Abstract
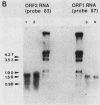
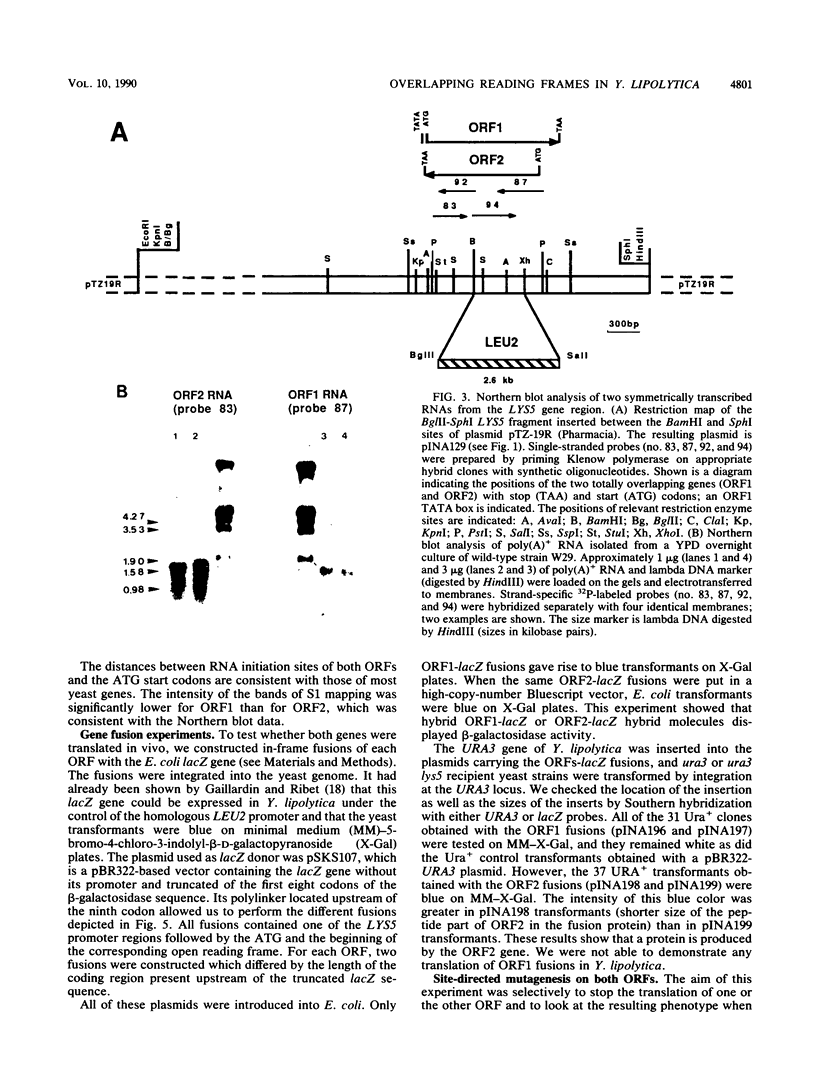
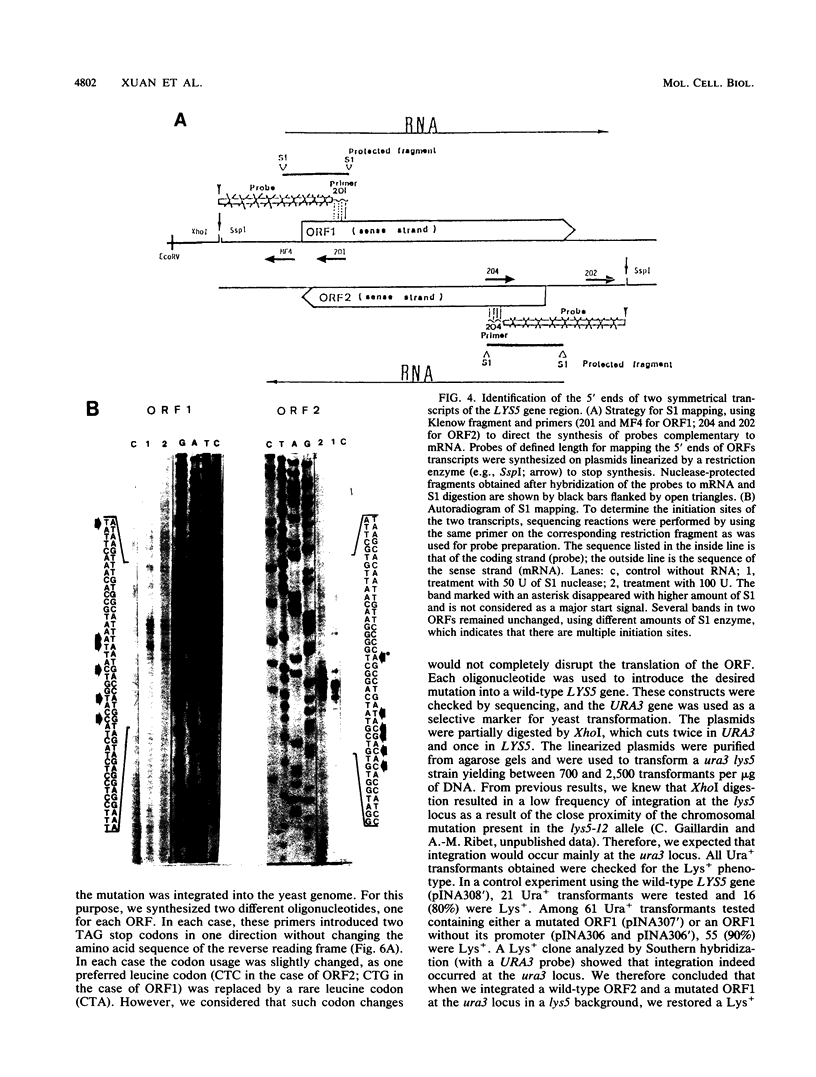
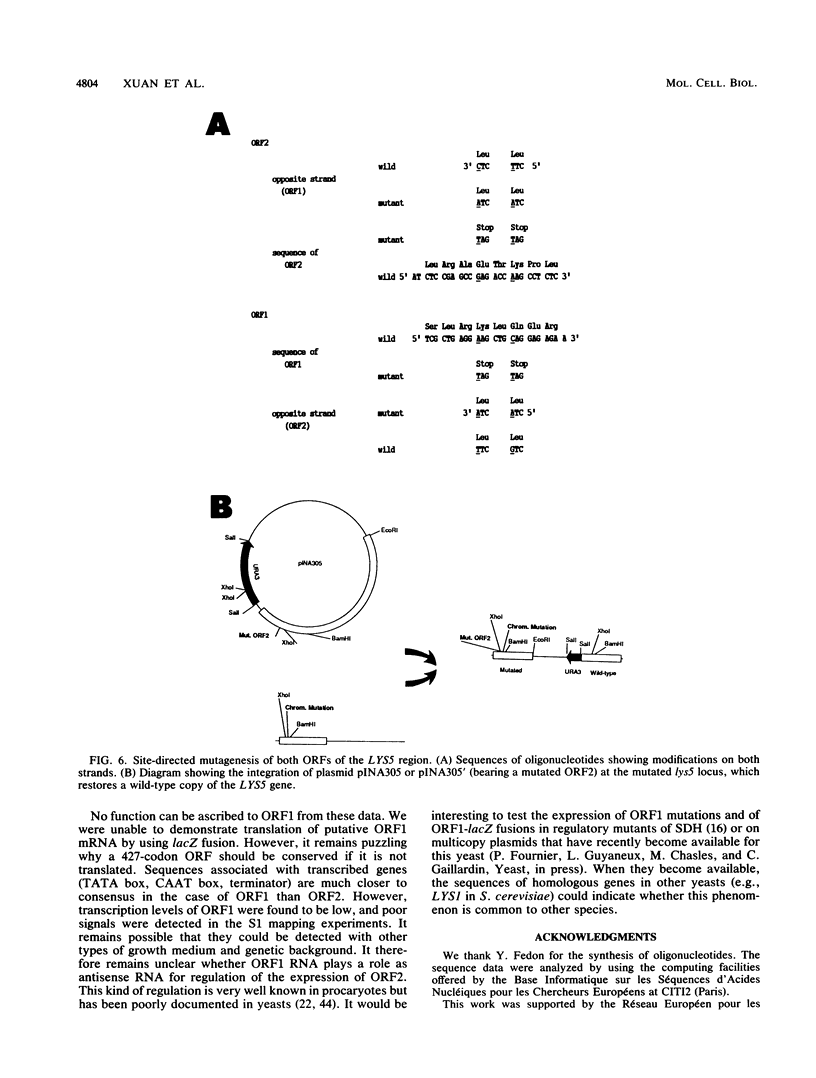
Mutants affected at the LYS5 locus of Yarrowia lipolytica lack detectable dehydrogenase (SDH) activity. The LYS5 gene has previously been cloned, and we present here the sequence of the 2.5-kilobase-pair (kb) DNA fragment complementing the lys5 mutation. Two large antiparallel open reading frames (ORF1 and ORF2) were observed, flanked by potential transcription signals. Both ORFs appear to be transcribed, but several lines of evidence suggest that only ORF2 is translated and encodes SDH. (i) The global amino acid compositions of Saccharomyces cerevisiae SDH and of the putative ORF2 product are similar and that of ORF1 is dissimilar. (ii) An in-frame translational fusion of ORF2 with the Escherichia coli lacZ gene was introduced into yeast cells and resulted in a beta-galactosidase activity regulated similarly to SDH; no beta-galactosidase activity was obtained with an in-frame fusion of ORF1 with lacZ. (iii) The introduction of a stop codon at the beginning of ORF2 prevented SDH expression in yeast cells, whereas no phenotypic effect was observed when ORF1 translation was blocked.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adelman J. P., Bond C. T., Douglass J., Herbert E. Two mammalian genes transcribed from opposite strands of the same DNA locus. Science. 1987 Mar 20;235(4795):1514–1517. doi: 10.1126/science.3547652. [DOI] [PubMed] [Google Scholar]
- Arndt K., Fink G. R. GCN4 protein, a positive transcription factor in yeast, binds general control promoters at all 5' TGACTC 3' sequences. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8516–8520. doi: 10.1073/pnas.83.22.8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrell B. G., Air G. M., Hutchison C. A., 3rd Overlapping genes in bacteriophage phiX174. Nature. 1976 Nov 4;264(5581):34–41. doi: 10.1038/264034a0. [DOI] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Bhattacharjee J. K. alpha-Aminoadipate pathway for the biosynthesis of lysine in lower eukaryotes. Crit Rev Microbiol. 1985;12(2):131–151. doi: 10.3109/10408418509104427. [DOI] [PubMed] [Google Scholar]
- Bost K. L., Smith E. M., Blalock J. E. Regions of complementarity between the messenger RNAs for epidermal growth factor, transferrin, interleukin-2 and their respective receptors. Biochem Biophys Res Commun. 1985 May 16;128(3):1373–1380. doi: 10.1016/0006-291x(85)91092-7. [DOI] [PubMed] [Google Scholar]
- Bost K. L., Smith E. M., Blalock J. E. Similarity between the corticotropin (ACTH) receptor and a peptide encoded by an RNA that is complementary to ACTH mRNA. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1372–1375. doi: 10.1073/pnas.82.5.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Carter P., Bedouelle H., Winter G. Improved oligonucleotide site-directed mutagenesis using M13 vectors. Nucleic Acids Res. 1985 Jun 25;13(12):4431–4443. doi: 10.1093/nar/13.12.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Davidow L. S., Kaczmarek F. S., DeZeeuw J. R., Conlon S. W., Lauth M. R., Pereira D. A., Franke A. E. The Yarrowia lipolytica LEU2 gene. Curr Genet. 1987;11(5):377–383. doi: 10.1007/BF00378180. [DOI] [PubMed] [Google Scholar]
- Davidow L. S., O'Donnell M. M., Kaczmarek F. S., Pereira D. A., DeZeeuw J. R., Franke A. E. Cloning and sequencing of the alkaline extracellular protease gene of Yarrowia lipolytica. J Bacteriol. 1987 Oct;169(10):4621–4629. doi: 10.1128/jb.169.10.4621-4629.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson M. J., Tuite M. F., Roberts N. A., Kingsman A. J., Kingsman S. M., Perkins R. E., Conroy S. C., Fothergill L. A. Conservation of high efficiency promoter sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 1982 Apr 24;10(8):2625–2637. doi: 10.1093/nar/10.8.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson J., Ho J. Y., Peterson T. A., Reed S. I. Nucleotide sequence of the yeast cell division cycle start genes CDC28, CDC36, CDC37, and CDC39, and a structural analysis of the predicted products. Nucleic Acids Res. 1986 Aug 26;14(16):6681–6697. doi: 10.1093/nar/14.16.6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillardin C., Fournier P., Sylvestre G., Heslot H. Mutants of Saccharomycopsis lipolytica defective in lysine catabolism. J Bacteriol. 1976 Jan;125(1):48–57. doi: 10.1128/jb.125.1.48-57.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillardin C., Ribet A. M. LEU2 directed expression of beta-galactosidase activity and phleomycin resistance in Yarrowia lipolytica. Curr Genet. 1987;11(5):369–375. doi: 10.1007/BF00378179. [DOI] [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Green P. J., Pines O., Inouye M. The role of antisense RNA in gene regulation. Annu Rev Biochem. 1986;55:569–597. doi: 10.1146/annurev.bi.55.070186.003033. [DOI] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters: positive and negative elements. Cell. 1984 Apr;36(4):799–800. doi: 10.1016/0092-8674(84)90028-x. [DOI] [PubMed] [Google Scholar]
- Görcs T. J., Gottschall P. E., Coy D. H., Arimura A. Possible recognition of the GnRH receptor by an antiserum against a peptide encoded by nucleotide sequence complementary to mRNA of a GnRH precursor peptide. Peptides. 1986 Nov-Dec;7(6):1137–1145. doi: 10.1016/0196-9781(86)90144-0. [DOI] [PubMed] [Google Scholar]
- Hahn S., Pinkham J., Wei R., Miller R., Guarente L. The HAP3 regulatory locus of Saccharomyces cerevisiae encodes divergent overlapping transcripts. Mol Cell Biol. 1988 Feb;8(2):655–663. doi: 10.1128/mcb.8.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Keene M. A., Fechtel K., Fristrom J. W. Gene within a gene: nested Drosophila genes encode unrelated proteins on opposite DNA strands. Cell. 1986 Jan 17;44(1):33–42. doi: 10.1016/0092-8674(86)90482-4. [DOI] [PubMed] [Google Scholar]
- Kim S., Mellor J., Kingsman A. J., Kingsman S. M. Multiple control elements in the TRP1 promoter of Saccharomyces cerevisiae. Mol Cell Biol. 1986 Dec;6(12):4251–4258. doi: 10.1128/mcb.6.12.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986 Jan 31;44(2):283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Kramer B., Kramer W., Fritz H. J. Different base/base mismatches are corrected with different efficiencies by the methyl-directed DNA mismatch-repair system of E. coli. Cell. 1984 Oct;38(3):879–887. doi: 10.1016/0092-8674(84)90283-6. [DOI] [PubMed] [Google Scholar]
- Kreike J., Schulze M., Ahne F., Lang B. F. A yeast nuclear gene, MRS1, involved in mitochondrial RNA splicing: nucleotide sequence and mutational analysis of two overlapping open reading frames on opposite strands. EMBO J. 1987 Jul;6(7):2123–2129. doi: 10.1002/j.1460-2075.1987.tb02479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normark S., Bergström S., Edlund T., Grundström T., Jaurin B., Lindberg F. P., Olsson O. Overlapping genes. Annu Rev Genet. 1983;17:499–525. doi: 10.1146/annurev.ge.17.120183.002435. [DOI] [PubMed] [Google Scholar]
- Ogawa H., Fujioka M. Purification and characterization of saccharopine dehydrogenase from baker's yeast. J Biol Chem. 1978 May 25;253(10):3666–3670. [PubMed] [Google Scholar]
- Ramos F., Dubois E., Piérard A. Control of enzyme synthesis in the lysine biosynthetic pathway of Saccharomyces cerevisiae. Evidence for a regulatory role of gene LYS14. Eur J Biochem. 1988 Jan 15;171(1-2):171–176. doi: 10.1111/j.1432-1033.1988.tb13773.x. [DOI] [PubMed] [Google Scholar]
- Rossmann M. G., Moras D., Olsen K. W. Chemical and biological evolution of nucleotide-binding protein. Nature. 1974 Jul 19;250(463):194–199. doi: 10.1038/250194a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira S. K., Chou J., Richaud F. V., Casadaban M. J. New versatile plasmid vectors for expression of hybrid proteins coded by a cloned gene fused to lacZ gene sequences encoding an enzymatically active carboxy-terminal portion of beta-galactosidase. Gene. 1983 Nov;25(1):71–82. doi: 10.1016/0378-1119(83)90169-5. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Tuohy T. M., Mosurski K. R. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986 Jul 11;14(13):5125–5143. doi: 10.1093/nar/14.13.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer C. A., Gietz R. D., Hodgetts R. B. Overlapping transcription units in the dopa decarboxylase region of Drosophila. Nature. 1986 Jul 17;322(6076):279–281. doi: 10.1038/322279a0. [DOI] [PubMed] [Google Scholar]
- Wierenga R. K., Terpstra P., Hol W. G. Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol. 1986 Jan 5;187(1):101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- Xiao W., Rank G. H. Generation of an ilv bradytrophic phenocopy in yeast by antisense RNA. Curr Genet. 1988 Apr;13(4):283–289. doi: 10.1007/BF00424421. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]