Abstract
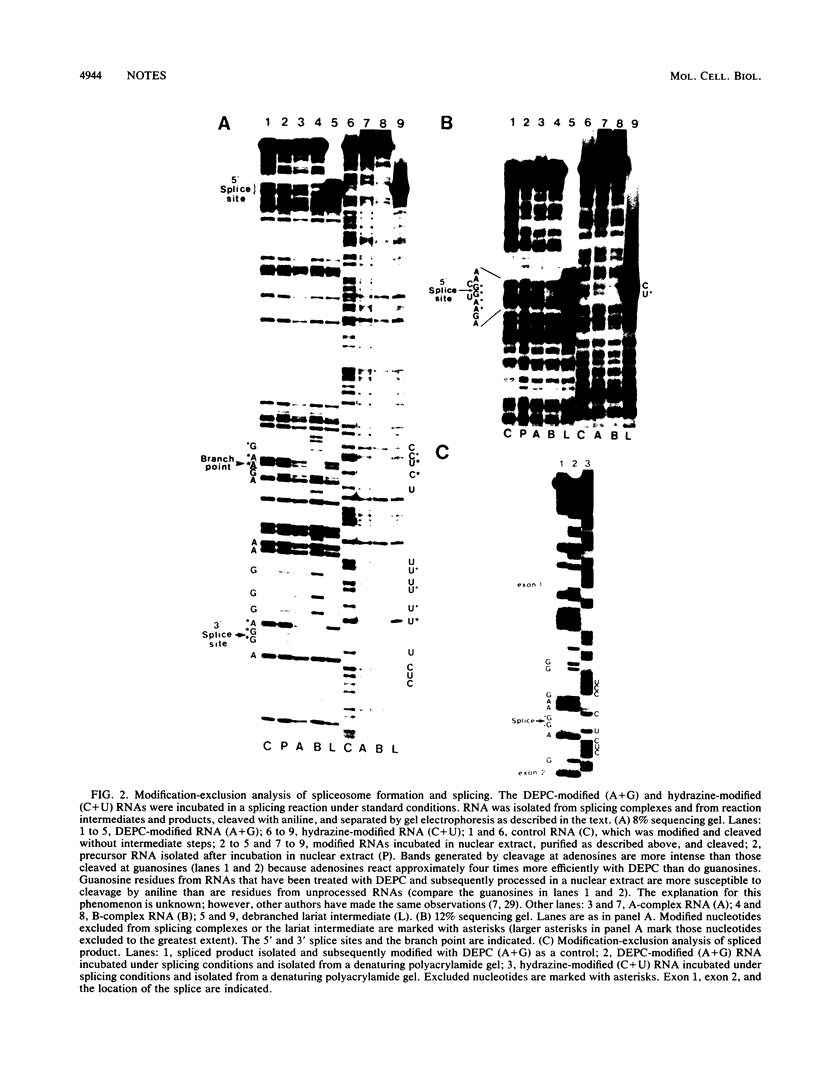
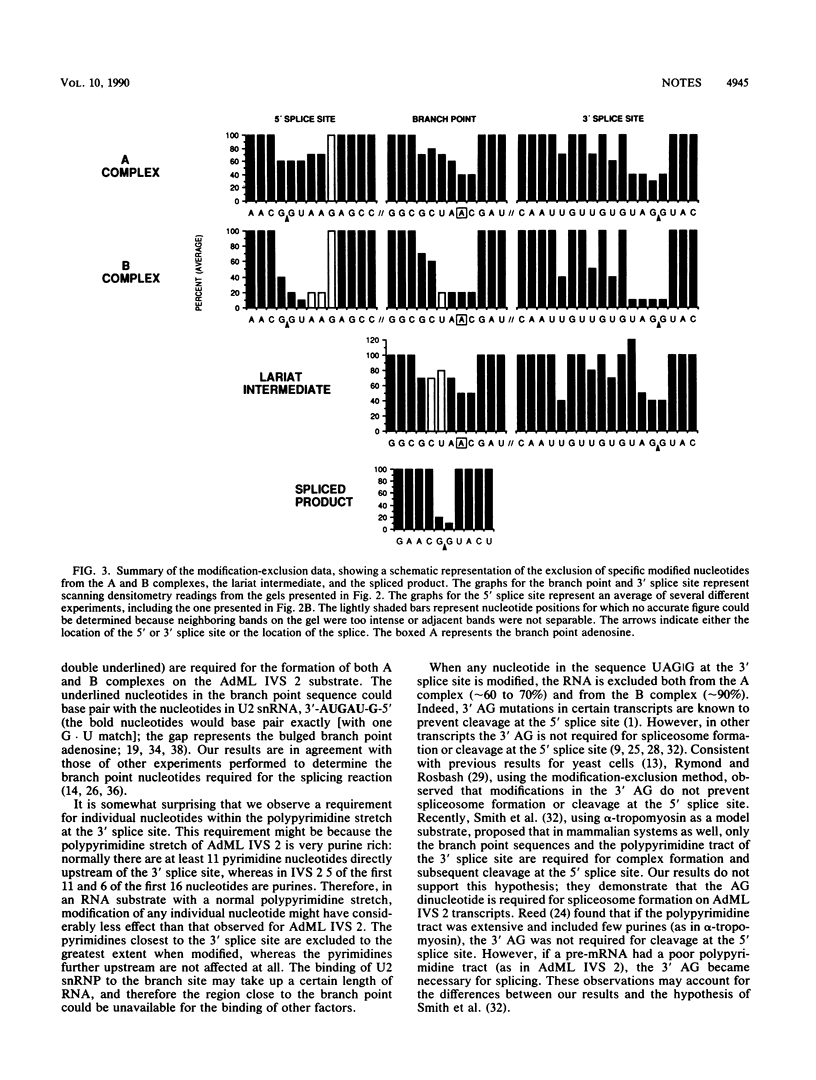
The stepwise assembly of splicing complexes and the subsequent splicing reaction were analyzed by the RNA modification-exclusion technique, which generates the equivalent of a complete set of point mutations in a single reaction. We found that although the sequences surrounding the 5' splice site, the branch point, and the 3' splice site, including the 3' AG, were required for presplicing complex formation, modified nucleotides at these positions were not completely excluded. The same sequences were required for splicing complex formation; however, modified nucleotides in these sequences were excluded to a much greater extent.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aebi M., Hornig H., Padgett R. A., Reiser J., Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell. 1986 Nov 21;47(4):555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- Barabino S. M., Sproat B. S., Ryder U., Blencowe B. J., Lamond A. I. Mapping U2 snRNP--pre-mRNA interactions using biotinylated oligonucleotides made of 2'-OMe RNA. EMBO J. 1989 Dec 20;8(13):4171–4178. doi: 10.1002/j.1460-2075.1989.tb08602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody E., Abelson J. The "spliceosome": yeast pre-messenger RNA associates with a 40S complex in a splicing-dependent reaction. Science. 1985 May 24;228(4702):963–967. doi: 10.1126/science.3890181. [DOI] [PubMed] [Google Scholar]
- Chabot B., Steitz J. A. Multiple interactions between the splicing substrate and small nuclear ribonucleoproteins in spliceosomes. Mol Cell Biol. 1987 Jan;7(1):281–293. doi: 10.1128/mcb.7.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S. C., Abelson J. Spliceosome assembly in yeast. Genes Dev. 1987 Nov;1(9):1014–1027. doi: 10.1101/gad.1.9.1014. [DOI] [PubMed] [Google Scholar]
- Christofori G., Frendewey D., Keller W. Two spliceosomes can form simultaneously and independently on synthetic double-intron messenger RNA precursors. EMBO J. 1987 Jun;6(6):1747–1755. doi: 10.1002/j.1460-2075.1987.tb02427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway L., Wickens M. Analysis of mRNA 3' end formation by modification interference: the only modifications which prevent processing lie in AAUAAA and the poly(A) site. EMBO J. 1987 Dec 20;6(13):4177–4184. doi: 10.1002/j.1460-2075.1987.tb02764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frendewey D., Keller W. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell. 1985 Aug;42(1):355–367. doi: 10.1016/s0092-8674(85)80131-8. [DOI] [PubMed] [Google Scholar]
- Frendewey D., Krämer A., Keller W. Different small nuclear ribonucleoprotein particles are involved in different steps of splicing complex formation. Cold Spring Harb Symp Quant Biol. 1987;52:287–298. doi: 10.1101/sqb.1987.052.01.034. [DOI] [PubMed] [Google Scholar]
- Freyer G. A., Arenas J., Perkins K. K., Furneaux H. M., Pick L., Young B., Roberts R. J., Hurwitz J. In vitro formation of a lariat structure containing a G2'-5'G linkage. J Biol Chem. 1987 Mar 25;262(9):4267–4273. [PubMed] [Google Scholar]
- Green M. R. Pre-mRNA splicing. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- Guthrie C., Patterson B. Spliceosomal snRNAs. Annu Rev Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- Hornig H., Aebi M., Weissmann C. Effect of mutations at the lariat branch acceptor site on beta-globin pre-mRNA splicing in vitro. Nature. 1986 Dec 11;324(6097):589–591. doi: 10.1038/324589a0. [DOI] [PubMed] [Google Scholar]
- Humphrey T., Christofori G., Lucijanic V., Keller W. Cleavage and polyadenylation of messenger RNA precursors in vitro occurs within large and specific 3' processing complexes. EMBO J. 1987 Dec 20;6(13):4159–4168. doi: 10.1002/j.1460-2075.1987.tb02762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska M. M., Sharp P. A. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell. 1986 Sep 12;46(6):845–855. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- Konarska M. M., Sharp P. A. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell. 1987 Jun 19;49(6):763–774. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- Krämer A., Keller W. Purification of a protein required for the splicing of pre-mRNA and its separation from the lariat debranching enzyme. EMBO J. 1985 Dec 16;4(13A):3571–3581. doi: 10.1002/j.1460-2075.1985.tb04119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K. K., Green M. R. Mammalian U2 snRNP has a sequence-specific RNA-binding activity. Genes Dev. 1989 Oct;3(10):1562–1571. doi: 10.1101/gad.3.10.1562. [DOI] [PubMed] [Google Scholar]
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikielny C. W., Rosbash M. Specific small nuclear RNAs are associated with yeast spliceosomes. Cell. 1986 Jun 20;45(6):869–877. doi: 10.1016/0092-8674(86)90561-1. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. Intron sequences involved in lariat formation during pre-mRNA splicing. Cell. 1985 May;41(1):95–105. doi: 10.1016/0092-8674(85)90064-9. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. The role of the mammalian branchpoint sequence in pre-mRNA splicing. Genes Dev. 1988 Oct;2(10):1268–1276. doi: 10.1101/gad.2.10.1268. [DOI] [PubMed] [Google Scholar]
- Reed R. The organization of 3' splice-site sequences in mammalian introns. Genes Dev. 1989 Dec;3(12B):2113–2123. doi: 10.1101/gad.3.12b.2113. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Green M. R. An RNA processing activity that debranches RNA lariats. Science. 1985 Jul 12;229(4709):135–140. doi: 10.1126/science.2990042. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Green M. R. Role of the 3' splice site consensus sequence in mammalian pre-mRNA splicing. Nature. 1985 Oct 24;317(6039):732–734. doi: 10.1038/317732a0. [DOI] [PubMed] [Google Scholar]
- Rymond B. C., Rosbash M. A chemical modification/interference study of yeast pre-mRNA spliceosome assembly and splicing. Genes Dev. 1988 Apr;2(4):428–439. doi: 10.1101/gad.2.4.428. [DOI] [PubMed] [Google Scholar]
- Siliciano P. G., Guthrie C. 5' splice site selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev. 1988 Oct;2(10):1258–1267. doi: 10.1101/gad.2.10.1258. [DOI] [PubMed] [Google Scholar]
- Smith C. W., Porro E. B., Patton J. G., Nadal-Ginard B. Scanning from an independently specified branch point defines the 3' splice site of mammalian introns. Nature. 1989 Nov 16;342(6247):243–247. doi: 10.1038/342243a0. [DOI] [PubMed] [Google Scholar]
- Séraphin B., Kretzner L., Rosbash M. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5' cleavage site. EMBO J. 1988 Aug;7(8):2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Manley J. L. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev. 1989 Oct;3(10):1553–1561. doi: 10.1101/gad.3.10.1553. [DOI] [PubMed] [Google Scholar]
- Yuo C. Y., Weiner A. M. A U1 small nuclear ribonucleoprotein particle with altered specificity induces alternative splicing of an adenovirus E1A mRNA precursor. Mol Cell Biol. 1989 Aug;9(8):3429–3437. doi: 10.1128/mcb.9.8.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y. A., Goldstein A. M., Weiner A. M. UACUAAC is the preferred branch site for mammalian mRNA splicing. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2752–2756. doi: 10.1073/pnas.86.8.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986 Sep 12;46(6):827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in human U2 snRNA can suppress a branch site mutation. Genes Dev. 1989 Oct;3(10):1545–1552. doi: 10.1101/gad.3.10.1545. [DOI] [PubMed] [Google Scholar]