Background: Subcellular localization of splicing factor Tra2β is closely related to its function.
Results: NLSs in RS domains are required for Tra2β nuclear localization, while serine phosphorylation of the NLSs promotes Tra2β cytoplasmic accumulation.
Conclusion: Serine phosphorylation has a competitive effect against the NLS directed-nuclear location of Tra2β.
Significance: New insight into the molecular basis for phosphorylation-regulated Tra2β subcellular localization.
Keywords: Molecular Biology, Nuclear Transport, Phosphorylation, Serine, Subcellular Fractionation, NLSs, Tra2β, Nuclear Speckles, Subcellular Localization
Abstract
The serine/arginine-rich (SR) proteins are one type of major actors in regulation of pre-mRNA splicing. Their functions are closely related to the intracellular spatial organization. The RS domain and phosphorylation status of SR proteins are two critical factors in determining the subcellular distribution. Mammalian Transformer-2β (Tra2β) protein, a member of SR proteins, is known to play multiple important roles in development and diseases. In the present study, we characterized the subcellular and subnuclear localization of Tra2β protein and its related mechanisms. The results demonstrated that in the brain the nuclear and cytoplasmic localization of Tra2β were correlated with its phosphorylation status. Using deletional mutation analysis, we showed that the nuclear localization of Tra2β was determined by multiple nuclear localization signals (NLSs) in the RS domains. The point-mutation analysis disclosed that phosphorylation of serine residues in the NLSs inhibited the function of NLS in directing Tra2β to the nucleus. In addition, we identified at least two nuclear speckle localization signals within the RS1 domain, but not in the RS2 domain. The nuclear speckle localization signals determined the localization of RS1 domain-contained proteins to the nuclear speckle. The function of the signals did not depend on the presence of serine residues. The results provide new insight into the mechanisms by which the subcellular and subnuclear localization of Tra2β proteins are regulated.
Introduction
In eukaryotes, the expression of most genes is regulated by alternative pre-mRNA splicing, through which introns are selectively removed to generate multiple transcript variants from a single gene (1, 2). The importance of splicing is underlined by the increasing number of diseases associated with mis-splicing (3).
The alternative pre-mRNA splicing is highly regulated by a multitude of RNA cis-acting elements and many trans-acting protein factors. Serine/arginine-rich (SR)3 proteins are described as a highly conserved family of essential splicing factors. These proteins contain at least one RNA recognition motifs (RRMs) at the N terminus and an arginine/serine-rich (RS) domain at the C terminus. The RRMs bind to RNA sequence in a coordinated pattern to determine splicing specificity and commit pre-mRNA substrates to the splicing pathway. The RS domains contain multiple RS dipeptide repeats and mediate specific protein-protein interactions in a number of spliceosomal assembly steps (4, 5).
SR proteins are nuclear phosphoproteins. The spatial organization is closely related to their functions. These proteins are concentrated, together with other splicing factors, in nuclear subregions termed speckles. The speckles correspond to interchromatin granule clusters (IGCs), where are the sites of storage or assembly of splicing factors (6). The nuclear organization of splicing factors is dynamic, as shown by the fact that inhibition of transcriptional and/or splicing activity causes reorganization of the nuclear speckles (7). SR proteins and other splicing factors are recruited from the IGCs to the perichromatin fibrils (PFs) where active transcription and splicing occur (8, 9). Because SR proteins are known to affect splice site selection in a concentration-dependent manner, such dynamic spatial organization of splicing factors among different nuclear pools may provide a mechanism to regulate alternative splicing. Indeed, it was demonstrated recently that alternative splicing in vivo can be modulated by decreased nuclear SR protein levels (10). In addition, several SR proteins (ASF/SF2, 9G8, and SRp20), were reported to continuously shuttled between the nucleus and the cytoplasm (11), playing coordinated roles in multiple post-transcriptional events (12–14).
The role of RS domains in directing the nuclear and subnuclear localization may vary among different SR proteins. The RS domain of several SR proteins, SC35, SRp20, and Drosophila Transformer, has been shown to serve as both a nuclear localization signal (NLS) and subnuclear localization signal (15, 16). Generally, the reversible phosphorylation at multiple serine residues within the RS domain affects the subnuclear distribution of SR proteins (17–20). However, there is exception. For example, the RS domain of SF2/ASF is neither necessary nor sufficient for targeting to the nuclear speckles, although it acts as a nuclear localization signal (21). Thus, the precise structural basis for RS domains and the role of its phosphorylation status in determining the intracellular and subnuclear distribution remain to be characterized for each SR protein.
The mammalian transformer-2β (Tra2β) belongs to the SR-like protein family and has an RRM and two RS domains. One RS domain is located at the N terminus and the other at the C terminus, separated by an RRM (22, 23). Tra2β is highly expressed in brain tissues and subject to developmental regulation in a tissue- and temporal-specific pattern (24). Tra2β controls the pre-mRNA splicing of the survival motor neuron (SMN) and tau genes (25–28). Aberrant splicing of the genes is related to spinal muscular atrophy (SMA) and frontotemporal dementia (FTD), respectively. Besides in the central nervous system, the abnormal splicing events elicited by dysfunction of Tra2β have also been observed in cancer (29), stroke (30) and vascular smooth muscle diversification (31).
In contrast to the importance of Tra2β in the diseases associated with mis-splicing, the precise mechanisms underlying the Tra2β nuclear function are poorly understood. To address this issue, this study characterized the structure and phosphorylation of the RS domains of Tra2β and their functions in the nuclear and nuclear speckle localization.
EXPERIMENTAL PROCEDURES
Preparation of Expression Plasmids
To express the GFP-fused Tra2β, cDNA fragments encoding the full-length human Tra2β protein (NM_004593, 122–988 nt) were inserted into the pEGFP-C2 vector, in which transcription is driven by the CMV promoter and the coding sequence was in frame with the C-terminal of GFP. We used mutagenesis kit (TOYOBO) to create various GFP-fused Tra2β truncations and mutations. PCR products were completely sequenced, and all chimeric protein cDNAs were sequenced at the junction sites to confirm in-frame ligations.
Cell Culture and DNA Transfection
Human neuroblastoma SH-SY5Y cells were grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal calf serum at 37 °C with 5% CO2. Transfection was performed using the FuGENE HD Transfection Reagent (Roche) following the supplier's protocol. Briefly, cells were transfected with 0.2 μg of plasmid DNA per well of 24-well plate (60–80% confluent cells), in the presence of 0.6 μl of FuGENE HD Reagent.
Indirect Cell Immunofluorescence
Cells grown on coverslips were fixed for immunofluorescence assays between 10 and 12 h after transfection to prevent the formation of aggregates. The cells were washed with phosphate-buffered saline (PBS) and incubated with 4% paraformaldehyde for 30 min, followed by incubation for 5 min in 0.3% Triton X-100 (in PBS) to permeabilize the cells. The fixed cells were incubated in blocking buffer (5% BSA) for 1 h at room temperature, followed by incubation with anti-SC35 monoclonal antibody (1:2000, Sigma), washed three times with PBS, and incubated for 1 h at room temperature with Alexa Fluor 568-conjugated donkey anti-mouse IgG (1:500, Invitrogen). After DAPI (4′,6-diamidino-2-phenylindole) staining, the coverslips were mounted onto glass slides using fluoromount medium (Sigma). Fluorescence was detected by confocal laser-scanning microscopy (TCS SP2; Leica, Mannheim, Germany). To get more clear and detailed images of speckle pattern in the subnuclear foci, we lowered the brightness of the photos and the weaker signals in the cytoplasm became invisible in these photos.
Immunohistochemical Staining and Fluorescence Immunolabeling
For localization of endogenous Tra2β protein, C57BL/6 mice were deeply anesthetized. Then intracardial perfusion was performed with saline followed by 4% paraformaldehyde. The brains were removed and coronal sections were cut with a freezing microtome (Leica) at a thickness of 30 μm. Sections were preincubated with 0.3% H2O2 for 30 min, followed by incubation in blocking buffer containing 5% BSA and 0.3% Triton X-100 in PBS for 1 h at 37 °C, then incubated overnight at 4 °C with rabbit anti-Tra2β (1:500, Sigma). After three washes in PBS, the sections were incubated with biotinylated secondary antibodies (1:200, Vector Laboratories) for 45 min at 37 °C, followed by avidin-biotin-peroxidase (1:200, Vectastain Elite ABC kit, Vector Laboratories) for 30 min at 37 °C. Immunoreactivity was visualized with 0.05% diaminobenzidine (DAB) (Sigma). After washing, the sections were conterstained with hematoxylin. Negative controls received the same treatment except that the primary antibodies were omitted, and showed no specific staining.
For immunofluorescence staining, the sections were incubated with rabbit anti Tra2β overnight at 4 °C. Sections were then incubated with Alexa Fluor 568-conjugated donkey anti-rabbit IgG (1:500, Invitrogen) at 37 °C for 1 h to reveal the positive signals. After washing, the sections were counterstained with DAPI and mounted on glass slides and coverslipped using fluoromount medium.
Protein Dephosphorylation
Brain tissues or cells were lysed in RIPA buffer containing protease inhibitors. Protein (200 μg) extracts were incubated with 50 units of calf intestinal alkaline phosphatase (CIP; New England Biolabs) for 30 min at 37 °C in phosphatase buffer (50 mm Tris-HCl, pH 7.9, 10 mm MgCl2, 100 mm NaCl, and 1 mm DTT). The reaction was stopped by the addition of SDS sample buffer, and the proteins were resolved by SDS-PAGE. The protein extracts incubated for 30 min at 37 °C in the same buffer without the CIP was used as the negative control (CIP-). For the mock control (mock), brain tissues or cells were lysed in RIPA buffer containing both protease inhibitor and phosphatase inhibitor, but not treated in phosphatase buffer at 37 °C.
Cytoplasmic and Nuclear Protein Extraction
Nuclear and cytoplasmic fractions of brain tissues or cells were separated following the procedure described in the Cytoplasmic and Nuclear Protein Extraction Kit (Fermentas). Briefly, fresh tissue samples or harvested SH-SY5Y cells were homogenized gently in PBS, followed by centrifugation at 250 × g for 5 min. The remaining cells were resuspended in cell lysis buffer and incubated on ice for 10 min. Cells were lysed by gentle pipetting, and the cytoplasmic fraction was separated from intact nuclei by centrifugation at 500 × g for 7 min. Cytoplasmic fractions were centrifuged at 20,000 × g for 15 min, and the supernatant was collected and stored. Nuclei were washed three times, and the pellets were collected.
Western Blot Analysis
Protein samples were separated by 12% SDS-PAGE for further Western blot analysis as described previously (32). Rabbit anti Tra2β (1:500, Sigma), monoclonal mouse anti-GFP (1:5000, Invitrogen), mouse anti-TBP (1:500, Abcam), mouse anti-β-actin (1:5000, Sigma), mouse anti-GAPDH (1:5000, Kangcheng, Shanghai) antibodies were used as primary antibodies and horseradish peroxidase (HRP)- or alkaline phosphatase (AP)-conjugated goat anti-rabbit or horse anti-mouse IgG (1:1000, Kangcheng, Shanghai) as secondary antibodies. HRP signals were developed by using enhanced chemiluminescence (ECL) reagent and exposure to x-ray film. AP signals were developed by using 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium (BCIP/NBT) substrates. Different exposure time or staining time was used for each membrane to avoid overexposure or overstaining of the bands. The image analysis was performed using Bio-Rad Image Instrument with Multi-Analyst software.
Morphology Analysis and Data Quantification
All experiments were repeated at least three times independently. For measurement of the percentage of GFP fusion proteins within the nuclei, GFP signal total intensity (mean intensity × total area) was analyzed in ImageProPlus software. The relative nuclear GFP signal intensity was calculated by normalizing the GFP signal colocalized with DAPI (nuclei) to the total GFP signal in the whole cell. Data were expressed as means ± S.E. and statistical significance was determined by one-way ANOVA.
RESULTS
Phosphorylation Status Is Correlated with Nuclear/Cytoplasmic Localization of Tra2β in the Brain
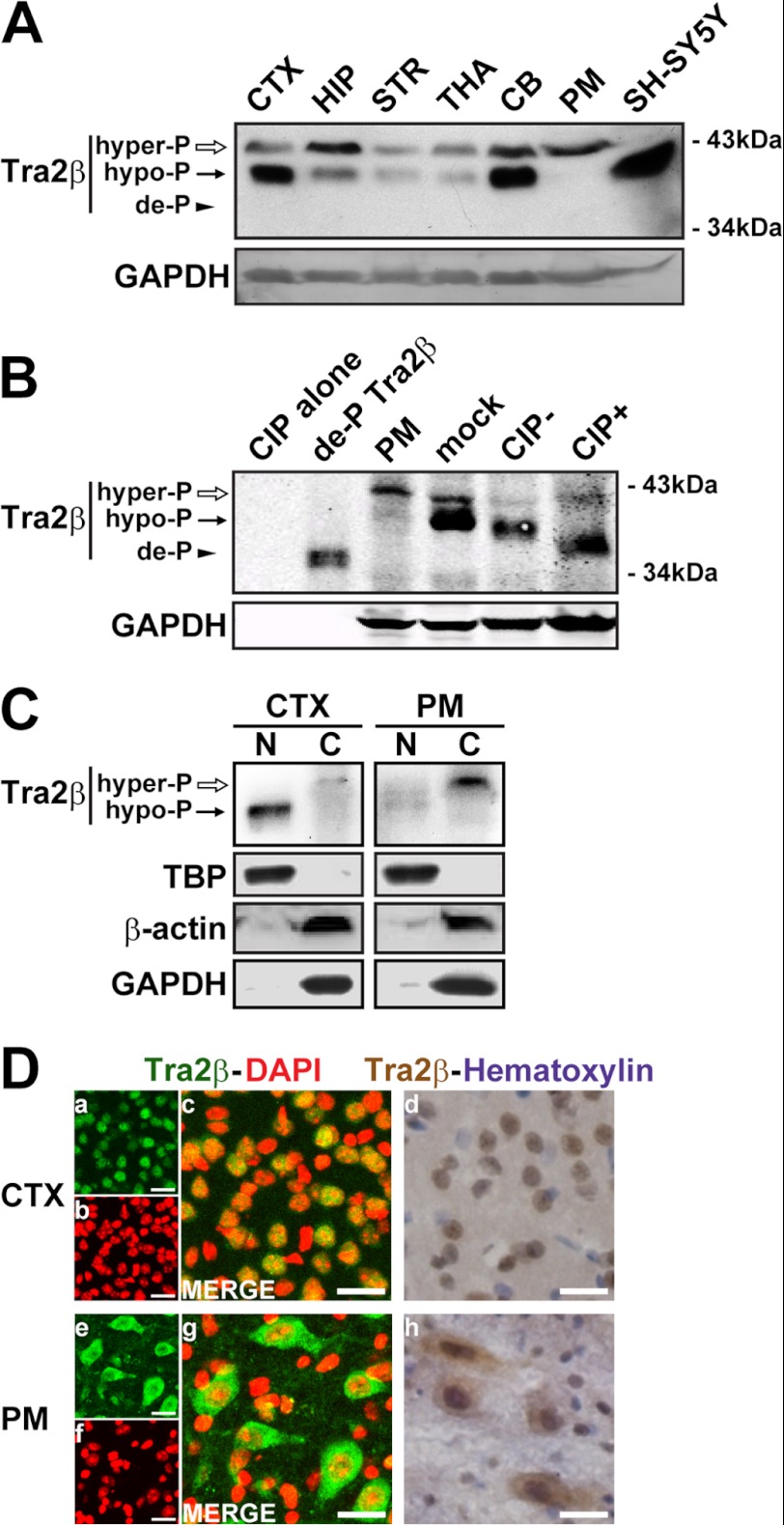
The transcripts of Tra2β gene are expressed ubiquitously in the brain (24, 33). However, the detailed localization of Tra2β proteins in the brain has not yet been clearly characterized. To address this issue, we used an antibody specific to Tra2β to investigate the expression pattern of Tra2β in various mouse brain regions, including the cerebral cortex (CTX), hippocampus (HIP), striatum (STR), thalamus (THA), cerebellum (CB), pons and medulla (PM). The specificity and efficiency of this commercial available antibody was proven by the Tra2β overexpression and RNAi assay (data not shown). As shown in Fig. 1A, two bands (MW 42 and 40 kDa, respectively) were found in almost all the investigated brain regions. Both bands do not match with predicted 35 kDa based on the amino acid residues derived from the Tra2β mRNA sequence. It indicates the post-translational modification of Tra2β in the brain, consistent with the reported previously. After calf intestinal alkaline phosphatase (CIP) treatment, the 42 kDa band became weaker, the 40 kDa band disappeared and a 35 kDa band corresponding to the de-phosphorylated Tra2β appeared (Fig. 1B). The CIP treatment indicated that the 42 kDa band was the Tra2β phosphorylated at higher extent (hyper-P), in relative to the 40 kDa band (hypo-P). Western blot analysis also showed that the ratio of the hyper-P and hypo-P Tra2β proteins varied in different brain regions (Fig. 1A). In the CTX, the hypo-P Tra2β was predominant, and detected mainly in the nuclear fraction. However, in the PM, Tra2β was predominantly hyper-P, and detected strongly in the cytoplasm fraction, almost invisible in the nuclear fraction (Fig. 1C). To further characterize the subcellular localization of Tra2β in different brain regions, the immunohistochemistry (IHC) and immunofluorescence (IF) analysis was performed on brain slices. As shown in Fig. 1D, Tra2β was localized predominantly in the nucleus in the CTX, while predominantly in the cytoplasm in the PM. These results suggest that the distinct subcellular localizations of Tra2β are likely correlated to its phosphorylation status in various brain regions.
FIGURE 1.
Expression, phosphorylation, and subcellular localization of Tra2β proteins in mouse brain. A, representative Western blot images showing the expression of Tra2β proteins in various brain regions. Total protein extracts of various brain regions were used. CTX: cerebral cortex, HIP: hippocampus, STR: striatum, THA: thalamus, CB: cerebellum, PM: pons and medulla. SH-SY5Y: SH-SY5Y cells. GAPDH was used as the loading control. B, total protein extracts of mouse brain with (+) or without (−) CIP treatment were analyzed. For CIP+, protein extracts of CTX were treated with CIP in 37 °C for 30 min; For CIP−, protein extracts of CTX were treated without CIP in 37 °C for 30 min; mock, protein extracts of CTX without treatment in 37 °C; CIP alone, phosphatase reaction buffer and CIP alone, without protein extracts; de-P Tra2β, the prokaryotic expressed and purified GST-Tra2β fusion protein in which the GST tag was cut off by thrombin, was used as de-P Tra2β positive control; PM, protein extracts from pons and medulla, as hyper-P Tra2β positive control. The hyper-phosphorylated (hyper-P), hypo-phosphorylated (hypo-P), and de-phorsphorylated (de-P) Tra2β protein bands were indicated by white arrows, black arrows, and arrowhead, respectively. GAPDH was used as the loading control. C, Western blot images of Tra2β protein in the nuclear (N) and cytoplasmic (C) fractions of the CTX and PM. TBP (TATA box-binding protein) was used as the loading control of nuclear proteins, β-actin and GAPDH were used as the loading controls of cytoplasmic proteins. D, left: confocal microscopy showing immunofluorescent signals of Tra2β (green) and DAPI (red) in the CTX and PM (a–c, e–g). Right: IHC staining showing signals of Tra2β (brown) and hematoxylin-counterstaining (violet) in the CTX and PM (d and h). Scale bars: 20 μm.
Either RS Domain Is Necessary and Sufficient for the Nuclear Localization of Tra2β
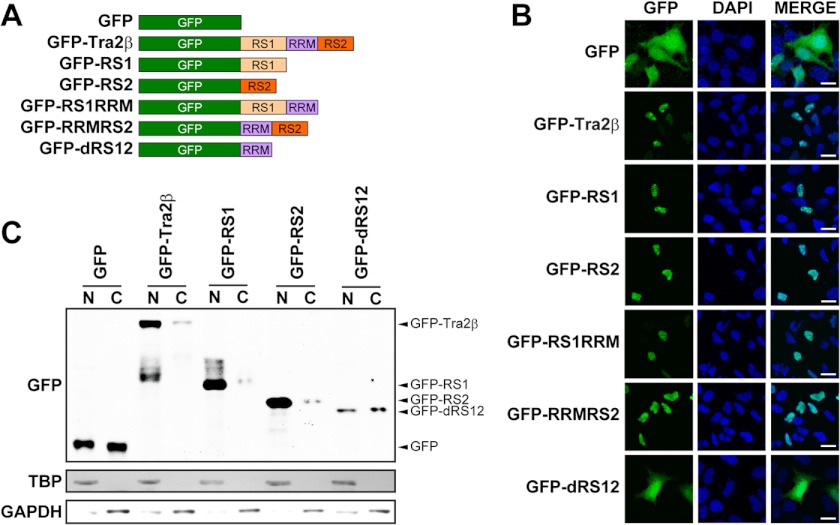
Tra2β contains two RS domains, the N-terminal RS1 domain and the C-terminal RS2 domain, and an RNA recognition motif (RRM). To study the role of RS domains in the subcellular distribution of Tra2β, we prepared several DNA constructs to express the green fluorescent protein (GFP) containing the full-length of Tra2β protein or various Tra2β domains at the C-terminal (Fig. 2A). A human neuroblastoma SH-SY5Y cell line was used for transient overexpression of these fusion proteins, because our preliminary experiments showed that in SH-SY5Y cells Tra2β was predominantly hypo-p (Fig. 1A) and localized exclusively in the nucleus (data not shown). As shown in Fig. 2B, the GFP-Tra2β fusion protein was detected exclusively in the nucleus, while GFP alone did not localize to a specific compartment and was detected in both the cytoplasm and nucleus, indicating that GFP did not affect the nuclear localization of Tra2β. Fig. 2 showed that all the fusion proteins containing either RS1 or RS2 domain were localized in the nucleus, regardless of the presence of RRM (GFP-RS1RRM & GFP-RRMRS2) or not (GFP-RS1 & GFP-RS2). Meanwhile, the GFP-dRS12 fusion protein containing the RRM only, lacking both RS1 and RS2 domains, was detected in both the cytoplasm and nucleus. The effect of RS and RRM domains on nuclear/cytoplasmic localization of Tra2β in COS-1 cells gave similar results (as shown in Fig. 4). To further confirm these findings, subcellular fractionation of SH-SY5Y cells with the expression of different fusion proteins was carried out. The results showed that GFP-Tra2β, GFP-RS1, GFP-RS2 proteins were predominantly detected in the nuclear fraction. In comparison, GFP and GFP-dRS12 proteins were equally in the nuclear and cytoplasmic fraction (Fig. 2C). The results indicate that the RS domain directs the GFP protein into nucleus, while the RRM domain does not. In addition, either RS1 or RS2 domain is necessary and sufficient for the nuclear localization.
FIGURE 2.
Effects of RS and RRM domains on nuclear/cytoplasmic localization of Tra2β in SH-SY5Y cells. A, illustrations of the DNA construct to express the recombinant fusion proteins. The GFP reporter gene was fused in-frame with the full-length Tra2β (GFP-Tra2β), the N-terminal RS1 domain (GFP-RS1) or the C-terminal RS2 domain (GFP-RS2); or the full-length Tra2β lacking the RS2 domain (GFP-RS1RRM), lacking the RS1 domain (GFP-RRMRS2) or lacking both RS1 and RS2 domains (GFP-dRS12). B, fluorescent signals of GFP (or GFP fusion proteins) and nuclear staining (DAPI, blue) in the cells with expression of various GFP fusion proteins. Scale bars: 20 μm. C, Western blot images of the nuclear (N) and cytoplasmic (C) fractions of the SH-SY5Y cells with expression of various GFP fusion proteins. TBP was used as the loading control of nuclear proteins, and GAPDH was used as the loading control of cytoplasmic proteins.
FIGURE 4.
Effects of RS and RRM domains, NLSs, and phosphorylation on nuclear/cytoplasmic localization of Tra2β in COS-1 cells. Fluorescent signals of GFP (or GFP fusion proteins) and nuclear staining (DAPI, blue) in the COS-1 cells with expression of various GFP fusion proteins. Scale bars: 50 μm.
Nuclear Localization Signals (NLSs) in RS Domains Are Required for the Nuclear Localization of Tra2β
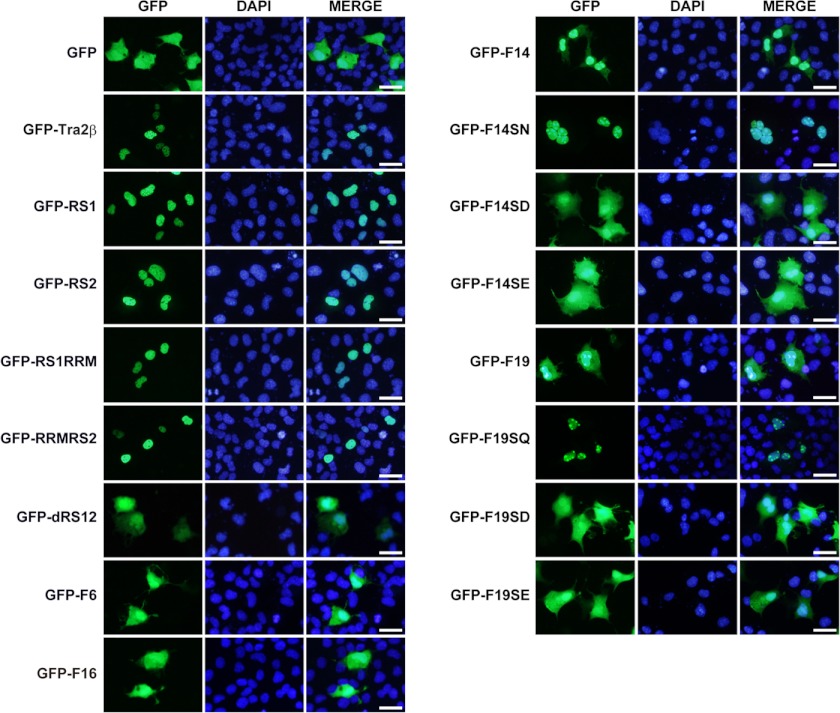
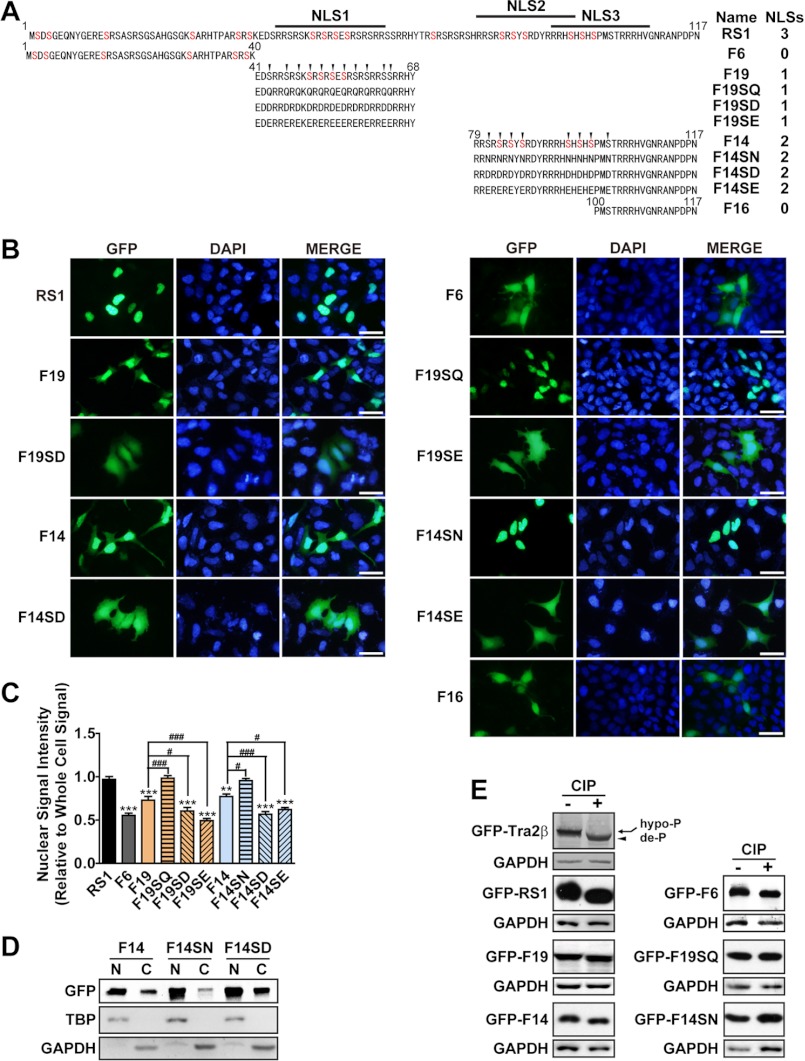
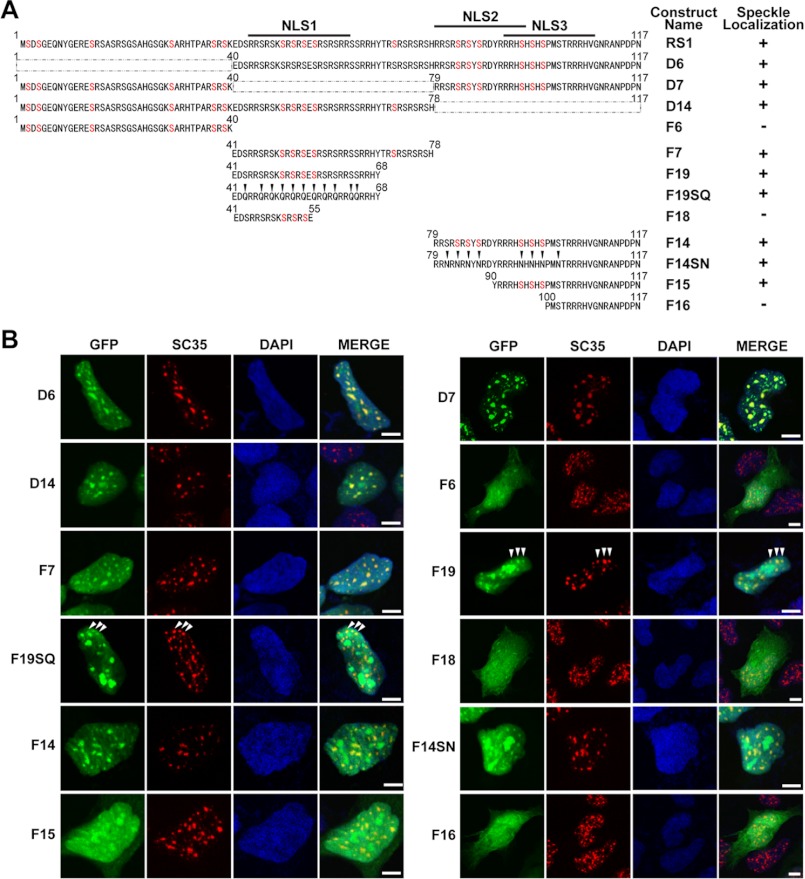
The RS domain contains multiple serine-arginine dipeptide repeats. The arginine clusters usually form NLSs (34, 35). The sequence analysis revealed three potential NLSs within the RS1. To determine the importance of these NLSs for the nuclear localization, a series of fragments of the RS1 domain were fused to GFP. As illustrated in Fig. 3A, the RS1 fragment contains the full-length RS1 domain; the F6 and F16 fragment contains no NLS; the F14 fragment contains two NLSs (NLSs 2 and 3) and the F19 fragment contains one NLS (NLS 1). The results in Fig. 3, B and C showed that the nuclear localization of GFP-F6 was ∼50%, much lower than that of GFP-RS1 (almost 100% localized in the nucleus). Similar to GFP-F6, GFP-F16 also had a dispersed distribution in the cytoplasm and nucleus. It indicates that the lacking of all three NLSs elicited the GFP-F6 and GFP-F16 to be distributed evenly between the nucleus and cytoplasm, losing the nuclear-specific localization. In contrast, the nuclear localization of GFP-F14 and GFP-F19 was higher than 70%, indicating the importance of NLSs for the nuclear localization of Tra2β.
FIGURE 3.
Effects of NLSs and phosphorylation on nuclear/cytoplasmic localization of Tra2β. A, illustrations of the DNA construct to express the GFP-RS1 truncations and mutants. The GFP reporter gene was fused in-frame with the full-length RS1 domain (GFP-RS1) or its serial truncations or mutants. The amino acid sequences of each construct were presented. Each construct was named as indicated in the Name column. The number of NLSs in each construct was indicated in the NLSs column. The serine residues which have been reported to be modified by phosphorylation in vivo were labeled in red. The F6 fragment contained 1–40aa of the RS1; the F19 fragment contained 41–68aa of the RS1; the F14 fragment contained 79–117aa of the RS1; The F16 fragment contained 100–117aa of the RS1. All the serine (S) of the F19 fragment were mutated into glutamine (Q), aspartic acid (D), or glutamic acid (E) in the F19SQ, F19SD, F19SE, respectively; all the serines (S) of the F14 fragment were mutated into asparagine (N), aspartic acid (D), or glutamic acid (E) in the F14SN, F14SD, F14SE, respectively. All the mutated serines were indicated by the arrows. B, fluorescent microscopy showing the localization of GFP-RS1 truncations and mutations in SH-SY5Y cells. DAPI (blue) was used for nuclear staining. Scale bars: 50 μm. C, statistic data showing the relative intensity of nuclear GFP signals to total GFP signals in the whole SH-SY5Y cell. Data were expressed in mean ± S.E. Significant differences were indicated: *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus RS1; #, p < 0.05; ##, p < 0.01; ###, p < 0.001 versus corresponding wild-type F19 or F14. D, fractionation of the nuclear and cytoplasmic extraction of F14, its non-phosphomimic mutant F14SN, and phosphomimic mutant F14SD. TBP and GAPDH were used as nuclear and cytoplasmic fraction loading controls, respectively. E, Western blot images verifying the phosphorylation status of the GFP-Tra2β protein or GFP-fused Tra2β fragments and their mutants in SH-SY5Y cells. The GFP-fused Tra2β protein or Tra2β fragments treated with (+) or without (−) CIP were shown. Shift of the band to lower MW after CIP treatment means that the fragment was phosphorylated and could be de-phosphorylated by CIP treatment in vitro. Anti-GFP antibody was used in Western blot. GAPDH was used as the loading control.
Phosphorylation of Serine Residues within the NLS Promotes the Cytoplasmic Localization of Tra2β
All SR proteins, including Tra2β, are heavily phosphorylated proteins that can be recognized by the monoclonal antibody mAb104 (36). Analysis showed that the RS domain of Tra2β is phosphorylated at multiple serine sites in vivo (37–41). As illustrated in Fig. 3A, F14 and F19 fragments have several serine residues which have been reported to be modified by phosphorylation in vivo (38, 39). To characterize the role of the phosphorylation of serine residues in the NLS-directed nuclear localization, all the serine residues within the F14 and F19 fragments were mutagenized to asparagine or glutamine (non-phosphomimic, F14SN, and F19SQ), or to aspartic acid and glutamic acid (phosphomimic, F14SD, F14SE, F19SD, and F19SE) (Fig. 3A).
As shown in Fig. 3, B and C, non-phosphomimic mutants GFP-F14SN showed a significant increase in nuclear localization in SH-SY5Y cells (almost 100% localized in the nucleus), compared with wild-type GFP-F14 (77% localized in the nucleus). In contrast, the phosphomimic mutants GFP-F14SD and GFP-F14SE were detected evenly in the nucleus and cytoplasm (∼50% localized in the nucleus). Similarly, non-phosphomimic mutants of F19 fragment also showed an exclusive nuclear localization (GFP-F19SQ), while phosphomimic mutants (GFP-F19SD and GFP-F19SE) showed an increase of cytoplasmic localization. Similar results were also observed in COS-1 cells, as shown in Fig. 4. To further confirm these findings, nuclear/cytoplasmic extraction was performed to separate proteins from these subcellular compartments. As shown in Fig. 3D, the cytoplasmic localization was significantly reduced by non-phosphomimic mutation in the GFP-F14 fragment (F14SN), while increased by phosphomimic mutation in the GFP-F14 fragment (F14SD). These results suggest that the serine and arginine residues within the RS repeats play different roles in the subcellular localization of Tra2β. The arginine clusters within the NLSs are required for the nuclear localization, while phosphorylation of serine residues enhances the cytoplasmic accumulation of Tra2β.
We also examined whether the serine residues are modified by phosphorylation. As shown in Fig. 3E, the relative mobility of these fusion proteins increased (GFP-Tra2β, GFP-RS1, GFP-F6, GFP-F14, GFP-F19) when they were treated with CIP. The level of increased mobility was related to the number of serine residues in each fragment and was eliminated by mutation of serine residues (GFP-F14SN, GFP-F19SQ). The results suggest that these wild-type fusion proteins are indeed phosphorylated at the serine residues in vivo.
RS1 Domain Determines the Nuclear Speckle Localization of Tra2β, But RS2 Does Not
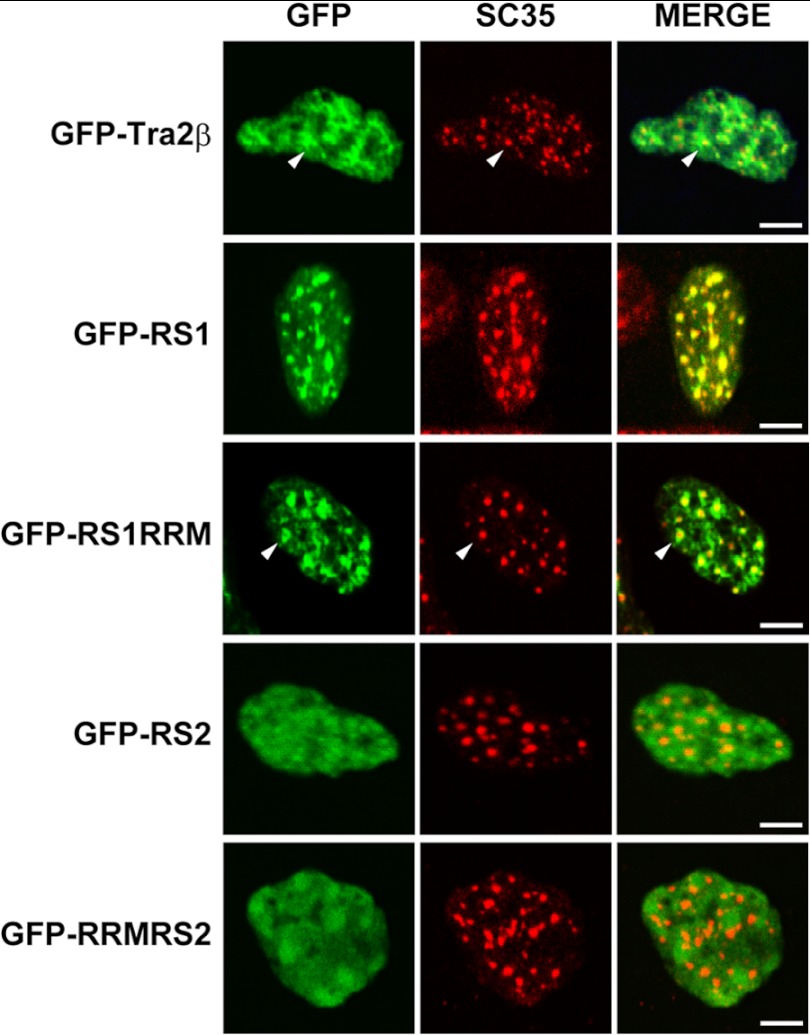
Tra2β is the mammalian homolog of Drosophila Tra protein. Unlike Drosophila Tra protein and other SR proteins that have only one RS domain, Tra2β has two RS domains at its N and C terminals, respectively (22, 23). The functional difference of the two RS domains is largely unknown. Studies have revealed that at steady states, a variety of SR proteins including Tra2β are localized in nuclear speckles. The speckles are the sites for storage and reassembly of splicing factors and supply the splicing factors to nearby PFs where active transcription and splicing occur (8, 9). In this study, using SC35 as a marker of nuclear speckles, we observed the effect of RS1 and RS2 domains on subnuclear distribution of Tra2β. As shown in Fig. 5, GFP-Tra2β and GFP-RS1RRM proteins were co-localized with SC35 in the nuclear speckles and also dispersed in the nucleoplasm. GFP-RS1 was highly concentrated within nuclear speckles. In comparison, GFP-RS2 and GFP-RRMRS2 had a diffused distribution throughout the nucleoplasm. The results indicate that the N-terminal RS1 domain was necessary and sufficient for the nuclear speckle localization of Tra2β. Studies in another series of constructs in which GFP were replaced by HA tags also showed that the RS1 domain determines the nuclear speckle localization of Tra2β protein, but RS2 domain does not (data not shown). Thus, the results suggest that although RS1 and RS2 domains both function as nuclear localization signals as demonstrated above, they have different abilities in directing Tra2β into subnuclear foci.
FIGURE 5.
Effects of RS1 and RS2 domains on subnuclear localization of Tra2β. Confocal microscopy images showing the immunofluorescent signals of GFP or nuclear speckle marker (SC35) and the overlay (MERGE) in the SH-SY5Y cells with expression of various GFP fusion proteins. Scale bars: 5 μm.
RS1 Contains at Least Two Sequences Determining the Nuclear Speckle Localization of Tra2β; Mutation of Serine Residues within the Sequences Does Not Affect the Localization
The specific sequence directing the nuclear speckle localization has been reported in Drosophila Tra protein. This sequence is located in the RS domain of Drosophila Tra protein, containing a nucleoplasmin-like bipartite NLS and a repeating arginine/serine dipeptide adjacent to a short stretch of basic amino acid (16). Through bioinformatic analysis, we identified several similar sequences in the RS1 domains of Tra2β protein. Thus, we proposed that RS1 domain possibly mediates the nuclear speckle localization of Tra2β protein through the same mechanism as Drosophila Tra protein. To dissect which sequence mediate the nuclear speckle localization of Tra2β, a fine mapping was carried out by preparing various DNA constructs to express the GFP-RS1 truncations and mutants, as illustrated in Fig. 6A. Results as shown in Fig. 6B demonstrated that the full-length RS1 domain, fragment F7 (aa 41–78) and F14 (aa 79–117) were co-localized with nuclear speckle marker SC35, while the fragment F6 (aa 1–40) was not. Deletion of aa 1–40 (D6), aa 41–78 (D7) or aa 79–117 (D14) did not disturb the nuclear speckle distribution of RS1. The results suggest that fragment D7 and D14 each has at least one speckle location signal. To identify the speckle location signal within the fragment aa 41–78 (F7), we compared the speckle localization of the fragments F7 (aa 41–78), F19 (aa 41–68), and F18 (aa 41–55). The results showed that both GFP-F7 and GFP-F19 were localized to the speckle, in contrast, GFP-F18 was not. It indicates that the sequence aa 56–68 likely has the signals for subnuclear localization of RS1 to speckle domains. Fig. 6 also showed that the GFP-F14 fragment (aa 79–117) and GFP-F15 fragment (aa 90–117) were distributed in nuclear speckles; a smaller fragment GFP-F16 (aa 100–117) was no longer localized to nuclear speckles. Thus, the sequence aa 90–99 was suggested to contain a motif required for subnuclear localization of RS1 to speckle domains.
FIGURE 6.
Nuclear speckle distribution patterns of GFP-RS1 truncations and mutants in SH-SY5Y cells. A, illustrations of GFP-RS1 truncations and mutants. The GFP reporter gene was fused in-frame with the full-length RS1 domain (GFP-RS1) or its serial truncations. The name of each construct was indicated in the Construct Name column. Localization to the nuclear speckles was scored positive in the Speckle Localization column when the staining signal of GFP-RS1 truncations and mutants was co-localized with that of SC35. The D6 fragment lacked 1–40aa of RS1; The D7 fragment lacked 42–78aa of RS1; D14 fragment lacked 79–117aa of RS1; F6 fragment contained 1–40aa of RS1; F7 fragment contained 41–78aa of RS1; F19 fragment contained 41–68aa of RS1; F18 fragment contained 41–55aa of RS1; F14 fragment contained 79–117aa of RS1; F15 fragment contained 90–117aa of RS1; F16 fragment contained 100–117aa of RS1. All the serines (S) of F19 fragment are mutated into glutamine (Q) in F19SQ; all the serines (S) of F14 fragment are mutated into asparagine (N) in F14SN. B, confocal microscopy showing immunofluorescent signals of GFP (green), SC35 (red), DAPI (blue), and MERGE (overlay) in cells with expression of GFP-RS1 truncations and mutants. Scale bars: 5 μm.
The similar results were also obtained by the experiments using a series of HA-tagged RS1 fragments (data not shown). Together, our results indicated that unlike Drosophila Tra protein containing only one copy of the nuclear speckle signal, Tra2β had at least two copies of this motif. These redundant signals implied a much tighter control of the subnuclear distribution of Tra2β than Drosophila Tra.
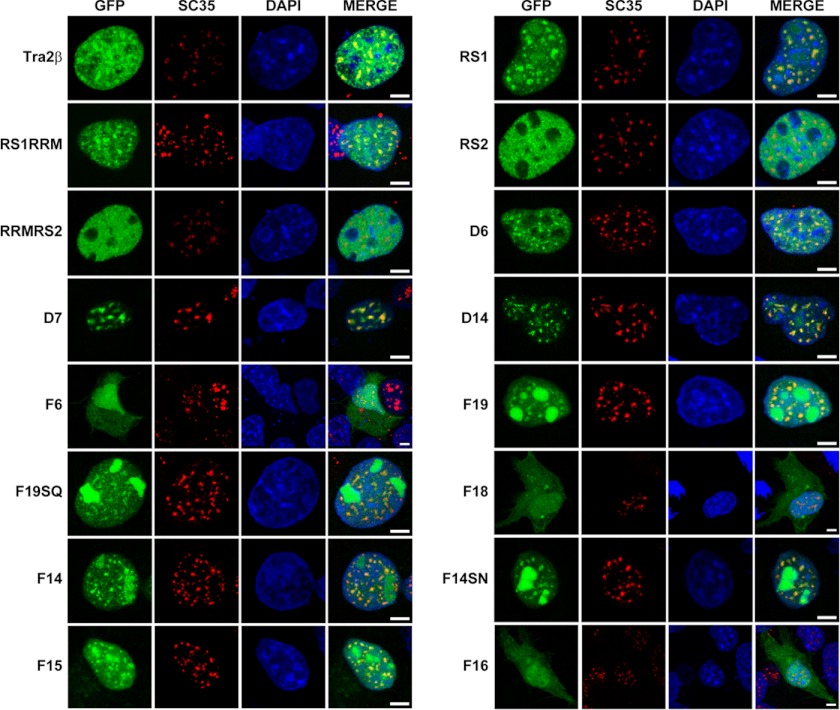
In the present study, we also examined the role of phosphorylation of serine residues in the speckle localization of the RS1 domain. As shown in Fig. 6B, when all serine residues within the nuclear speckle signals were mutated (GFP-F14SN and GFP-F19SQ), the co-localization of the fusion proteins with SC35 were still obvious, although with a very slight decrease, suggesting that elimination of phosphorylation on serine residues has no obvious effect on the function of nuclear speckle signals. Similar results were also obtained by the experiments in COS-1 cells (Fig. 7).
FIGURE 7.
Nuclear speckle distribution patterns of RS1, RS2 domain, truncations and mutants of GFP-RS1 in COS-1 cells. Confocal microscopy showing immunofluorescent signals of GFP (green), SC35 (red), DAPI (blue), and MERGE (overlay) in cells with expression of various GFP fusion proteins. Scale bars: 5 μm.
DISCUSSION
Tra2β is involved in multiple biological processes and various diseases. Characterization of Tra2β subcellular and subnuclear localization and its related mechanisms will provide a molecular basis for fully understanding the Tra2β-involved events. Here we disclosed that Tra2β protein was not localized exclusively in the nucleus, but also localized in the cytoplasm in many brain regions. It indicates a shuttling mechanism occurred in Tra2β subcellular distribution. SR protein shuttling plays multiple roles in facilitating mRNA transport across the nuclear pore, or having cytoplasmic functions, such as translational regulation, mRNA stability, and mRNA localization (11–14). The cytoplasmic localization of Tra2β may reflect a similar role possibly happened in its cytoplasmic functions. In addition, the regulation of nucleo-cytoplasmic shuttling of Tra2β may provide a mechanism for controlling the concentration of Tra2β in relative to other splicing factors in the nucleus. In this regard, it has been shown that the p38 kinase induced the translocation of the splicing repressor hnRNP A1 from the nucleus to the cytoplasm in response to osmotic stress, resulting in a shift in the splice site selection of the E1A reporter (42). In addition, in the ischemic brain, Tra2β was accumulated in cytoplasm and highly phosphorylated, accompanied with a change in the splice-site selection of its target mRNA (30). Thus, the Tra2β shuttling may provide a way to regulate its functional response to physiopathological stimuli by localization.
A nuclear localization signal or sequence (NLS) is an amino acid sequence which tags a protein for import into the cell nucleus by nuclear transport. Typically, this signal consists of one or more short sequences of positively charged lysines (K) or arginines (R) exposed on the protein surface. Well-studied examples of classical NLSs are the sequence K-K/R-X-K/R for monopartite NLSs of the SV40-Large T antigen (34), and the bipartite NLS of nucleoplasmin, which contains two basic clusters spaced by ∼10 less conserved residues (35). By bioinformatic analysis, we identified several nucleoplasmin-like NLSs located in the RS1 and RS2 domains. The experimental results demonstrated that these NLSs function as determining signals for nuclear localization of Tra2β proteins.
The presence of multiple NLS sequences in RS domain-containing splicing factors has been demonstrated (16). However, it remains unclear whether those redundant NLS sequences in native proteins function in a parallel or synergistic fashion. The present study demonstrated that the decrease in numbers of NLS did not leads to a looser control for nuclear import through comparing RS1 with F14SN or F19SQ fragment, indicating a parallel function of multiple NLSs of Tra2β.
Nuclear import of proteins is generally mediated by transport receptors that recognize specific NLSs. Two human importin β family proteins, transportin-SR1 (TRN-SR1) and transportin-SR2 (TRN-SR2) have been shown to mediate nuclear import of the SR proteins via the RS domain (43, 44). The nuclear import of SR proteins is usually directed by TRN-SR1 and TRN-SR2 in a phosphorylation-dependent manner (45, 46). As an exception, the import of Tra2β by TRN-SR1 is reportedly not depended on phosphorylation (46). Our results provide new experimental evidence to support the report, showing that the NLS-contained GFP-F14SN and GFP-F19SQ fragments can be imported into the nucleus even if phosphorylation were blocked by mutations of serine residues.
In addition, we showed that phosphorylation of serine residues in RS domains promotes cytosolic localization of Tra2β, although the NLS -directed nuclear import of Tra2β is phosphorylation independent. It is suggested that phosphorylation of serine residues may provide a balancing mechanism against the NLS-directed nuclear localization. The balance between the number of NLS and phosphorylation likely determines the subcellular localization of Tra2β proteins in vivo. The less the phosphorylation, the more proteins are localized in the nuclear and the less in the cytoplasm. It implies that the Tra2β concentration in relative to other SR proteins in the nucleus could be determined by the net effects of kinase and phosphatase activities in the cell. This mechanism allows cells to control the relative nuclear concentrations of Tra2β proteins, thereby regulating the gene expression at the splicing level in response to signaling.
We also demonstrated that localization of Tra2β to nuclear speckles depended upon the RS1 domain, not the RS2 domain. The functional importance of RS1 domain in spatial organization is supported by a recent finding that removal of the RS1 domain completely disabled Tra2β-mediated splicing activation of the physiological target exons (47). In addition, we observed that GFP-Tra2β and GFP-RS1RRM were concentrated in nuclear speckles with a diffusive distribution in the nearby nucleoplasm (Fig. 5). This is consistent with the characteristic speckle distribution of SR splicing factors. Nuclear speckles have been shown to serve as the storage/re-cycling sites for splicing factors, delivering the splicing factors to the nearby active sites of transcription and/or active sites of spliceosome assembly and splicing (9). Therefore, the diffused distribution of Tra2β and RS1RRM near the nuclear speckles is probably attributed to the presence of Tra2β and RS1RRM at the active sites of transcription and splicing. This possibility is supported by the observation that GFP-RS1, which lacks the RNA binding domain, is exclusively aggregated in condensed and enlarged nuclear speckles.
The Tra2β gene itself is alternatively spliced to five mRNA isoforms encoding at least 2 protein isoforms, Tra2β1 and Tra2βΔRS1 (23, 24, 48). The major isoform Tra2β1 encodes full-length Tra2β protein. Tra2βΔRS1 isoform encodes a truncated protein containing the RRM, glycine linker and the RS2 domain. Tra2βΔRS1 expression is tissue-specific in both flies and mammals, and up-regulated by expression of Clk kinases and neural stimulation (26, 49, 50). So far, no distinct function has been assigned to the Tra2βΔRS1 isoform compared with the full-length Tra2β, although this isoform is conserved in both vertebrates and invertebrates. Based on our observation showing the functional importance of RS1 in correct spatial organization, we propose a possible mechanistic explanation for the function of Tra2βΔRS1. Because the Tra2βΔRS1 contains a functional RRM sequence, the splicing repression could be happened due to the competitive inhibition through its binding to the same RNA targets, preventing the Tra2β being assembled into specific complexes. Thus, theoretically the Tra2βΔRS1 protein might operate as a splicing repressor isoform against major Tra2β isoform Tra2β1 (47), depending on the type of tissues where its expression level is high enough to act as a repressor.
Li and Bingham (1991) showed that the nuclear speckle localization signal of the Drosophila Tra was an amino acid sequence within the RS domain (15). It contains a nucleoplasmin-like bipartite nuclear localization signal (NLS) and a repeating arginine/serine (RS) dipeptide adjacent to a short stretch of basic amino acid (16). Here, by serial deletions, we identified at least two such speckle localization signals that exist in the amino acid sequences of the 56–68 aa and 90–100 aa within the RS1 domain of mammalian Tra2β. The reservation of either of them ensures the recombinant proteins to be remained in the nuclear speckles. These redundant nuclear speckle localization signals provide a structural basis for a tighter control of subnuclear localization for mammalian Tra2β, compared with Drosophila Tra.
We also studied the effect of phosphorylation on the nuclear speckle localization of Tra2β. It was demonstrated that mutation of serine residues has no obvious effect on the nuclear speckle localization. The results imply that mechanisms other than directly altering the phosphorylation of those serine residues may regulate the speckle localization of Tra2β. For example, kinases and phosphatases may influence the distribution of SR proteins indirectly by regulating the function of some SR partner proteins via phosphorylation modification.
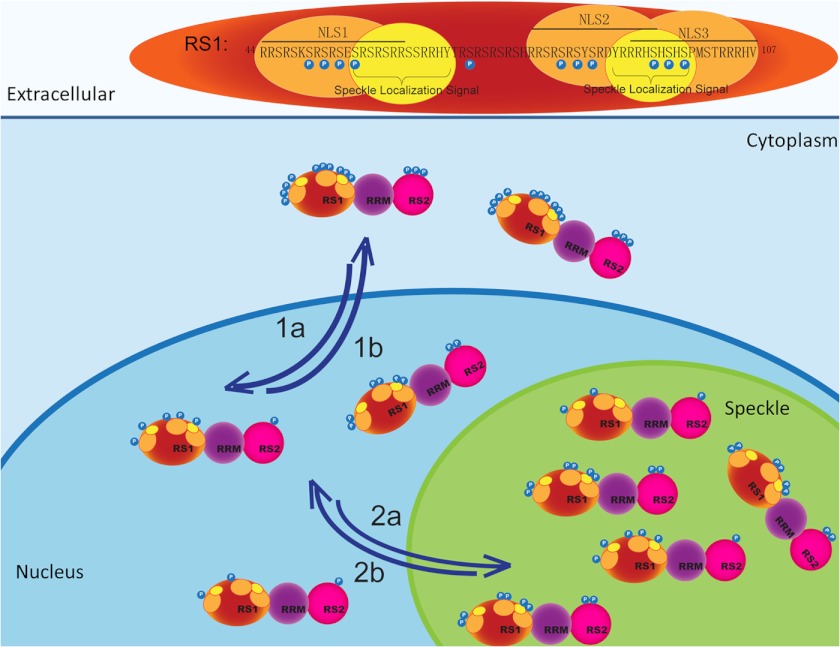
In summary, the present study provides the evidence important for elucidating the structural mechanisms for the function of Tra2β, with the respect to the subcellular and subnuclear localization. Based on our observations, a model regarding the role of NLSs and phosphorylation of serine residues in subcellular and subnuclear localization of Tra2β is proposed as illustrated in Fig. 8. In the model, we propose that the subcellular localization of Tra2β is determined by the net effect of two factors, phosphorylation of serine residues (promoting the cytoplasmic Tra2β localization) and NLSs (promoting the nuclear Tra2β localization). Besides, there are at least two speckle localization signals within the RS1 domain of Tra2β, which guide the import of Tra2β into the nuclear speckles in a phosphorylation-independent manner. As a functional consequence, through this mechanistic model, the subcellular and subnuclear localization of Tra2β will affect the relative nuclear concentrations of Tra2β to other splicing factors; thereby affect the regulation of the gene expression at the splicing level in response to signaling.
FIGURE 8.
A hypothetical model depicting the role of NLSs and phosphorylation of serine residues in subcellular and subnuclear localization of Tra2β. The RS1 domain of Tra2β is shown (top) with the regulatory phosphorylation site (blue), NLS (red), and nuclear speckle signal (yellow) highlighted. The NLS mediates nuclear import of Tra2β (1a). Accumulation of phosphorylation leads to more cytoplasmic localization of Tra2β (1b). Nuclear speckle signals mediate the nuclear speckle import of Tra2β, no matter the serine residues are phosphorylated or not (2a). Mechanisms other than directly altering the phosphorylation of those serine residues may negatively regulate the speckle import of Tra2β (2b).
Acknowledgment
We thank Ke Qiao from the Key Laboratory of Medical Molecular Virology for excellent technical expertise in confocal microscopy.
This work was supported by grants from the National Foundation of Natural Sciences of China (No. 30770661, 30971464, 81030020), and the Shanghai Leading Academic Discipline Project (No. B111).
- SR
- serine/arginine-rich
- Tra2β
- mammalian transformer-2β
- NLS
- nuclear localization signal
- RRM
- RNA recognition motif
- CIP
- calf intestinal alkaline phosphatase
- BCIP/NBT
- 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium.
REFERENCES
- 1. Black D. L. (2000) Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell 103, 367–370 [DOI] [PubMed] [Google Scholar]
- 2. Graveley B. R. (2001) Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 17, 100–107 [DOI] [PubMed] [Google Scholar]
- 3. Faustino N. A., Cooper T. A. (2003) Pre-mRNA splicing and human disease. Genes Dev. 17, 419–437 [DOI] [PubMed] [Google Scholar]
- 4. Fu X. D. (1995) The superfamily of arginine/serine-rich splicing factors. RNA 1, 663–680 [PMC free article] [PubMed] [Google Scholar]
- 5. Manley J. L., Tacke R. (1996) SR proteins and splicing control. Genes Dev. 10, 1569–1579 [DOI] [PubMed] [Google Scholar]
- 6. Spector D. L. (1993) Macromolecular domains within the cell nucleus. Annu. Rev. Cell Biol. 9, 265–315 [DOI] [PubMed] [Google Scholar]
- 7. O'Keefe R. T., Mayeda A., Sadowski C. L., Krainer A. R., Spector D. L. (1994) Disruption of pre-mRNA splicing in vivo results in reorganization of splicing factors. J. Cell Biol. 124, 249–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jiménez-Garcia L. F., Spector D. L. (1993) In vivo evidence that transcription and splicing are coordinated by a recruiting mechanism. Cell 73, 47–59 [DOI] [PubMed] [Google Scholar]
- 9. Huang S., Spector D. L. (1996) Intron-dependent recruitment of pre-mRNA splicing factors to sites of transcription. J. Cell Biol. 133, 719–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Misteli T., Spector D. L. (1997) Protein phosphorylation and the nuclear organization of pre-mRNA splicing. Trends Cell Biol. 7, 135–138 [DOI] [PubMed] [Google Scholar]
- 11. Cáceres J. F., Screaton G. R., Krainer A. R. (1998) A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 12, 55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang Y., Gattoni R., Stévenin J., Steitz J. A. (2003) SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell 11, 837–843 [DOI] [PubMed] [Google Scholar]
- 13. Lemaire R., Prasad J., Kashima T., Gustafson J., Manley J. L., Lafyatis R. (2002) Stability of a PKCI-1-related mRNA is controlled by the splicing factor ASF/SF2: a novel function for SR proteins. Genes Dev. 16, 594–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sanford J. R., Gray N. K., Beckmann K., Cáceres J. F. (2004) A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 18, 755–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li H., Bingham P. M. (1991) Arginine/serine-rich domains of the su(wa) and tra RNA processing regulators target proteins to a subnuclear compartment implicated in splicing. Cell 67, 335–342 [DOI] [PubMed] [Google Scholar]
- 16. Hedley M. L., Amrein H., Maniatis T. (1995) An amino acid sequence motif sufficient for subnuclear localization of an arginine/serine-rich splicing factor. Proc. Natl. Acad. Sci. U.S.A. 92, 11524–11528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gui J. F., Lane W. S., Fu X. D. (1994) A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature 369, 678–682 [DOI] [PubMed] [Google Scholar]
- 18. Colwill K., Pawson T., Andrews B., Prasad J., Manley J. L., Bell J. C., Duncan P. I. (1996) The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 15, 265–275 [PMC free article] [PubMed] [Google Scholar]
- 19. Wang H. Y., Lin W., Dyck J. A., Yeakley J. M., Songyang Z., Cantley L. C., Fu X. D. (1998) SRPK2: a differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J. Cell Biol. 140, 737–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Misteli T., Spector D. L. (1996) Serine/threonine phosphatase 1 modulates the subnuclear distribution of pre-mRNA splicing factors. Mol. Biol. Cell 7, 1559–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cáceres J. F., Misteli T., Screaton G. R., Spector D. L., Krainer A. R. (1997) Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 138, 225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Beil B., Screaton G., Stamm S. (1997) Molecular cloning of htra2-β-1 and htra2-β-2, two human homologs of tra-2 generated by alternative splicing. DNA Cell Biol. 16, 679–690 [DOI] [PubMed] [Google Scholar]
- 23. Nayler O., Cap C., Stamm S. (1998) Human transformer-2-β gene (SFRS10): complete nucleotide sequence, chromosomal localization, and generation of a tissue-specific isoform. Genomics 53, 191–202 [DOI] [PubMed] [Google Scholar]
- 24. Chen X., Guo L., Lin W., Xu P. (2003) Expression of Tra2β isoforms is developmentally regulated in a tissue- and temporal-specific pattern. Cell Biol. Int. 27, 491–496 [DOI] [PubMed] [Google Scholar]
- 25. Hofmann Y., Lorson C. L., Stamm S., Androphy E. J., Wirth B. (2000) Htra2-β1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2). Proc. Natl. Acad. Sci. U.S.A. 97, 9618–9623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Glatz D. C., Rujescu D., Tang Y., Berendt F. J., Hartmann A. M., Faltraco F., Rosenberg C., Hulette C., Jellinger K., Hampel H., Riederer P., Möller H. J., Andreadis A., Henkel K., Stamm S. (2006) The alternative splicing of tau exon 10 and its regulatory proteins CLK2 and TRA2-β1 changes in sporadic Alzheimer's disease. J. Neurochem. 96, 635–644 [DOI] [PubMed] [Google Scholar]
- 27. D'Souza I., Schellenberg G. D. (2006) Arginine/serine-rich protein interaction domain-dependent modulation of a tau exon 10 splicing enhancer: altered interactions and mechanisms for functionally antagonistic FTDP-17 mutations Δ280K AND N279K. J. Biol. Chem. 281, 2460–2469 [DOI] [PubMed] [Google Scholar]
- 28. Jiang Z., Tang H., Havlioglu N., Zhang X., Stamm S., Yan R., Wu J. Y. (2003) Mutations in tau gene exon 10 associated with FTDP-17 alter the activity of an exonic splicing enhancer to interact with Tra2β. J. Biol. Chem. 278, 18997–19007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Watermann D. O., Tang Y., Zur Hausen A., Jäger M., Stamm S., Stickeler E. (2006) Splicing factor Tra2-β1 is specifically induced in breast cancer and regulates alternative splicing of the CD44 gene. Cancer Res. 66, 4774–4780 [DOI] [PubMed] [Google Scholar]
- 30. Daoud R., Mies G., Smialowska A., Oláh L., Hossmann K. A., Stamm S. (2002) Ischemia induces a translocation of the splicing factor tra2-β1 and changes alternative splicing patterns in the brain. J. Neurosci. 22, 5889–5899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shukla S., Fisher S. A. (2008) Tra2β as a novel mediator of vascular smooth muscle diversification. Circ. Res. 103, 485–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Han Y., Chen X., Shi F., Li S., Huang J., Xie M., Hu L., Hoidal J. R., Xu P. (2007) CPG15, a new factor upregulated after ischemic brain injury, contributes to neuronal network re-establishment after glutamate-induced injury. J. Neurotrauma 24, 722–731 [DOI] [PubMed] [Google Scholar]
- 33. Segade F., Hurlé B., Claudio E., Ramos S., Lazo P. S. (1996) Molecular cloning of a mouse homologue for the Drosophila splicing regulator Tra2. FEBS Lett. 387, 152–156 [DOI] [PubMed] [Google Scholar]
- 34. Kalderon D., Roberts B. L., Richardson W. D., Smith A. E. (1984) A short amino acid sequence able to specify nuclear location. Cell 39, 499–509 [DOI] [PubMed] [Google Scholar]
- 35. Dingwall C., Robbins J., Dilworth S. M., Roberts B., Richardson W. D. (1988) The nucleoplasmin nuclear location sequence is larger and more complex than that of SV-40 large T antigen. J. Cell Biol. 107, 841–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zahler A. M., Lane W. S., Stolk J. A., Roth M. B. (1992) SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 6, 837–847 [DOI] [PubMed] [Google Scholar]
- 37. Daub H., Olsen J. V., Bairlein M., Gnad F., Oppermann F. S., Körner R., Greff Z., Kéri G., Stemmann O., Mann M. (2008) Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol. Cell 31, 438–448 [DOI] [PubMed] [Google Scholar]
- 38. Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P. (2008) A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 40. Wang B., Malik R., Nigg E. A., Körner R. (2008) Evaluation of the low-specificity protease elastase for large-scale phosphoproteome analysis. Anal. Chem. 80, 9526–9533 [DOI] [PubMed] [Google Scholar]
- 41. Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., Hu Y., Tan Z., Stokes M., Sullivan L., Mitchell J., Wetzel R., Macneill J., Ren J. M., Yuan J., Bakalarski C. E., Villen J., Kornhauser J. M., Smith B., Li D., Zhou X., Gygi S. P., Gu T. L., Polakiewicz R. D., Rush J., Comb M. J. (2007) Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131, 1190–1203 [DOI] [PubMed] [Google Scholar]
- 42. van der Houven van Oordt W., Diaz-Meco M. T., Lozano J., Krainer A. R., Moscat J., Cáceres J. F. (2000) The MKK(3/6)-p38-signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation. J. Cell Biol. 149, 307–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kataoka N., Bachorik J. L., Dreyfuss G. (1999) Transportin-SR, a nuclear import receptor for SR proteins. J. Cell Biol. 145, 1145–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lai M. C., Lin R. I., Huang S. Y., Tsai C. W., Tarn W. Y. (2000) A human importin-beta family protein, transportin-SR2, interacts with the phosphorylated RS domain of SR proteins. J. Biol. Chem. 275, 7950–7957 [DOI] [PubMed] [Google Scholar]
- 45. Lai M. C., Lin R. I., Tarn W. Y. (2001) Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc. Natl. Acad. Sci. U.S.A. 98, 10154–10159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yun C. Y., Velazquez-Dones A. L., Lyman S. K., Fu X. D. (2003) Phosphorylation-dependent and -independent nuclear import of RS domain-containing splicing factors and regulators. J. Biol. Chem. 278, 18050–18055 [DOI] [PubMed] [Google Scholar]
- 47. Grellscheid S., Dalgliesh C., Storbeck M., Best A., Liu Y., Jakubik M., Mende Y., Ehrmann I., Curk T., Rossbach K., Bourgeois C. F., Stévenin J., Grellscheid D., Jackson M. S., Wirth B., Elliott D. J. (2011) Identification of evolutionarily conserved exons as regulated targets for the splicing activator tra2β in development. PLoS Genet. 7, e1002390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Daoud R., Da Penha Berzaghi M., Siedler F., Hübener M., Stamm S. (1999) Activity-dependent regulation of alternative splicing patterns in the rat brain. Eur. J. Neurosci. 11, 788–802 [DOI] [PubMed] [Google Scholar]
- 49. Mattox W., Palmer M. J., Baker B. S. (1990) Alternative splicing of the sex determination gene transformer-2 is sex-specific in the germ line but not in the soma. Genes Dev. 4, 789–805 [DOI] [PubMed] [Google Scholar]
- 50. Stoilov P., Daoud R., Nayler O., Stamm S. (2004) Human tra2-β1 autoregulates its protein concentration by influencing alternative splicing of its pre-mRNA. Hum. Mol. Genet. 13, 509–524 [DOI] [PubMed] [Google Scholar]