Abstract
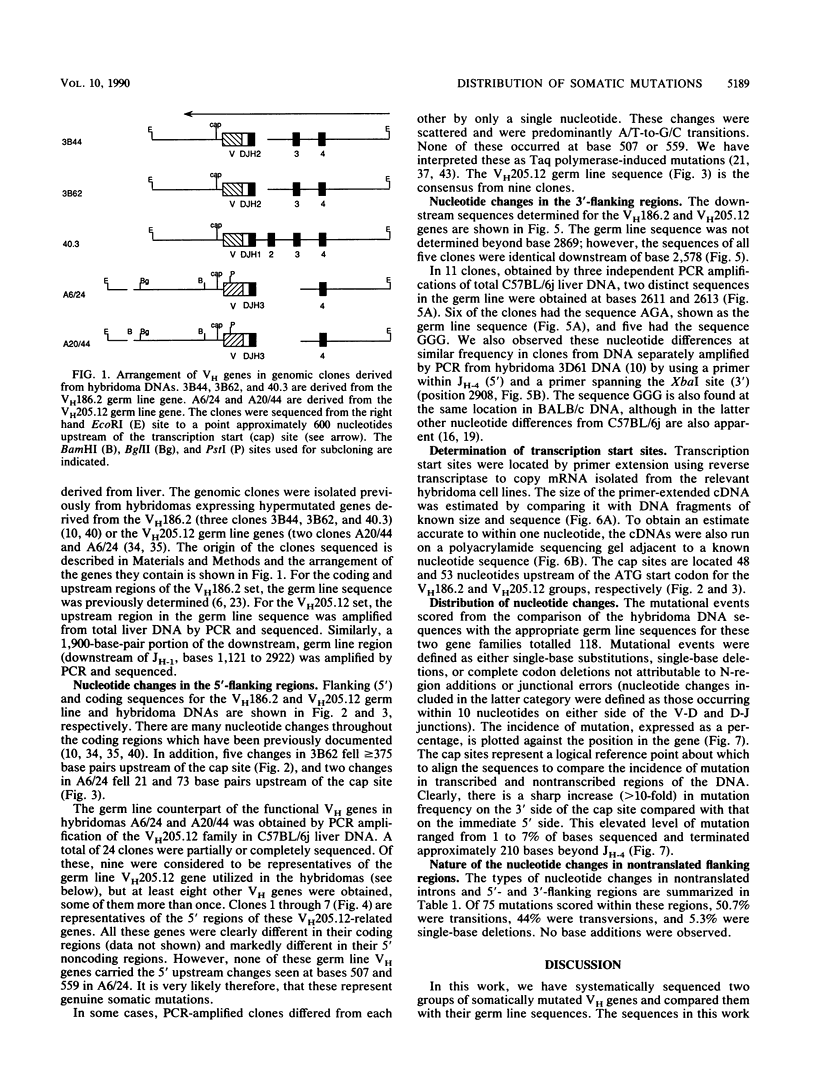
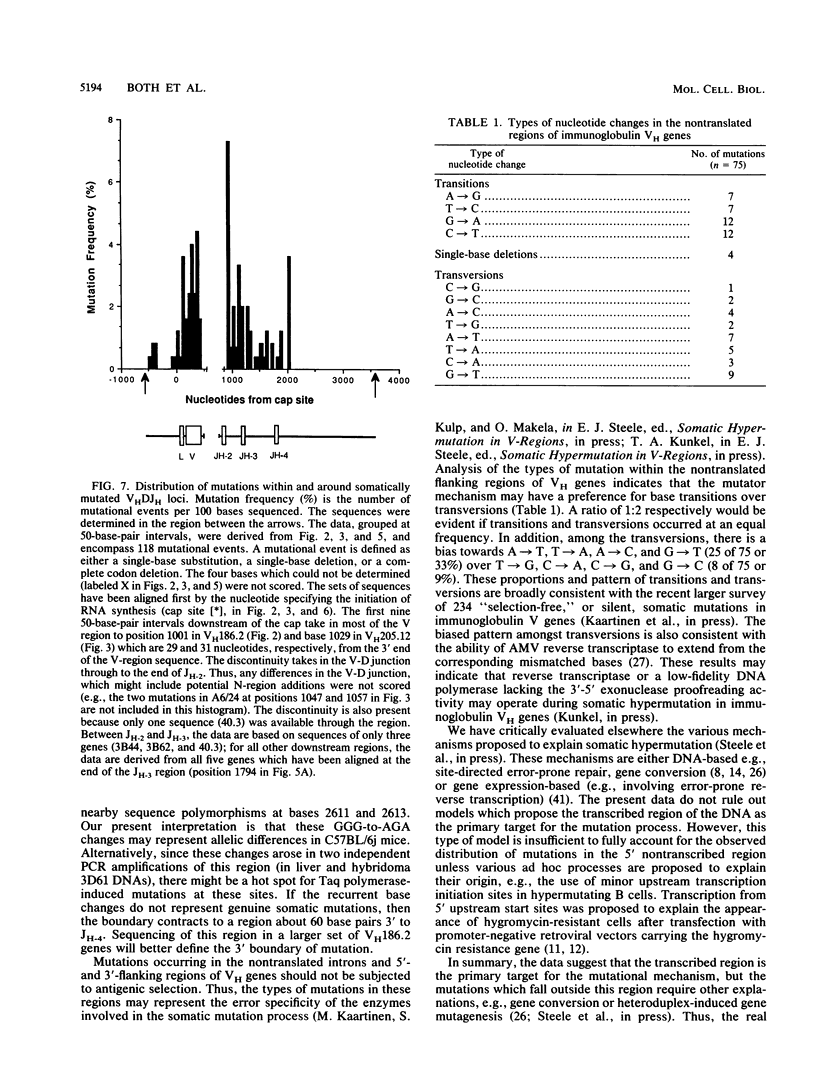
The mechanism of somatic hypermutation in the variable region of immunoglobulin genes expressed in mammalian B cells is a major unexplained phenomenon in the generation of diversity in the immune system. To evaluate possible mechanisms, the distribution of somatic mutations was examined for a group of five cloned, rearranged, somatically mutated VH genes generated in C57BL/6j mice. These mutated VH genes were sequenced and compared with their germ line counterparts from a point approximately 550 base pairs upstream of the transcription start site to an EcoRI site some 1,200 base pairs downstream of JH-4. The location of the transcription start (cap) sites was also precisely determined. Most (greater than or equal to 94%) of the 118 mutations scored occurred between the transcription start site and the distal end of JH-4. However, seven mutations occurred upstream of the transcribed region, and at least four were found downstream of JH-4. The target region for the mutator mechanism therefore clearly extends into the 3' nontranslated and 5' nontranscribed regions. Thus, models which propose the transcribed region of the DNA as the sole substrate for the mutation process are not ruled out but are inadequate to explain the upstream distribution of somatic mutations.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen D., Simon T., Sablitzky F., Rajewsky K., Cumano A. Antibody engineering for the analysis of affinity maturation of an anti-hapten response. EMBO J. 1988 Jul;7(7):1995–2001. doi: 10.1002/j.1460-2075.1988.tb03038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard D. W., Bothwell A. Mutational analysis of the immunoglobulin heavy chain promoter region. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9626–9630. doi: 10.1073/pnas.83.24.9626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Both G. W., Mattick J. S., Bellamy A. R. Serotype-specific glycoprotein of simian 11 rotavirus: coding assignment and gene sequence. Proc Natl Acad Sci U S A. 1983 May;80(10):3091–3095. doi: 10.1073/pnas.80.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothwell A. L., Paskind M., Reth M., Imanishi-Kari T., Rajewsky K., Baltimore D. Heavy chain variable region contribution to the NPb family of antibodies: somatic mutation evident in a gamma 2a variable region. Cell. 1981 Jun;24(3):625–637. doi: 10.1016/0092-8674(81)90089-1. [DOI] [PubMed] [Google Scholar]
- Boyle D. B., Coupar B. E., Gibbs A. J., Seigman L. J., Both G. W. Fowlpox virus thymidine kinase: nucleotide sequence and relationships to other thymidine kinases. Virology. 1987 Feb;156(2):355–365. doi: 10.1016/0042-6822(87)90415-6. [DOI] [PubMed] [Google Scholar]
- Brenner S., Milstein C. Origin of antibody variation. Nature. 1966 Jul 16;211(5046):242–243. doi: 10.1038/211242a0. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Cumano A., Rajewsky K. Clonal recruitment and somatic mutation in the generation of immunological memory to the hapten NP. EMBO J. 1986 Oct;5(10):2459–2468. doi: 10.1002/j.1460-2075.1986.tb04522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornburg R., Temin H. M. Retroviral vector system for the study of cDNA gene formation. Mol Cell Biol. 1988 Jun;8(6):2328–2334. doi: 10.1128/mcb.8.6.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty J. P., Temin H. M. A promoterless retroviral vector indicates that there are sequences in U3 required for 3' RNA processing. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1197–1201. doi: 10.1073/pnas.84.5.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkner F. G., Zachau H. G. Correct transcription of an immunoglobulin kappa gene requires an upstream fragment containing conserved sequence elements. Nature. 1984 Jul 5;310(5972):71–74. doi: 10.1038/310071a0. [DOI] [PubMed] [Google Scholar]
- Gearhart P. J., Bogenhagen D. F. Clusters of point mutations are found exclusively around rearranged antibody variable genes. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3439–3443. doi: 10.1073/pnas.80.11.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies S. D., Morrison S. L., Oi V. T., Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983 Jul;33(3):717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- Golding G. B., Gearhart P. J., Glickman B. W. Patterns of somatic mutations in immunoglobulin variable genes. Genetics. 1987 Jan;115(1):169–176. doi: 10.1093/genetics/115.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski J., Rollini P., Mach B. Somatic mutations of immunoglobulin variable genes are restricted to the rearranged V gene. Science. 1983 Jun 10;220(4602):1179–1181. doi: 10.1126/science.6857243. [DOI] [PubMed] [Google Scholar]
- Gough N. M., Bernard O. Sequences of the joining region genes for immunoglobulin heavy chains and their role in generation of antibody diversity. Proc Natl Acad Sci U S A. 1981 Jan;78(1):509–513. doi: 10.1073/pnas.78.1.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T., Nikaido T., Miyata T., Moriwaki K., Honjo T. The nucleotide sequences of rearranged and germline immunoglobulin VH genes of a mouse myeloma MC101 and evolution of VH genes in mouse. J Biol Chem. 1982 Jan 10;257(1):277–285. [PubMed] [Google Scholar]
- Keohavong P., Thilly W. G. Fidelity of DNA polymerases in DNA amplification. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9253–9257. doi: 10.1073/pnas.86.23.9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Davis M., Sinn E., Patten P., Hood L. Antibody diversity: somatic hypermutation of rearranged VH genes. Cell. 1981 Dec;27(3 Pt 2):573–581. doi: 10.1016/0092-8674(81)90399-8. [DOI] [PubMed] [Google Scholar]
- Krawinkel U., Zoebelein G., Bothwell A. L. Palindromic sequences are associated with sites of DNA breakage during gene conversion. Nucleic Acids Res. 1986 May 12;14(9):3871–3882. doi: 10.1093/nar/14.9.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Exonucleolytic proofreading. Cell. 1988 Jun 17;53(6):837–840. doi: 10.1016/s0092-8674(88)90189-4. [DOI] [PubMed] [Google Scholar]
- Liu Y. J., Joshua D. E., Williams G. T., Smith C. A., Gordon J., MacLennan I. C. Mechanism of antigen-driven selection in germinal centres. Nature. 1989 Dec 21;342(6252):929–931. doi: 10.1038/342929a0. [DOI] [PubMed] [Google Scholar]
- Maizels N. Might gene conversion be the mechanism of somatic hypermutation of mammalian immunoglobulin genes? Trends Genet. 1989 Jan;5(1):4–8. doi: 10.1016/0168-9525(89)90004-8. [DOI] [PubMed] [Google Scholar]
- Mendelman L. V., Petruska J., Goodman M. F. Base mispair extension kinetics. Comparison of DNA polymerase alpha and reverse transcriptase. J Biol Chem. 1990 Feb 5;265(4):2338–2346. [PubMed] [Google Scholar]
- Parslow T. G., Blair D. L., Murphy W. J., Granner D. K. Structure of the 5' ends of immunoglobulin genes: a novel conserved sequence. Proc Natl Acad Sci U S A. 1984 May;81(9):2650–2654. doi: 10.1073/pnas.81.9.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pech M., Höchtl J., Schnell H., Zachau H. G. Differences between germ-line and rearranged immunoglobulin V kappa coding sequences suggest a localized mutation mechanism. Nature. 1981 Jun 25;291(5817):668–670. doi: 10.1038/291668a0. [DOI] [PubMed] [Google Scholar]
- Reanney D. Genetic noise in evolution? 1984 Jan 26-Feb 1Nature. 307(5949):318–319. doi: 10.1038/307318a0. [DOI] [PubMed] [Google Scholar]
- Roes J., Hüppi K., Rajewsky K., Sablitzky F. V gene rearrangement is required to fully activate the hypermutation mechanism in B cells. J Immunol. 1989 Feb 1;142(3):1022–1026. [PubMed] [Google Scholar]
- Sablitzky F., Rajewsky K. Molecular basis of an isogeneic anti-idiotypic response. EMBO J. 1984 Dec 1;3(12):3005–3012. doi: 10.1002/j.1460-2075.1984.tb02247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablitzky F., Wildner G., Rajewsky K. Somatic mutation and clonal expansion of B cells in an antigen-driven immune response. EMBO J. 1985 Feb;4(2):345–350. doi: 10.1002/j.1460-2075.1985.tb03635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sakano H., Maki R., Kurosawa Y., Roeder W., Tonegawa S. Two types of somatic recombination are necessary for the generation of complete immunoglobulin heavy-chain genes. Nature. 1980 Aug 14;286(5774):676–683. doi: 10.1038/286676a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selsing E., Storb U. Somatic mutation of immunoglobulin light-chain variable-region genes. Cell. 1981 Jul;25(1):47–58. doi: 10.1016/0092-8674(81)90230-0. [DOI] [PubMed] [Google Scholar]
- Siekevitz M., Kocks C., Rajewsky K., Dildrop R. Analysis of somatic mutation and class switching in naive and memory B cells generating adoptive primary and secondary responses. Cell. 1987 Mar 13;48(5):757–770. doi: 10.1016/0092-8674(87)90073-0. [DOI] [PubMed] [Google Scholar]
- Tindall K. R., Kunkel T. A. Fidelity of DNA synthesis by the Thermus aquaticus DNA polymerase. Biochemistry. 1988 Aug 9;27(16):6008–6013. doi: 10.1021/bi00416a027. [DOI] [PubMed] [Google Scholar]
- Weiss S., Wu G. E. Somatic point mutations in unrearranged immunoglobulin gene segments encoding the variable region of lambda light chains. EMBO J. 1987 Apr;6(4):927–932. doi: 10.1002/j.1460-2075.1987.tb04840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]