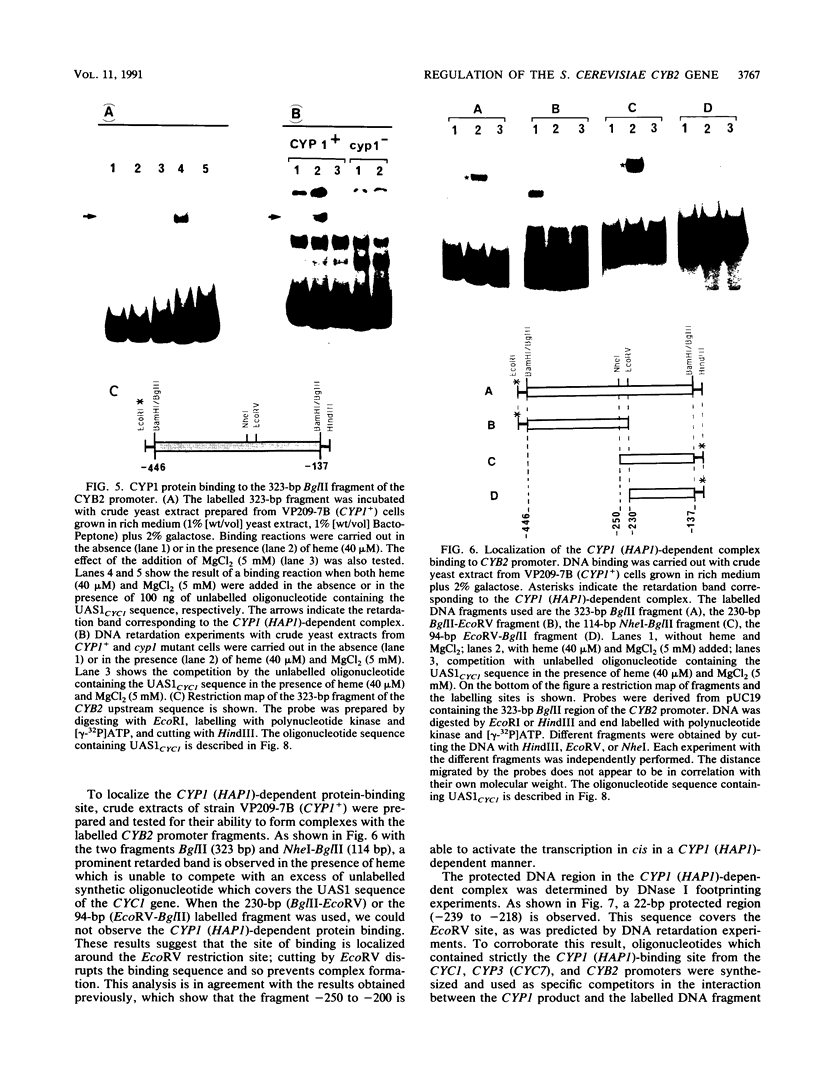
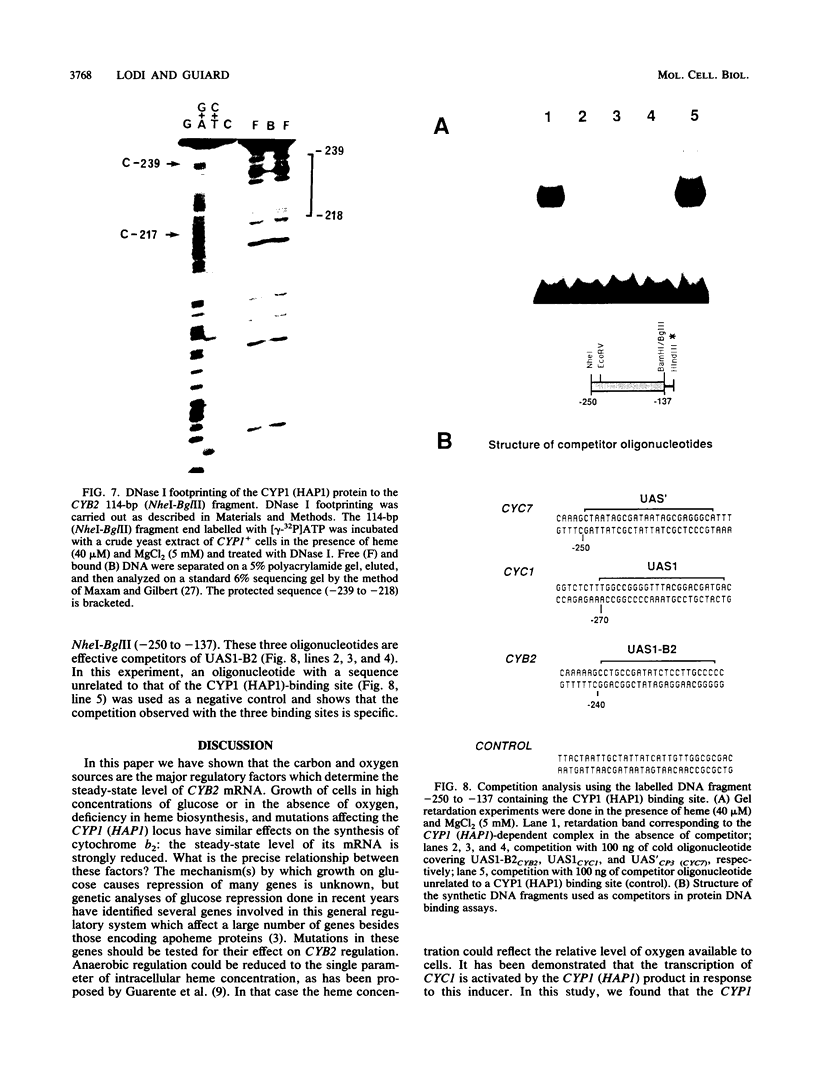
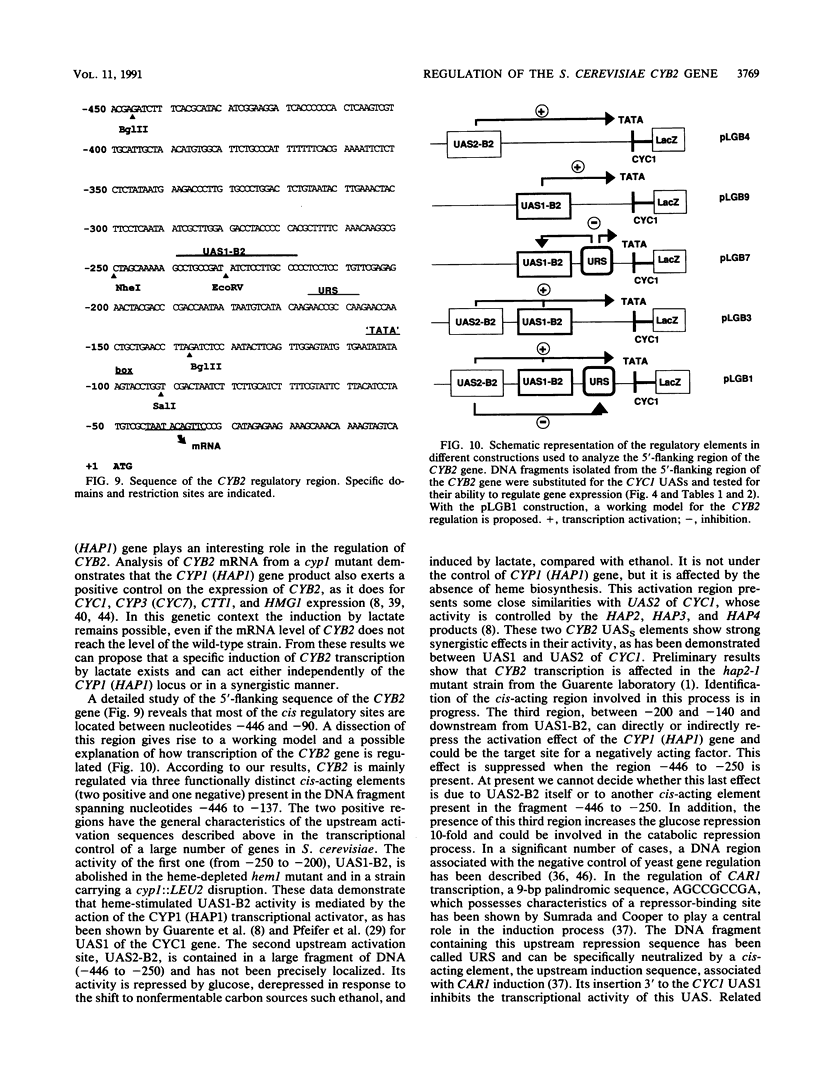
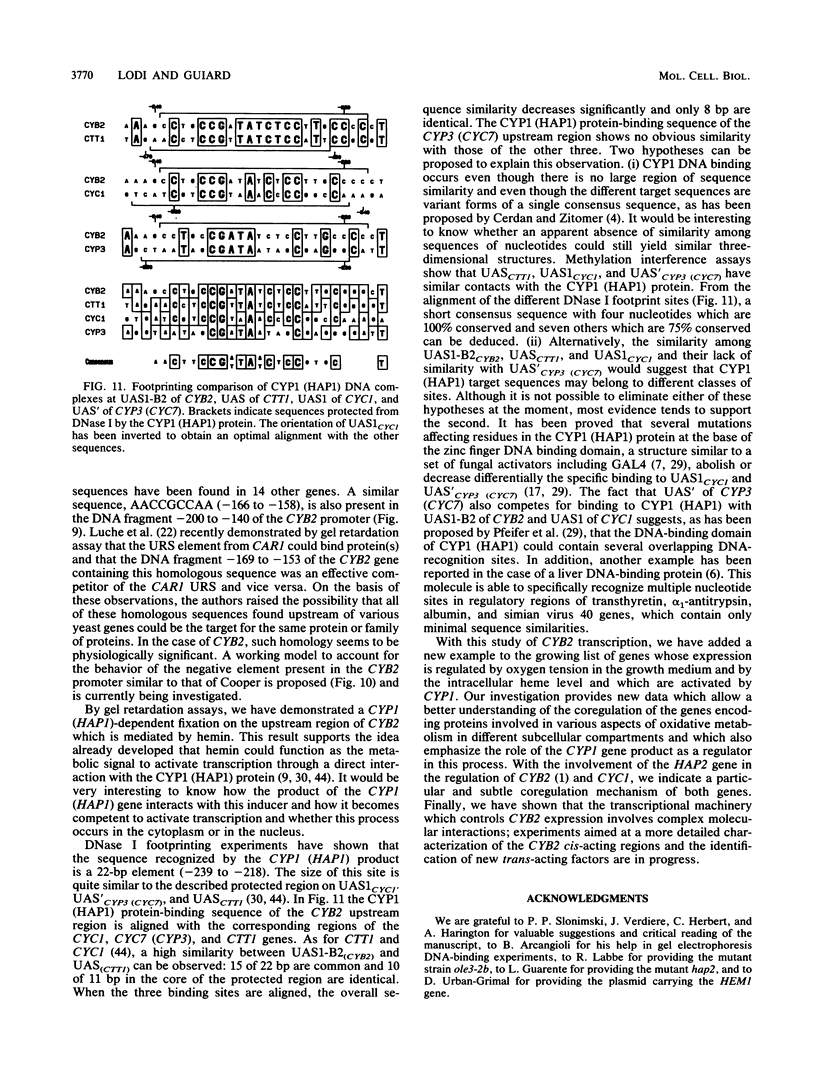
Abstract
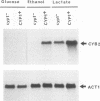
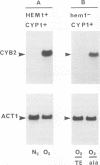
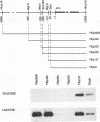
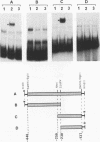
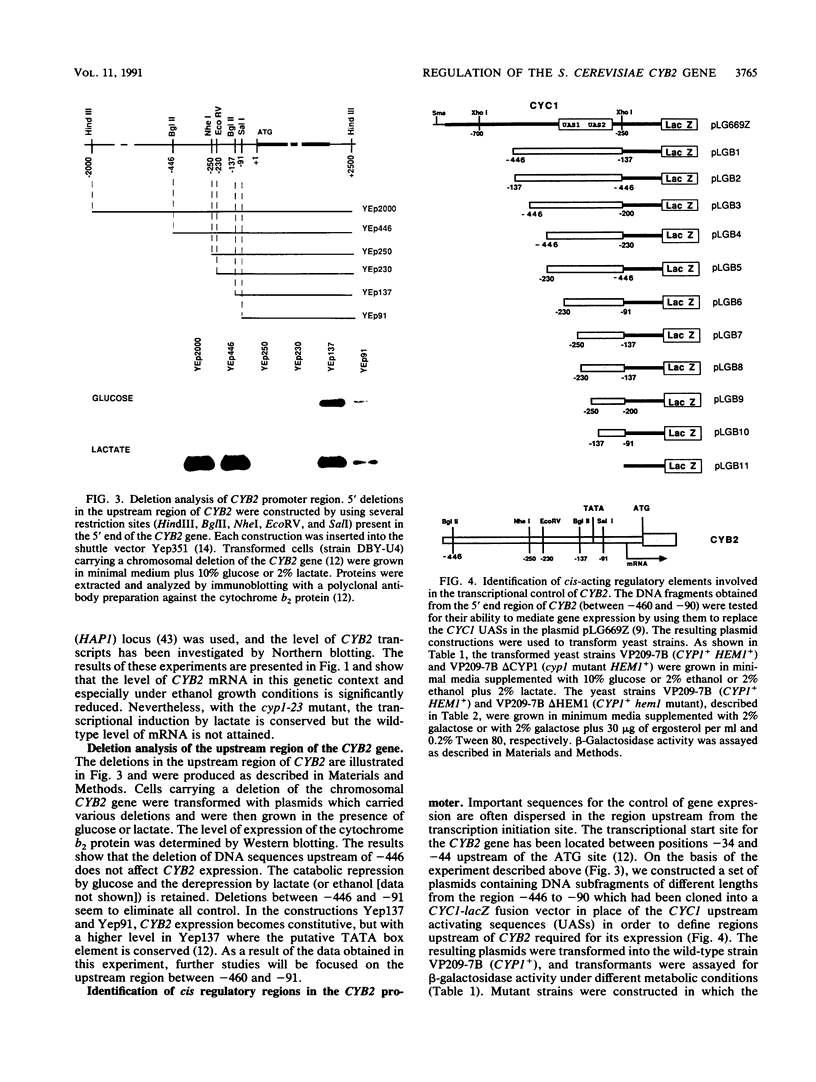
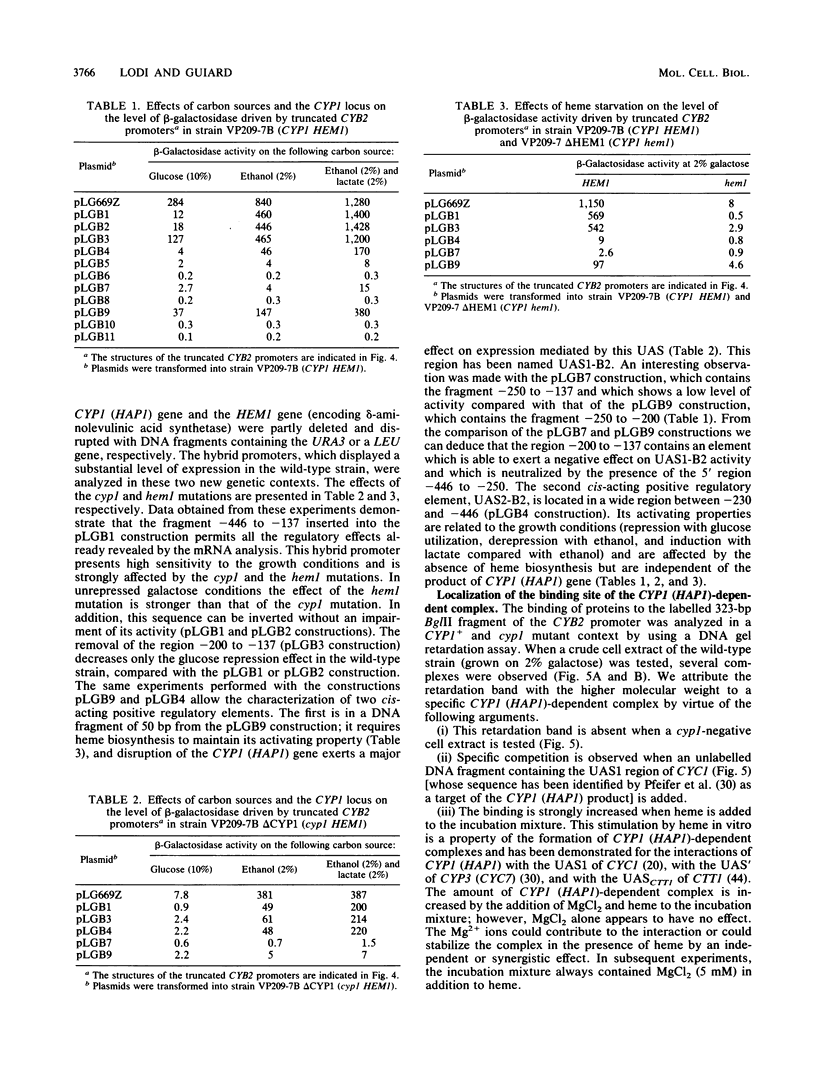
Expression of the Saccharomyces cerevisiae gene encoding cytochrome b2 (EC 1.2.2.3), CYB2, was investigated by direct analysis of mRNA transcripts and by measurement of the expression of lacZ fused to the CYB2 control regions. These studies indicated that regulation of the CYB2 gene is subject to several metabolic controls at the transcriptional level: inhibition due to glucose fermentation, induction by lactate, and inhibition in anaerobiosis or in absence of heme biosynthesis. Furthermore, we have shown that the CYB2 promoter contains one cis negative regulatory region and two heme-dependent positive regions, one of which is controlled by the transcriptional regulator CYP1 (HAP1) which is involved in the modulation of the expression of several oxygen-regulated genes. The CYP1 (HAP1)-binding sequence was located by gel retardation and DNase I footprinting experiments and compared with the binding sequences previously characterized in detail (UAS1CYC1, UAS'CYP3 (CYC7), and UASCTT1).
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beato M. Gene regulation by steroid hormones. Cell. 1989 Feb 10;56(3):335–344. doi: 10.1016/0092-8674(89)90237-7. [DOI] [PubMed] [Google Scholar]
- Capeillere-Blandin C. Transient kinetics of the one-electron transfer reaction between reduced flavocytochrome b2 and oxidized cytochrome c. Evidence for the existence of a protein complex in the reaction. Eur J Biochem. 1982 Nov 15;128(2-3):533–542. doi: 10.1111/j.1432-1033.1982.tb06998.x. [DOI] [PubMed] [Google Scholar]
- Carlson M. Regulation of sugar utilization in Saccharomyces species. J Bacteriol. 1987 Nov;169(11):4873–4877. doi: 10.1128/jb.169.11.4873-4877.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerdan M. E., Zitomer R. S. Oxygen-dependent upstream activation sites of Saccharomyces cerevisiae cytochrome c genes are related forms of the same sequence. Mol Cell Biol. 1988 Jun;8(6):2275–2279. doi: 10.1128/mcb.8.6.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavilier L., Péré-Aubert G., Somlo M., Slonimski P. P. Réseau d'interactions entre des génes non liés : régulation synergique ou antagoniste de la synthèse de l'iso-1-cytochrome c, de l'iso-2-cytochrome c et du cytochrome b2. Biochimie. 1976;58(1-2):155–172. doi: 10.1016/s0300-9084(76)80366-5. [DOI] [PubMed] [Google Scholar]
- Costa R. H., Grayson D. R., Xanthopoulos K. G., Darnell J. E., Jr A liver-specific DNA-binding protein recognizes multiple nucleotide sites in regulatory regions of transthyretin, alpha 1-antitrypsin, albumin, and simian virus 40 genes. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3840–3844. doi: 10.1073/pnas.85.11.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creusot F., Verdière J., Gaisne M., Slonimski P. P. CYP1 (HAP1) regulator of oxygen-dependent gene expression in yeast. I. Overall organization of the protein sequence displays several novel structural domains. J Mol Biol. 1988 Nov 20;204(2):263–276. doi: 10.1016/0022-2836(88)90574-8. [DOI] [PubMed] [Google Scholar]
- Guarente L., Lalonde B., Gifford P., Alani E. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell. 1984 Feb;36(2):503–511. doi: 10.1016/0092-8674(84)90243-5. [DOI] [PubMed] [Google Scholar]
- Guarente L., Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983 Apr;32(4):1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Guarente L., Ptashne M. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2199–2203. doi: 10.1073/pnas.78.4.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Yocum R. R., Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiard B. Structure, expression and regulation of a nuclear gene encoding a mitochondrial protein: the yeast L(+)-lactate cytochrome c oxidoreductase (cytochrome b2). EMBO J. 1985 Dec 1;4(12):3265–3272. doi: 10.1002/j.1460-2075.1985.tb04076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl F. U., Ostermann J., Guiard B., Neupert W. Successive translocation into and out of the mitochondrial matrix: targeting of proteins to the intermembrane space by a bipartite signal peptide. Cell. 1987 Dec 24;51(6):1027–1037. doi: 10.1016/0092-8674(87)90589-7. [DOI] [PubMed] [Google Scholar]
- Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986 Sep;2(3):163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Hörtner H., Ammerer G., Hartter E., Hamilton B., Rytka J., Bilinski T., Ruis H. Regulation of synthesis of catalases and iso-1-cytochrome c in Saccharomyces cerevisiae by glucose, oxygen and heme. Eur J Biochem. 1982 Nov;128(1):179–184. doi: 10.1111/j.1432-1033.1982.tb06949.x. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. S., Guarente L. Mutations that alter transcriptional activation but not DNA binding in the zinc finger of yeast activator HAPI. Nature. 1989 Nov 9;342(6246):200–203. doi: 10.1038/342200a0. [DOI] [PubMed] [Google Scholar]
- Kim K. S., Pfeifer K., Powell L., Guarente L. Internal deletions in the yeast transcriptional activator HAP1 have opposite effects at two sequence elements. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4524–4528. doi: 10.1073/pnas.87.12.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LABEYRIE F., SLONIMSKI P. P. MODE D'ACTION DES LACTICODESHYDROG'ENASES LI'EES AUX SYST'EMES FLAVINIQUE ET CYTOCHROMIQUE. Bull Soc Chim Biol (Paris) 1964;46:1793–1828. [PubMed] [Google Scholar]
- Lalonde B., Arcangioli B., Guarente L. A single Saccharomyces cerevisiae upstream activation site (UAS1) has two distinct regions essential for its activity. Mol Cell Biol. 1986 Dec;6(12):4690–4696. doi: 10.1128/mcb.6.12.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry C. V., Weiss J. L., Walthall D. A., Zitomer R. S. Modulator sequences mediate oxygen regulation of CYC1 and a neighboring gene in yeast. Proc Natl Acad Sci U S A. 1983 Jan;80(1):151–155. doi: 10.1073/pnas.80.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luche R. M., Sumrada R., Cooper T. G. A cis-acting element present in multiple genes serves as a repressor protein binding site for the yeast CAR1 gene. Mol Cell Biol. 1990 Aug;10(8):3884–3895. doi: 10.1128/mcb.10.8.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccecchini M. L., Rudin Y., Blobel G., Schatz G. Import of proteins into mitochondria: precursor forms of the extramitochondrially made F1-ATPase subunits in yeast. Proc Natl Acad Sci U S A. 1979 Jan;76(1):343–347. doi: 10.1073/pnas.76.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Matner R. R., Sherman F. Differential accumulation of two apo-iso-cytochromes c in processing mutants of yeast. J Biol Chem. 1982 Aug 25;257(16):9811–9821. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Montgomery D. L., Boss J. M., McAndrew S. J., Marr L., Walthall D. A., Zitomer R. S. The molecular characterization of three transcriptional mutations in the yeast iso-2-cytochrome c gene. J Biol Chem. 1982 Jul 10;257(13):7756–7761. [PubMed] [Google Scholar]
- Pfeifer K., Kim K. S., Kogan S., Guarente L. Functional dissection and sequence of yeast HAP1 activator. Cell. 1989 Jan 27;56(2):291–301. doi: 10.1016/0092-8674(89)90903-3. [DOI] [PubMed] [Google Scholar]
- Pfeifer K., Prezant T., Guarente L. Yeast HAP1 activator binds to two upstream activation sites of different sequence. Cell. 1987 Apr 10;49(1):19–27. doi: 10.1016/0092-8674(87)90751-3. [DOI] [PubMed] [Google Scholar]
- Prezant T., Pfeifer K., Guarente L. Organization of the regulatory region of the yeast CYC7 gene: multiple factors are involved in regulation. Mol Cell Biol. 1987 Sep;7(9):3252–3259. doi: 10.1128/mcb.7.9.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J., Sherman F. Genes affecting the expression of cytochrome c in yeast: genetic mapping and genetic interactions. Genetics. 1980 Apr;94(4):871–889. doi: 10.1093/genetics/94.4.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SOMLO M. INDUCTION DES LACTICO-CYTOCHROME C REDUCTASES (D- ET L-) DE LA LEVURE AEROBIE PAR LES LACTATES (D- ET L-) Biochim Biophys Acta. 1965 Feb 15;97:183–201. [PubMed] [Google Scholar]
- Sherman F., Stewart J. W. Genetics and biosynthesis of cytochrome c. Annu Rev Genet. 1971;5:257–296. doi: 10.1146/annurev.ge.05.120171.001353. [DOI] [PubMed] [Google Scholar]
- Spevak W., Hartig A., Meindl P., Ruis H. Heme control region of the catalase T gene of the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1986 Apr;203(1):73–78. doi: 10.1007/BF00330386. [DOI] [PubMed] [Google Scholar]
- Sumrada R. A., Cooper T. G. Ubiquitous upstream repression sequences control activation of the inducible arginase gene in yeast. Proc Natl Acad Sci U S A. 1987 Jun;84(12):3997–4001. doi: 10.1073/pnas.84.12.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsness M., Schafer W., D'Ari L., Rine J. Positive and negative transcriptional control by heme of genes encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Dec;9(12):5702–5712. doi: 10.1128/mcb.9.12.5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdière J., Creusot F., Guarente L., Slonimski P. P. The overproducing CYP1 and the underproducing hap1 mutations are alleles of the same gene which regulates in trans the expression of the structural genes encoding iso-cytochromes c. Curr Genet. 1986;10(5):339–342. doi: 10.1007/BF00418404. [DOI] [PubMed] [Google Scholar]
- Verdière J., Creusot F., Guérineau M. Regulation of the expression of iso 2-cytochrome c gene in S. cerevisiae: cloning of the positive regulatory gene CYP1 and identification of the region of its target sequence on the structural gene CYP3. Mol Gen Genet. 1985;199(3):524–533. doi: 10.1007/BF00330769. [DOI] [PubMed] [Google Scholar]
- Verdière J., Gaisne M., Guiard B., Defranoux N., Slonimski P. P. CYP1 (HAP1) regulator of oxygen-dependent gene expression in yeast. II. Missense mutation suggests alternative Zn fingers as discriminating agents of gene control. J Mol Biol. 1988 Nov 20;204(2):277–282. doi: 10.1016/0022-2836(88)90575-x. [DOI] [PubMed] [Google Scholar]
- Verdière J., Petrochilo E. CYP3--15: an up-promoter mutation at the ISO-2 cytochrome c structural gene locus and its interaction with other independent regulatory mutations of cytochrome c synthesis in Saccharomyces cerevisiae. Mol Gen Genet. 1979 Sep;175(2):209–216. doi: 10.1007/BF00425538. [DOI] [PubMed] [Google Scholar]
- Winkler H., Adam G., Mattes E., Schanz M., Hartig A., Ruis H. Co-ordinate control of synthesis of mitochondrial and non-mitochondrial hemoproteins: a binding site for the HAP1 (CYP1) protein in the UAS region of the yeast catalase T gene (CTT1). EMBO J. 1988 Jun;7(6):1799–1804. doi: 10.1002/j.1460-2075.1988.tb03011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods R. A., Sanders H. K., Briquet M., Foury F., Drysdale B. E., Mattoon J. R. Regulation of mitochondrial biogenesis: enzymatic changes in cytochrome-deficient yeast mutants requiring delta-aminolevulinic acid. J Biol Chem. 1975 Dec 10;250(23):9090–9098. [PubMed] [Google Scholar]
- Wright C. F., Zitomer R. S. A positive regulatory site and a negative regulatory site control the expression of the Saccharomyces cerevisiae CYC7 gene. Mol Cell Biol. 1984 Oct;4(10):2023–2030. doi: 10.1128/mcb.4.10.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]