Abstract
The RPC31 gene encoding the C31 subunit of Saccharomyces cerevisiae RNA polymerase C (III) has been isolated, starting from a C-terminal fragment cloned on a lambda gt11 library. It is unique on the yeast genome and lies on the left arm of chromosome XIV, very close to a NotI site. Its coding sequence perfectly matches the amino acid sequence of two oligopeptides prepared from purified C31. It is also identical to the ACP2 gene previously described as encoding an HMG1-like protein (W. Haggren and D. Kolodrubetz, Mol. Cell. Biol. 8:1282-1289, 1988). Thus, ACP2 and RPC31 are allelic and encode a subunit of RNA polymerase C. The c31 protein has a highly acidic C-terminal tail also found in several other chromatin-interacting proteins, including animal HMG1. Outside this domain, however, there is no appreciable homology to any known protein. The growth phenotypes of a gene deletion, of insertions, and of nonsense mutations indicate that the C31 protein is strictly required for cell growth and that most of the acidic domain is essential for its function. Random mutagenesis failed to yield temperature-sensitive mutants, but a slowly growing mutant was constructed by partial suppression of a UAA nonsense allele of RPC31. Its reduced rate of tRNA synthesis in vivo relative to 5.8S rRNA supports the hypothesis that the C31 protein is a functional subunit of RNA polymerase C.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison L. A., Moyle M., Shales M., Ingles C. J. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985 Sep;42(2):599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- Baldari C., Cesareni G. Plasmids pEMBLY: new single-stranded shuttle vectors for the recovery and analysis of yeast DNA sequences. Gene. 1985;35(1-2):27–32. doi: 10.1016/0378-1119(85)90154-4. [DOI] [PubMed] [Google Scholar]
- Bianchi M. E., Beltrame M., Paonessa G. Specific recognition of cruciform DNA by nuclear protein HMG1. Science. 1989 Feb 24;243(4894 Pt 1):1056–1059. doi: 10.1126/science.2922595. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Burton Z., Burgess R. R., Lin J., Moore D., Holder S., Gross C. A. The nucleotide sequence of the cloned rpoD gene for the RNA polymerase sigma subunit from E coli K12. Nucleic Acids Res. 1981 Jun 25;9(12):2889–2903. doi: 10.1093/nar/9.12.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. S., Saltzman A. G., Hiipakka R. A., Huang I. Y., Liao S. S. Prostatic spermine-binding protein. Cloning and nucleotide sequence of cDNA, amino acid sequence, and androgenic control of mRNA level. J Biol Chem. 1987 Feb 25;262(6):2826–2831. [PubMed] [Google Scholar]
- Clark D. J., Kimura T. Electrostatic mechanism of chromatin folding. J Mol Biol. 1990 Feb 20;211(4):883–896. doi: 10.1016/0022-2836(90)90081-V. [DOI] [PubMed] [Google Scholar]
- Elledge S. J., Davis R. W. A family of versatile centromeric vectors designed for use in the sectoring-shuffle mutagenesis assay in Saccharomyces cerevisiae. Gene. 1988 Oct 30;70(2):303–312. doi: 10.1016/0378-1119(88)90202-8. [DOI] [PubMed] [Google Scholar]
- Falco S. C., Botstein D. A rapid chromosome-mapping method for cloned fragments of yeast DNA. Genetics. 1983 Dec;105(4):857–872. doi: 10.1093/genetics/105.4.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiduschek E. P., Tocchini-Valentini G. P. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- Ginsberg A. M., King B. O., Roeder R. G. Xenopus 5S gene transcription factor, TFIIIA: characterization of a cDNA clone and measurement of RNA levels throughout development. Cell. 1984 Dec;39(3 Pt 2):479–489. doi: 10.1016/0092-8674(84)90455-0. [DOI] [PubMed] [Google Scholar]
- Goebl M. G., Yochem J., Jentsch S., McGrath J. P., Varshavsky A., Byers B. The yeast cell cycle gene CDC34 encodes a ubiquitin-conjugating enzyme. Science. 1988 Sep 9;241(4871):1331–1335. doi: 10.1126/science.2842867. [DOI] [PubMed] [Google Scholar]
- Gudenus R., Mariotte S., Moenne A., Ruet A., Memet S., Buhler J. M., Sentenac A., Thuriaux P. Conditional mutants of RPC160, the gene encoding the largest subunit of RNA polymerase C in Saccharomyces cerevisiae. Genetics. 1988 Jul;119(3):517–526. doi: 10.1093/genetics/119.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggren W., Kolodrubetz D. The Saccharomyces cerevisiae ACP2 gene encodes an essential HMG1-like protein. Mol Cell Biol. 1988 Mar;8(3):1282–1289. doi: 10.1128/mcb.8.3.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann J. D., Chamberlin M. J. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- Hinkle D. C., Chamberlin M. J. Studies of the binding of Escherichia coli RNA polymerase to DNA. II. The kinetics of the binding reaction. J Mol Biol. 1972 Sep 28;70(2):187–195. doi: 10.1016/0022-2836(72)90532-3. [DOI] [PubMed] [Google Scholar]
- Huet J., Riva M., Sentenac A., Fromageot P. Yeast RNA polymerase C and its subunits. Specific antibodies as structural and functional probes. J Biol Chem. 1985 Dec 5;260(28):15304–15310. [PubMed] [Google Scholar]
- Huisman O., Raymond W., Froehlich K. U., Errada P., Kleckner N., Botstein D., Hoyt M. A. A Tn10-lacZ-kanR-URA3 gene fusion transposon for insertion mutagenesis and fusion analysis of yeast and bacterial genes. Genetics. 1987 Jun;116(2):191–199. doi: 10.1093/genetics/116.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessel M., Schulze F., Fibi M., Gruss P. Primary structure and nuclear localization of a murine homeodomain protein. Proc Natl Acad Sci U S A. 1987 Aug;84(15):5306–5310. doi: 10.1073/pnas.84.15.5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapholz S., Esposito R. E. Recombination and chromosome segregation during the single division meiosis in SPO12-1 and SPO13-1 diploids. Genetics. 1980 Nov;96(3):589–611. doi: 10.1093/genetics/96.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodrubetz D., Burgum A. Duplicated NHP6 genes of Saccharomyces cerevisiae encode proteins homologous to bovine high mobility group protein 1. J Biol Chem. 1990 Feb 25;265(6):3234–3239. [PubMed] [Google Scholar]
- Kolodziej P., Young R. A. RNA polymerase II subunit RPB3 is an essential component of the mRNA transcription apparatus. Mol Cell Biol. 1989 Dec;9(12):5387–5394. doi: 10.1128/mcb.9.12.5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe M., Binnie C., Schmidt R., Losick R. Cloned gene encoding the delta subunit of Bacillus subtilis RNA polymerase. Gene. 1988 Jul 15;67(1):13–19. doi: 10.1016/0378-1119(88)90003-0. [DOI] [PubMed] [Google Scholar]
- Mann C., Buhler J. M., Treich I., Sentenac A. RPC40, a unique gene for a subunit shared between yeast RNA polymerases A and C. Cell. 1987 Feb 27;48(4):627–637. doi: 10.1016/0092-8674(87)90241-8. [DOI] [PubMed] [Google Scholar]
- Marck C. 'DNA Strider': a 'C' program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1988 Mar 11;16(5):1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGoldrick E. M., Wheals A. E. Controlling the growth rate of Saccharomyces cerevisiae cells using the glucose analogue D-glucosamine. J Gen Microbiol. 1989 Sep;135(9):2407–2411. doi: 10.1099/00221287-135-9-2407. [DOI] [PubMed] [Google Scholar]
- Mortimer R. K., Schild D., Contopoulou C. R., Kans J. A. Genetic map of Saccharomyces cerevisiae, edition 10. Yeast. 1989 Sep-Oct;5(5):321–403. doi: 10.1002/yea.320050503. [DOI] [PubMed] [Google Scholar]
- Mémet S., Gouy M., Marck C., Sentenac A., Buhler J. M. RPA190, the gene coding for the largest subunit of yeast RNA polymerase A. J Biol Chem. 1988 Feb 25;263(6):2830–2839. [PubMed] [Google Scholar]
- Naumovski L., Chu G., Berg P., Friedberg E. C. RAD3 gene of Saccharomyces cerevisiae: nucleotide sequence of wild-type and mutant alleles, transcript mapping, and aspects of gene regulation. Mol Cell Biol. 1985 Jan;5(1):17–26. doi: 10.1128/mcb.5.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paonessa G., Frank R., Cortese R. Nucleotide sequence of rat liver HMG1 cDNA. Nucleic Acids Res. 1987 Nov 11;15(21):9077–9077. doi: 10.1093/nar/15.21.9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent S. A., Fenimore C. M., Bostian K. A. Vector systems for the expression, analysis and cloning of DNA sequences in S. cerevisiae. Yeast. 1985 Dec;1(2):83–138. doi: 10.1002/yea.320010202. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pentecost B. T., Wright J. M., Dixon G. H. Isolation and sequence of cDNA clones coding for a member of the family of high mobility group proteins (HMG-T) in trout and analysis of HMG-T-mRNA's in trout tissues. Nucleic Acids Res. 1985 Jul 11;13(13):4871–4888. doi: 10.1093/nar/13.13.4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
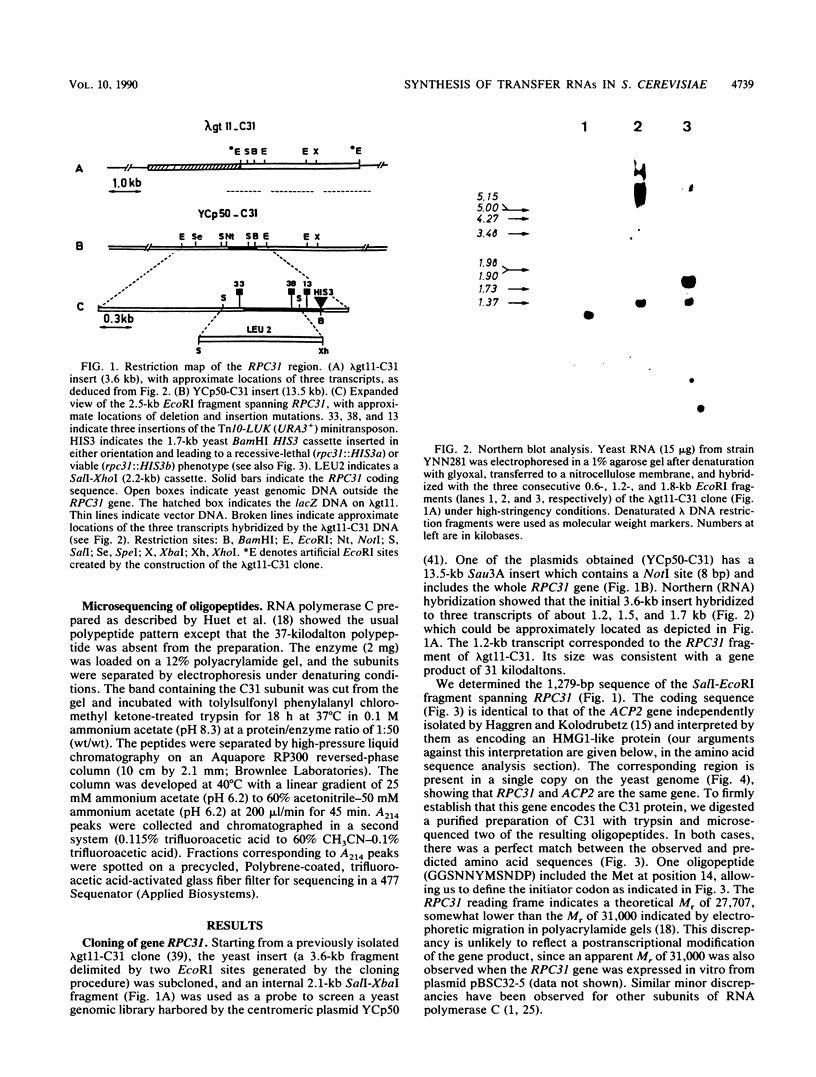
- Reynolds P., Higgins D. R., Prakash L., Prakash S. The nucleotide sequence of the RAD3 gene of Saccharomyces cerevisiae: a potential adenine nucleotide binding amino acid sequence and a nonessential acidic carboxyl terminal region. Nucleic Acids Res. 1985 Apr 11;13(7):2357–2372. doi: 10.1093/nar/13.7.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds P., Weber S., Prakash L. RAD6 gene of Saccharomyces cerevisiae encodes a protein containing a tract of 13 consecutive aspartates. Proc Natl Acad Sci U S A. 1985 Jan;82(1):168–172. doi: 10.1073/pnas.82.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva M., Buhler J. M., Sentenac A., Fromageot P., Hawthorne D. C. Natural variation in yeast RNA polymerase A. Formation of a mosaic RNA polymerase A in a meiotic segregant from an interspecific hybrid. J Biol Chem. 1982 Apr 25;257(8):4570–4577. [PubMed] [Google Scholar]
- Riva M., Memet S., Micouin J. Y., Huet J., Treich I., Dassa J., Young R., Buhler J. M., Sentenac A., Fromageot P. Isolation of structural genes for yeast RNA polymerases by immunological screening. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1554–1558. doi: 10.1073/pnas.83.6.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Fink G. R. KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell. 1987 Mar 27;48(6):1047–1060. doi: 10.1016/0092-8674(87)90712-4. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60(2-3):237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sawadogo M., Sentenac A. RNA polymerase B (II) and general transcription factors. Annu Rev Biochem. 1990;59:711–754. doi: 10.1146/annurev.bi.59.070190.003431. [DOI] [PubMed] [Google Scholar]
- Sentenac A. Eukaryotic RNA polymerases. CRC Crit Rev Biochem. 1985;18(1):31–90. doi: 10.3109/10409238509082539. [DOI] [PubMed] [Google Scholar]
- Sweetser D., Nonet M., Young R. A. Prokaryotic and eukaryotic RNA polymerases have homologous core subunits. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1192–1196. doi: 10.1073/pnas.84.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjian R., Losick R., Pero J., Hinnebush A. Purification and comparative properties of the delta and sigma subunits of RNA polymerase from Bacillus subtilis. Eur J Biochem. 1977 Mar 15;74(1):149–154. doi: 10.1111/j.1432-1033.1977.tb11376.x. [DOI] [PubMed] [Google Scholar]
- Traglia H. M., Atkinson N. S., Hopper A. K. Structural and functional analyses of Saccharomyces cerevisiae wild-type and mutant RNA1 genes. Mol Cell Biol. 1989 Jul;9(7):2989–2999. doi: 10.1128/mcb.9.7.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B. Mapping chromosomal genes of Saccharomyces cerevisiae using an improved genetic mapping method. Genetics. 1979 Jul;92(3):803–821. doi: 10.1093/genetics/92.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woychik N. A., Young R. A. RNA polymerase II subunit RPB4 is essential for high- and low-temperature yeast cell growth. Mol Cell Biol. 1989 Jul;9(7):2854–2859. doi: 10.1128/mcb.9.7.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi M., Nomura M. Cloning and sequence determination of the gene encoding the largest subunit of the fission yeast Schizosaccharomyces pombe RNA polymerase I. Gene. 1988 Dec 30;74(2):503–515. doi: 10.1016/0378-1119(88)90183-7. [DOI] [PubMed] [Google Scholar]