DNA during telophase and then began to re-enter the nucleus in cells progressing through telophase (Fig. 3A). Untagged A3G was cytoplasmic during interphase and excluded during telophase (Fig. 3B). The localization of untagged A3B and A3G mimics the localization of HA- and eGFP-tagged proteins (compare Fig. 3 and Figs. S5 and S6). These data support our findings with tagged proteins that, under normal conditions, A3B only co-localizes with DNA during interphase and that A3D, A3F and A3G are excluded in both interphase and throughout mitosis.
Figure 3. Untagged A3B and A3G are excluded from genomic DNA in the same manner as HA- and eGFP-tagged derivatives. (A) Images of HeLa cells transfected with untagged A3B labeled with anti-A3B and anti-rabbit FITC (top) then stained with Hoechst dye to illuminate the DNA (bottom). The indicated progression through telophase (early and late) was based on chromatin condensation. (B) Images of HeLa cells transfected with untagged A3G labeled with anti-A3G and anti-rabbit FITC (top) and DNA stain as before (bottom). Representative images are based on several telophase cells.
Determinants of double domain APOBEC3 localization
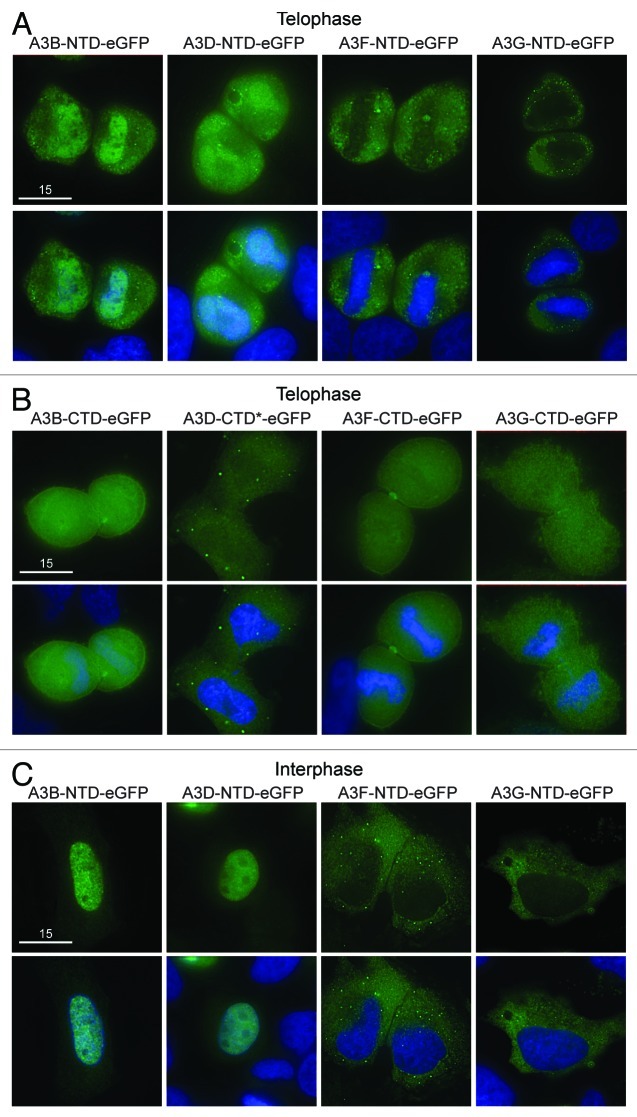
We hypothesized that APOBEC3 exclusion from mitotic DNA is governed by an internal regulatory element. To test the hypothesis that at least one of the two domains in the double domain proteins harbor subcellular localization determinants, we analyzed the localization of the N- and C-terminal domains of A3B, A3D, A3F and A3G separately. During prophase, metaphase and anaphase all of these proteins were excluded from condensed chromosomes, similar to AID and all the other APOBEC3s (Figs. 1 and 2, data not shown). During telophase, the N-terminal halves of A3B and A3D were both cell-wide and associated with DNA, while the N-terminal domains of A3G and A3F were excluded from the reassembling nuclear compartment (Fig. 4A). The C-terminal domains of A3B, A3D, A3F and A3G were cell-wide (Fig. 4B). As expected,39 the localization of the N-terminal domains during interphase was similar to the localization of the full-length proteins during interphase (i.e., A3B N-terminal and full-length were nuclear, while A3F and A3G N-terminal and full-length were cytoplasmic Fig. 4C). Interestingly, the N-terminal domain of A3D was an exception, as it appeared predominantly nuclear, whereas full-length A3D was cytoplasmic and its C-terminal domain was cell-wide (Fig. 4). This difference prompted us to test whether A3D shuttles from the nucleus into the cytoplasm through the CRM1 pathway using the inhibitor leptomycin B (lepB).51 As a control, AID was cytoplasmic, and lepB caused it to become more nuclear32,33 (Fig. S7). In contrast, A3D remained cytoplasmic in the presence of lepB, implying that it does not use the CRM1 pathway (Fig. S7). In addition, the ability of N-terminal A3B to co-localize with DNA during telophase was different from the delayed re-entry of full-length A3B, which remains excluded from DNA during telophase (Figs. 2B and 4A). Concordant with prior work,37,39 these data indicate that that A3F and A3G are likely to be excluded actively from DNA, since their N-terminal domains alone are excluded (Fig. 4A). A3B and A3D may rely on the size of the double domain or a motif created from the combination of both domains since in both cases the N-terminal half of the protein has access to DNA during telophase while the full-length protein is excluded (Fig. 2B and 4A). Interestingly, forcing two A3C proteins together in an A3C-A3C-eGFP chimera created a protein that was cytoplasmic in interphase and excluded from genomic DNA in the same way as A3B, A3D, A3F and A3G (Fig. S8). Thus, both cis determinants and size have the capacity to influence the telophase DNA exclusion phenotype seen for A3B, A3D, A3F and A3G.
Figure 4. Localization of single domain variants of A3B, A3D, A3F and A3G. (A) Images of HeLa cells in telophase expressing N-terminal or (B) C-terminal halves of the indicated APOBEC3-eGFP constructs. DAPI and merged images below. Wild-type A3D-CTD was toxic, so a catalytic mutant was used (A3D-CTD-E264A; indicated by *). All images are representative of several telophase cells. (C) Representative images of HeLa cells in interphase expressing the N-terminal halves of the indicated APOBEC3-eGFP constructs.
DNA deaminase activity does not change between interphase and mitosis
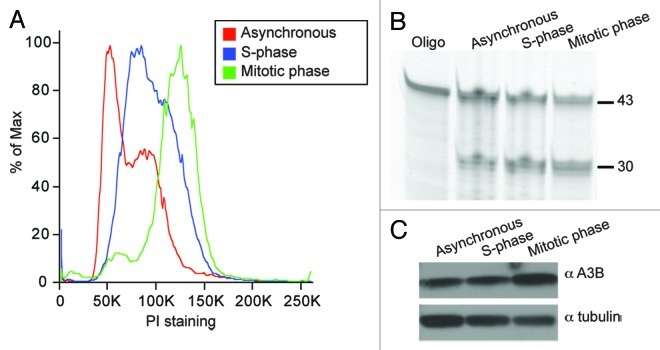
We hypothesized that in addition to physical exclusion of the APOBECs during mitosis, the instrinsic deaminase activity of these enzymes might be up- or downregulated during mitosis. First, we tested for changes in the deaminase activity of APOBECs that co-localize with DNA (A3A, A3B, A3C and A3H). We observed no difference in activity from transiently transfected untreated or mitotic cell lysates (Fig. S9). However, A3A and A3B blocked cells from exiting G1, preventing comparison of activity in untreated and mitotic cells (Fig. S9). To circumvent the arrest caused by enforced overexpression of A3A and A3B, we chose to focus on endogenous A3B activity. A3B mRNA is expressed highly in the osteosarcoma cell line U2OS, which grows normally and may have adapted to A3B expression (Burns, Leonard and Harris, unpublished data). We tested lysates from asynchronous cell populations, synchronized cells harvested in S phase, as well as nocodazole-treated synchronized cells in prometaphase52 (Fig. 5A). Both deaminase activity and A3B protein levels were similar under all conditions (Fig. 5B and C). These experiments show that endogenous A3B is active in during interphase and during mitosis, and they further indicate that its intrinsic DNA deaminase activity may not vary during the cell-cycle. These results suggest that genomic deamination is not prevented by cell cycle-specific regulation of APOBEC3 activity.
Figure 5. Similar A3B deaminase activity in untreated, S-phase and mitotic cell lysates. (A) Cell cycle profiles of asynchronous cells (red), S-phase cells (blue) and mitotic cells (green). See methods for synchronization details. (B) Deaminase assay using lysates from cell populations shown in (A). (C) Immunoblot of protein lysates from (A) probed with anti-A3B or anti-tubulin antibodies.
APOBEC3-induced cell cycle perturbations
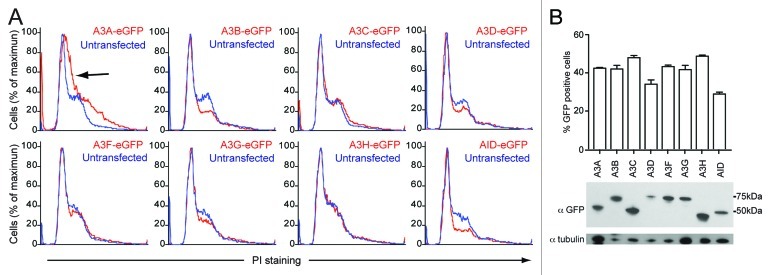
The cell cycle is a highly regulated developmental program with delicate checks and balances that prevent cells from dividing in the presence of DNA damage. We used these innate DNA damage-sensing properties to test for DNA damage caused by the APOBEC3 proteins. Tetracycline inducible HEK293 and HeLa cells show cell cycle disruptions after overexpression of A3A13,14 and A3B,14 and repair-deficient B cells have shown toxicity after AID induction.53 We therefore tested all APOBEC3s and AID for cell cycle effects by transient transfection in HEK293T and HeLa cells. Based on the mitotic images described above, we predicted that A3A, A3B and A3H were mostly likely to alter the cell cycle profile over time, and, based on our activity data, A3C would have little effect even though it distributes cell-wide. Representative profiles for each APOBEC3 in these two cell lines are shown for 48 h expression in HEK293T cells and 96 h expression in HeLa cells (Fig. 6A; Fig. S10A).
Figure 6. APOBEC3 effects on cell cycle progression in HEK293T cells. (A) DNA flow cytometry profiles of HEK293T cells transfected with APOBEC3-eGFP constructs. Representative profiles of triplicate independent PI-staining experiments are shown at 48 h. APOBEC3-eGFP expressing cell profiles (red) are overlaid on untransfected cells in the same population (blue). The shift toward S-phase in A3A-eGFP expressing cells is indicated (black arrow). (B) Expression of the indicated APOBEC3-eGFP proteins in HEK293T cells as shown by flow cytometry (top) and immunoblotting (bottom).
A3A caused a consistent shift of the G1 peak toward S in both cell lines and a broadening of the G2 population in HEK293T cells (arrows in Fig. 6A; Fig. S10A). In HEK293T cells, only A3B, A3D and AID caused decreases in mitotic cells (Fig. 6A), while in HeLa cells only A3B and AID caused a dramatic decreases in DNA content, an indication of apoptotic cells (Fig. S10A). Surprisingly, A3H did not cause a reproducible effect on the cell cycle profile. A3C, A3F and A3G did not cause dramatic changes to the shape or proportions of the cell cycle profile, indicating that transient overexpression is not the cause of these cell cycle perturbations. From these data it is clear the A3A, A3B and AID can affect cell cycle progression, as has been shown in different systems,13,14,53 and that A3D may also be able to activate cellular checkpoints despite low levels of expression (Fig. 6B; Fig. S10B). Similar cell cycle defects are dependent on the catalytic activity of both A3A and A3B and have been linked to genomic mutations, supporting our use of cell cycle perturbations as a measure of genomic DNA deamination.13,14
Discussion
We hypothesized that the mitotic breakdown of the nuclear envelope would allow APOBEC3s access to genomic cytosines for deamination. Since several APOBEC3s are positively charged and known to bind DNA,54,55 we expected to see the APOBEC3 proteins interacting with DNA during prophase upon dissolution of the nuclear envelope. Instead, we discovered that cytoplasmic APOBEC3 proteins do not have access to genomic DNA, even during mitosis. The mechanism preventing the APOBEC3s from interacting with genomic DNA during prophase, metaphase and anaphase is unclear but may be as simple as exclusion from condensed chromatin, since eGFP is excluded in a similar manner. However, this simplistic model does not explain all observations, as DNA exclusion during telophase may be dependent on a combination of N-terminal determinants and protein size. A3F and A3G, for example, have cis determinants, because their N-terminal domains alone are cytoplasmic, whereas A3B and A3D N-terminal proteins co-localize with DNA during telophase, while their full-length forms are excluded.
We were surprised that full-length A3D is cytoplasmic, but separately, each domain had access to the nucleus. Moreover, full-length A3D had the capacity to affect HEK293T and HeLa cell cycle profiles. We theorized that full-length A3D might shuttle between the cytoplasmic and nuclear compartment, but we did not detect CRM1-dependent shuttling. Although we have no evidence that the N- and C-terminal domains of A3D are expressed separately in our transient transfections, it is possible that separate N- and C-terminal domains may have a role to play in genomic mutation. Double-domain APOBEC3 proteins in other species (e.g., sheep and pig) can be expressed as either N- or C-terminal domains alone, as well as the full-length form.56,57 Human A3F also has alternative isoforms.58 It is therefore possible that other double domain APOBEC3 proteins may be expressed as single N- or C-terminal domain variants that could have subcellular distributions that differ from the full-length enzymes.
Importantly, we have shown that endogenous A3B is similarly active in interphase, S phase and mitotic cell lysates. Transient transfection experiments with A3C and A3H also indicated no difference in activity between extracts from interphase and mitotic cell populations. These results suggested that DNA deamination activity is not up- or downregulated during different stages of the cell cycle. Instead subcellular localization may direct a perpetually active APOBEC3 enzyme to its substrate. In this model, we expect that the majority of genomic deamination events by APOBEC3 proteins occur during interphase. Previous work has indicated that AID deaminates genomic DNA during G1 phase.59 Because we see exclusion of the APOBEC3 proteins during the majority of mitosis, it seems likely that genomic deamination generally occurs during interphase and may be specific to G1, although further work is needed to distinguish between G1, S and G2 deamination by endogenous APOBEC3 proteins in relevant primary cell types.
Use of cell cycle profiles as proxy for DNA deamination has precedent for both A3A and A3B and has been linked to the catalytic activity of these enzymes and their capacity to cause genomic mutations.13,14 We observed a cell cycle progression defect in HeLa and HEK293T cells expressing A3A, A3B, A3D and AID (and mildly for A3H), implying that deamination and the ensuing DNA damage response is occurring in these cells. Based on our results, we predict that A3A, A3B, A3D, A3H and AID may deaminate genomic DNA. While A3A is tightly regulated and is confined to cells of the myeloid lineage,60 other APOBEC3 proteins are expressed more broadly.61,62 Interestingly, A3B, A3D and A3H all have been inactivated or deleted to a certain extent in the human population.29,48,49,63-65 More than 90% of people of Oceanic heritage have an A3B deletion polymorphism that leaves the surrounding A3A and A3C genes intact but completely removes A3B.63 Chimpanzee A3D is much more active than human A3D, and this is largely dependent on a single amino acid difference.64,65 Likewise, there are several haplotypes of A3H, and the most prevalent among them are unstable.29,48,49 We used the human A3H haplotype II in these experiments, because it is stable and active against HIV-1.3,6,29 Thus, A3B, A3D and A3H may be sufficiently detrimental to genomic DNA and an individual’s well-being to warrant selective inactivation of the activity of these enzymes. Although the experiments we report are transient tests of the capability of the APOBEC3s to affect cell cycle progression, the data suggest that A3A, A3B, A3D and A3H may be interesting for further studies on genomic mutation. In fact, transient expression of A3A causes genomic mutations in the nuclear and mitochondrial genomes,12 and high levels of endogenous A3B induce genomic mutations in breast cancer cells and correlate with higher mutation loads in tumors.14
The long-term effect of chronic or episodic genomic deamination may be cancer predisposition. This is strongly supported by the link between AID expression and B cell cancer.7,8,66 Breast cancer genome sequencing has shown that breast tumors have large numbers of somatic mutations and also high percentages of cytosine to thymine (C to T) transitions and regions of clustered mutations.67,68 These mutation signatures from breast cancers have been linked to A3B signatures, especially in clustered regions, where single-stranded DNA may be available for deamination.14 Many other cancers also have a high proportion of C to T mutations, including brain, gastric, head and neck, ovarian, pancreatic and prostate cancers.69-73 Over the next few years, with the increasing availability of cancer genome sequences and additional molecular studies of the APOBEC3 proteins, we predict that minimally A3B14 and potentially other APOBEC3s will come to the center stage as our understanding of cancer mutagenesis develops.
Materials and Methods
Fixed cell microscopy experiments
Microscopy procedures have been described.30 Briefly, HeLa cells plated on glass coverslips (12-545-85; Fisher Scientific) in 6-well plates were transiently transfected with 400 ng each of eGFP, HA or untagged constructs (Transit-LT1; Mirus) and incubated for 48 h. The cells were fixed with 4% paraformaldehdye in phosphate buffered saline (PFA in PBS) for 20 min at room temperature. APOBEC3-HA and untagged expressing cells were incubated with primary antibody in blocking buffer. Rabbit polyclonal sera were identified as described.74 Hybridoma media was used for immunofluorescence. The anti-HA antibody (MMS-101P; Covance) was used at 1:200 and visualized with anti-mouse FITC (115095146; Jackson), while untagged A3B was identified with Rb10 14, untagged A3G was identified with Rb10 93, and these samples were visualized with anti-rabbit FITC (111095144; Jackson) (Fig. S6). All slides were treated with 0.1% Hoechst dye to stain the nuclei. The slides were mounted with 50% glycerol and imaged (Deltavision; Applied Precision). All images were deconvolved using SoftWorks (Applied Precision).
Live cell experiments
Movies were taken as described.30 Briefly, HeLa cells plated at 40,000 cells/well in 4-well chambers (Nunc) were transiently transfected with 200 ng of eGFP-tagged histone 2B and 400 ng of APOBEC3-mCherry. The cells expressed these constructs for 48 h before transfer to a heated chamber (37°C) on the microscope (Deltavision; Applied Precision). Images of dividing cells were taken every three minutes for 1–3 h. These images were deconvolved and used to create quicktime movies. Treatment with leptomycin B to inhibit CRM1 dependent export has been reported.30,75 Cells were treated with 40 ng/mL of leptomycin B dissolved in ethanol or ethanol diluted in media alone. After 3 h these cells were fixed for imaging.
DNA deaminase oligonucleotide cleavage assays
Synchronization protocols were modified for this experiment.52 HEK293T or U2OS cells were treated with 2 mM thymidine (Sigma-Aldrich) to cause G1/S-phase arrest. After 17 h the thymidine was removed and the cells were released in DMEM with fetal bovine serum (media). The HEK293T cells were then transfected with the indicated APOBEC3-eGFP constructs (Transit-LT1, Mirus). After 8 h the media was removed, and a second thymidine block was added. After another 17 h incubation the thymidine was removed, and the cells were released into media. After 4 h cells were harvested for S-phase. Alternatively, the cells were released for 2 h and then treated with 120 ng/mL of nocodazole (Sigma-Aldrich). After 16 h the nocodazole treated cells were harvested for metaphase. A fraction of the cells were analyzed for cell cycle profile (see below). The rest were pelleted, washed and resuspended in lysis buffer (25 mM Hepes, pH 7.4, 250 mM NaCl, 10% glycerol, 0.5% Triton X-100, 1 mM EDTA, 1 mM MgCl2, 1 mM ZnCl2) then sonicated three times for 3 sec. Samples were run on western blots and probed with anti-A3B (Rb10 14), anti-GFP (1:5,000, 632 381; BD Clontech) or anti-tubulin (1:20,000, MMS-407R; Covance). The DNA deamination assay has been described.60 Lysates were mixed with 6-FAM labeled 43 nucleotide containing oligo containing a TTCC deamination site for 30 min at 37°C before addition of uracil DNA glycosylase and NaOH to create and break an abasic site. The samples were run on 15% acrylimide-urea gels for separation and analyzed with a Fuji-FLA 5000 scanner.
Cell cycle profiling experiments
HeLa or HEK293T cells were plated into 6-well plates at 200,000 cells/well and transfected the next day with 300 (A3A, A3B, A3C, A3F, A3G) or 400 ng (A3D, A3H and AID) of eGFP constructs. The cells were harvested and fixed with 4% PFA for cell cycle analysis or lysed in loading buffer for western blots. The cell cycle samples were treated with 0.1% Triton X 100, 20 µg/mL propidium iodide and 40 µg/mL RNase A (Qiagen) in PBS for 30 min at room temperature before flow cytometry (BD Biosciences FACS Canto II). GFP-positive and GFP-negative live cells were analyzed for their PI staining profiles using FloJo and GraphPad Prism. The lysates were run on 4–15% polyacrylamide gels, transferred to pvdf and then blotted with anti-GFP (1:5,000, 632 381; BD Clontech) and anti-tubulin (1:20,000, MMS-407R; Covance).
Supplementary Material
Acknowledgments
We thank J. Mueller for the eGFP-H2B construct and U2OS cells, M.B. Burns and A. Green for contributions to antibody development, E. Refsland and A.M. Land for assistance with cell cycle profiles and M. Carpenter for advice with activity assays. This work was supported by grants from the National Institutes of Health R01 AI064046 and P01 GM091743. L. Lackey was supported in part by an NSF pre-doctoral fellowship and subsequently by a position on the Institute for Molecular Virology Training Grant NIH T32 AI083196.
Glossary
Abbreviations:
- AID
activation induced cytosine deaminase
- APOBEC3 (A3)
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3
- C
cytosine
- CTD
C-terminal domain
- eGFP
enhanced green fluorescent protein
- H2B
histone 2B
- kDa
kilodalton
- lepB
leptomycin B
- NTD
N-terminal domain
- T
thymine
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/cc/article/23713
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/23713
References
- 1.Conticello SG. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 2008;9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albin JS, Harris RS. Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert Rev Mol Med. 2010;12:e4. doi: 10.1017/S1462399409001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Larue RS, Lengyel J, Jónsson SR, Andrésdóttir V, Harris RS. Lentiviral Vif degrades the APOBEC3Z3/APOBEC3H protein of its mammalian host and is capable of cross-species activity. J Virol. 2010;84:8193–201. doi: 10.1128/JVI.00685-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malim MH, Emerman M. HIV-1 accessory proteins--ensuring viral survival in a hostile environment. Cell Host Microbe. 2008;3:388–98. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 5.Aguiar RS, Peterlin BM. APOBEC3 proteins and reverse transcription. Virus Res. 2008;134:74–85. doi: 10.1016/j.virusres.2007.12.022. [DOI] [PubMed] [Google Scholar]
- 6.Hultquist JF, Lengyel JA, Refsland EW, LaRue RS, Lackey L, Brown WL, et al. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J Virol. 2011;85:11220–34. doi: 10.1128/JVI.05238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okazaki IM, Hiai H, Kakazu N, Yamada S, Muramatsu M, Kinoshita K, et al. Constitutive expression of AID leads to tumorigenesis. J Exp Med. 2003;197:1173–81. doi: 10.1084/jem.20030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pasqualucci L, Bhagat G, Jankovic M, Compagno M, Smith P, Muramatsu M, et al. AID is required for germinal center-derived lymphomagenesis. Nat Genet. 2008;40:108–12. doi: 10.1038/ng.2007.35. [DOI] [PubMed] [Google Scholar]
- 9.Robbiani DF, Bothmer A, Callen E, Reina-San-Martin B, Dorsett Y, Difilippantonio S, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–38. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakamuta M, Chang BH, Zsigmond E, Kobayashi K, Lei H, Ishida BY, et al. Complete phenotypic characterization of apobec-1 knockout mice with a wild-type genetic background and a human apolipoprotein B transgenic background, and restoration of apolipoprotein B mRNA editing by somatic gene transfer of Apobec-1. J Biol Chem. 1996;271:25981–8. doi: 10.1074/jbc.271.42.25981. [DOI] [PubMed] [Google Scholar]
- 11.Yamanaka S, Balestra ME, Ferrell LD, Fan J, Arnold KS, Taylor S, et al. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc Natl Acad Sci U S A. 1995;92:8483–7. doi: 10.1073/pnas.92.18.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suspène R, Aynaud MM, Guétard D, Henry M, Eckhoff G, Marchio A, et al. Somatic hypermutation of human mitochondrial and nuclear DNA by APOBEC3 cytidine deaminases, a pathway for DNA catabolism. Proc Natl Acad Sci U S A. 2011;108:4858–63. doi: 10.1073/pnas.1009687108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landry S, Narvaiza I, Linfesty DC, Weitzman MD. APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO Rep. 2011;12:444–50. doi: 10.1038/embor.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013 doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- 16.Reich NC, Liu L. Tracking STAT nuclear traffic. Nat Rev Immunol. 2006;6:602–12. doi: 10.1038/nri1885. [DOI] [PubMed] [Google Scholar]
- 17.Pelzer C, Thome M. IKKα takes control of canonical NF-κB activation. Nat Immunol. 2011;12:815–6. doi: 10.1038/ni.2082. [DOI] [PubMed] [Google Scholar]
- 18.Yoneda Y. Nucleocytoplasmic protein traffic and its significance to cell function. Genes Cells. 2000;5:777–87. doi: 10.1046/j.1365-2443.2000.00366.x. [DOI] [PubMed] [Google Scholar]
- 19.Güttinger S, Laurell E, Kutay U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat Rev Mol Cell Biol. 2009;10:178–91. doi: 10.1038/nrm2641. [DOI] [PubMed] [Google Scholar]
- 20.Estrada-Gelonch A, Aramburu J, López-Rodríguez C. Exclusion of NFAT5 from mitotic chromatin resets its nucleo-cytoplasmic distribution in interphase. PLoS One. 2009;4:e7036. doi: 10.1371/journal.pone.0007036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaidi SK, Young DW, Pockwinse SM, Javed A, Lian JB, Stein JL, et al. Mitotic partitioning and selective reorganization of tissue-specific transcription factors in progeny cells. Proc Natl Acad Sci U S A. 2003;100:14852–7. doi: 10.1073/pnas.2533076100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niewiadomska AM, Tian C, Tan L, Wang T, Sarkis PT, Yu XF. Differential inhibition of long interspersed element 1 by APOBEC3 does not correlate with high-molecular-mass-complex formation or P-body association. J Virol. 2007;81:9577–83. doi: 10.1128/JVI.02800-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kinomoto M, Kanno T, Shimura M, Ishizaka Y, Kojima A, Kurata T, et al. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res. 2007;35:2955–64. doi: 10.1093/nar/gkm181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marin M, Golem S, Rose KM, Kozak SL, Kabat D. Human immunodeficiency virus type 1 Vif functionally interacts with diverse APOBEC3 cytidine deaminases and moves with them between cytoplasmic sites of mRNA metabolism. J Virol. 2008;82:987–98. doi: 10.1128/JVI.01078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muckenfuss H, Hamdorf M, Held U, Perkovic M, Löwer J, Cichutek K, et al. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem. 2006;281:22161–72. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- 26.Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, et al. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr Biol. 2006;16:480–5. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 27.Goila-Gaur R, Khan MA, Miyagi E, Kao S, Strebel K. Targeting APOBEC3A to the viral nucleoprotein complex confers antiviral activity. Retrovirology. 2007;4:61. doi: 10.1186/1742-4690-4-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bogerd HP, Wiegand HL, Doehle BP, Lueders KK, Cullen BR. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 2006;34:89–95. doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.OhAinle M, Kerns JA, Li MM, Malik HS, Emerman M. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe. 2008;4:249–59. doi: 10.1016/j.chom.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lackey L, Demorest ZL, Land AM, Hultquist JF, Brown WL, Harris RS. APOBEC3B and AID have similar nuclear import mechanisms. J Mol Biol. 2012;419:301–14. doi: 10.1016/j.jmb.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patenaude AM, Orthwein A, Hu Y, Campo VA, Kavli B, Buschiazzo A, et al. Active nuclear import and cytoplasmic retention of activation-induced deaminase. Nat Struct Mol Biol. 2009;16:517–27. doi: 10.1038/nsmb.1598. [DOI] [PubMed] [Google Scholar]
- 32.McBride KM, Barreto V, Ramiro AR, Stavropoulos P, Nussenzweig MC. Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase. J Exp Med. 2004;199:1235–44. doi: 10.1084/jem.20040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ito S, Nagaoka H, Shinkura R, Begum N, Muramatsu M, Nakata M, et al. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc Natl Acad Sci U S A. 2004;101:1975–80. doi: 10.1073/pnas.0307335101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Y, Sowden MP, Yang Y, Smith HC. Intracellular trafficking determinants in APOBEC-1, the catalytic subunit for cytidine to uridine editing of apolipoprotein B mRNA. Exp Cell Res. 2001;267:153–64. doi: 10.1006/excr.2001.5255. [DOI] [PubMed] [Google Scholar]
- 35.Blanc V, Kennedy S, Davidson NO. A novel nuclear localization signal in the auxiliary domain of apobec-1 complementation factor regulates nucleocytoplasmic import and shuttling. J Biol Chem. 2003;278:41198–204. doi: 10.1074/jbc.M302951200. [DOI] [PubMed] [Google Scholar]
- 36.Blanc V, Henderson JO, Kennedy S, Davidson NO. Mutagenesis of apobec-1 complementation factor reveals distinct domains that modulate RNA binding, protein-protein interaction with apobec-1, and complementation of C to U RNA-editing activity. J Biol Chem. 2001;276:46386–93. doi: 10.1074/jbc.M107654200. [DOI] [PubMed] [Google Scholar]
- 37.Bennett RP, Presnyak V, Wedekind JE, Smith HC. Nuclear Exclusion of the HIV-1 host defense factor APOBEC3G requires a novel cytoplasmic retention signal and is not dependent on RNA binding. J Biol Chem. 2008;283:7320–7. doi: 10.1074/jbc.M708567200. [DOI] [PubMed] [Google Scholar]
- 38.Stenglein MD, Harris RS. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J Biol Chem. 2006;281:16837–41. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- 39.Stenglein MD, Matsuo H, Harris RS. Two regions within the amino-terminal half of APOBEC3G cooperate to determine cytoplasmic localization. J Virol. 2008;82:9591–9. doi: 10.1128/JVI.02471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wichroski MJ, Robb GB, Rana TM. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog. 2006;2:e41. doi: 10.1371/journal.ppat.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 42.Gallois-Montbrun S, Holmes RK, Swanson CM, Fernández-Ocaña M, Byers HL, Ward MA, et al. Comparison of cellular ribonucleoprotein complexes associated with the APOBEC3F and APOBEC3G antiviral proteins. J Virol. 2008;82:5636–42. doi: 10.1128/JVI.00287-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gallois-Montbrun S, Kramer B, Swanson CM, Byers H, Lynham S, Ward M, et al. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J Virol. 2007;81:2165–78. doi: 10.1128/JVI.02287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonvin M, Achermann F, Greeve I, Stroka D, Keogh A, Inderbitzin D, et al. Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology. 2006;43:1364–74. doi: 10.1002/hep.21187. [DOI] [PubMed] [Google Scholar]
- 45.Nowarski R, Wilner OI, Cheshin O, Shahar OD, Kenig E, Baraz L, et al. APOBEC3G enhances lymphoma cell radioresistance by promoting cytidine deaminase-dependent DNA repair. Blood. 2012;120:366–75. doi: 10.1182/blood-2012-01-402123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brar SS, Watson M, Diaz M. Activation-induced cytosine deaminase (AID) is actively exported out of the nucleus but retained by the induction of DNA breaks. J Biol Chem. 2004;279:26395–401. doi: 10.1074/jbc.M403503200. [DOI] [PubMed] [Google Scholar]
- 47.Pak V, Heidecker G, Pathak VK, Derse D. The role of amino-terminal sequences in cellular localization and antiviral activity of APOBEC3B. J Virol. 2011;85:8538–47. doi: 10.1128/JVI.02645-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.OhAinle M, Kerns JA, Malik HS, Emerman M. Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H. J Virol. 2006;80:3853–62. doi: 10.1128/JVI.80.8.3853-3862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X, Abudu A, Son S, Dang Y, Venta PJ, Zheng YH. Analysis of human APOBEC3H haplotypes and anti-human immunodeficiency virus type 1 activity. J Virol. 2011;85:3142–52. doi: 10.1128/JVI.02049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin RM, Cardoso MC. Chromatin condensation modulates access and binding of nuclear proteins. FASEB J. 2010;24:1066–72. doi: 10.1096/fj.08-128959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolff B, Sanglier JJ, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–47. doi: 10.1016/S1074-5521(97)90257-X. [DOI] [PubMed] [Google Scholar]
- 52.Harper JV. Synchronization of cell populations in G1/S and G2/M phases of the cell cycle. Methods Mol Biol. 2005;296:157–66. doi: 10.1385/1-59259-857-9:157. [DOI] [PubMed] [Google Scholar]
- 53.Hasham MG, Donghia NM, Coffey E, Maynard J, Snow KJ, Ames J, et al. Widespread genomic breaks generated by activation-induced cytidine deaminase are prevented by homologous recombination. Nat Immunol. 2010;11:820–6. doi: 10.1038/ni.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen KM, Harjes E, Gross PJ, Fahmy A, Lu Y, Shindo K, et al. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature. 2008;452:116–9. doi: 10.1038/nature06638. [DOI] [PubMed] [Google Scholar]
- 55.Chelico L, Pham P, Calabrese P, Goodman MF. APOBEC3G DNA deaminase acts processively 3′ --> 5′ on single-stranded DNA. Nat Struct Mol Biol. 2006;13:392–9. doi: 10.1038/nsmb1086. [DOI] [PubMed] [Google Scholar]
- 56.LaRue RS, Jónsson SR, Silverstein KA, Lajoie M, Bertrand D, El-Mabrouk N, et al. The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol Biol. 2008;9:104. doi: 10.1186/1471-2199-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jónsson SR, Haché G, Stenglein MD, Fahrenkrug SC, Andrésdóttir V, Harris RS. Evolutionarily conserved and non-conserved retrovirus restriction activities of artiodactyl APOBEC3F proteins. Nucleic Acids Res. 2006;34:5683–94. doi: 10.1093/nar/gkl721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lassen KG, Wissing S, Lobritz MA, Santiago M, Greene WC. Identification of two APOBEC3F splice variants displaying HIV-1 antiviral activity and contrasting sensitivity to Vif. J Biol Chem. 2010;285:29326–35. doi: 10.1074/jbc.M110.154054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schrader CE, Guikema JE, Linehan EK, Selsing E, Stavnezer J. Activation-induced cytidine deaminase-dependent DNA breaks in class switch recombination occur during G1 phase of the cell cycle and depend upon mismatch repair. J Immunol. 2007;179:6064–71. doi: 10.4049/jimmunol.179.9.6064. [DOI] [PubMed] [Google Scholar]
- 60.Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat Struct Mol Biol. 2010;17:222–9. doi: 10.1038/nsmb.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Refsland EW, Stenglein MD, Shindo K, Albin JS, Brown WL, Harris RS. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. 2010;38:4274–84. doi: 10.1093/nar/gkq174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koning FA, Newman EN, Kim EY, Kunstman KJ, Wolinsky SM, Malim MH. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J Virol. 2009;83:9474–85. doi: 10.1128/JVI.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kidd JM, Newman TL, Tuzun E, Kaul R, Eichler EE. Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet. 2007;3:e63. doi: 10.1371/journal.pgen.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Duggal NK, Malik HS, Emerman M. The breadth of antiviral activity of Apobec3DE in chimpanzees has been driven by positive selection. J Virol. 2011;85:11361–71. doi: 10.1128/JVI.05046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dang Y, Abudu A, Son S, Harjes E, Spearman P, Matsuo H, et al. Identification of a single amino acid required for APOBEC3 antiretroviral cytidine deaminase activity. J Virol. 2011;85:5691–5. doi: 10.1128/JVI.00243-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ramiro AR, Jankovic M, Eisenreich T, Difilippantonio S, Chen-Kiang S, Muramatsu M, et al. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 2004;118:431–8. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 67.Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, et al. Breast Cancer Working Group of the International Cancer Genome Consortium Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–93. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, et al. Oslo Breast Cancer Consortium (OSBREAC) The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–4. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–7. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jones S, Wang TL, Shih IeM, Mao TL, Nakayama K, Roden R, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–31. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–7. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–20. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carpenter MA, Li M, Rathore A, Lackey L, Law EK, Land AM, et al. Methylcytosine and normal cytosine deamination by the foreign DNA restriction enzyme APOBEC3A. J Biol Chem. 2012;287:34801–8. doi: 10.1074/jbc.M112.385161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Demorest ZL, Li M, Harris RS. Phosphorylation directly regulates the intrinsic DNA cytidine deaminase activity of activation-induced deaminase and APOBEC3G protein. J Biol Chem. 2011;286:26568–75. doi: 10.1074/jbc.M111.235721. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.