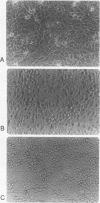
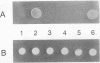
Abstract
The Krev-1 gene has been shown to suppress ras-mediated transformation in vitro. Both ras and Krev-1 proteins have identical effector domains (ras residues 32 to 40), which are required for biological activity and for the interaction of Ras p21 with Ras GTPase-activating protein (GAP). In this study, five amino acid residues flanking the ras effector domain, which are not conserved with the Krev-1 protein, were shown to be required for normal protein-protein interactions and biological activity. The substitution of Krev-1 p21 residues 26, 27, 30, 31, and 45 with the corresponding amino acid residues from Ras p21 resulted in a Krev-1 protein which had ras function in both mammalian and yeast biological assays. Replacement of these residues in Ras p21 with the corresponding Krev-1 p21 amino acids resulted in ras proteins which were impaired biologically or reduced in their affinity for in vitro GAP binding. Evaluation of these mutant ras proteins have implications for Ras p21-GAP interactions in vivo.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adari H., Lowy D. R., Willumsen B. M., Der C. J., McCormick F. Guanosine triphosphatase activating protein (GAP) interacts with the p21 ras effector binding domain. Science. 1988 Apr 22;240(4851):518–521. doi: 10.1126/science.2833817. [DOI] [PubMed] [Google Scholar]
- Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- Breviario D., Hinnebusch A., Cannon J., Tatchell K., Dhar R. Carbon source regulation of RAS1 expression in Saccharomyces cerevisiae and the phenotypes of ras2- cells. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4152–4156. doi: 10.1073/pnas.83.12.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broek D., Samiy N., Fasano O., Fujiyama A., Tamanoi F., Northup J., Wigler M. Differential activation of yeast adenylate cyclase by wild-type and mutant RAS proteins. Cell. 1985 Jul;41(3):763–769. doi: 10.1016/s0092-8674(85)80057-x. [DOI] [PubMed] [Google Scholar]
- Buss J. E., Quilliam L. A., Kato K., Casey P. J., Solski P. A., Wong G., Clark R., McCormick F., Bokoch G. M., Der C. J. The COOH-terminal domain of the Rap1A (Krev-1) protein is isoprenylated and supports transformation by an H-Ras:Rap1A chimeric protein. Mol Cell Biol. 1991 Mar;11(3):1523–1530. doi: 10.1128/mcb.11.3.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calés C., Hancock J. F., Marshall C. J., Hall A. The cytoplasmic protein GAP is implicated as the target for regulation by the ras gene product. Nature. 1988 Apr 7;332(6164):548–551. doi: 10.1038/332548a0. [DOI] [PubMed] [Google Scholar]
- Cannon J. F., Gibbs J. B., Tatchell K. Suppressors of the ras2 mutation of Saccharomyces cerevisiae. Genetics. 1986 Jun;113(2):247–264. doi: 10.1093/genetics/113.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vendittis E., Vitelli A., Zahn R., Fasano O. Suppression of defective RAS1 and RAS2 functions in yeast by an adenylate cyclase activated by a single amino acid change. EMBO J. 1986 Dec 20;5(13):3657–3663. doi: 10.1002/j.1460-2075.1986.tb04696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downward J., Graves J. D., Warne P. H., Rayter S., Cantrell D. A. Stimulation of p21ras upon T-cell activation. Nature. 1990 Aug 23;346(6286):719–723. doi: 10.1038/346719a0. [DOI] [PubMed] [Google Scholar]
- Ellis C., Moran M., McCormick F., Pawson T. Phosphorylation of GAP and GAP-associated proteins by transforming and mitogenic tyrosine kinases. Nature. 1990 Jan 25;343(6256):377–381. doi: 10.1038/343377a0. [DOI] [PubMed] [Google Scholar]
- Farnsworth C. L., Marshall M. S., Gibbs J. B., Stacey D. W., Feig L. A. Preferential inhibition of the oncogenic form of RasH by mutations in the GAP binding/"effector" domain. Cell. 1991 Feb 8;64(3):625–633. doi: 10.1016/0092-8674(91)90246-u. [DOI] [PubMed] [Google Scholar]
- Fasano O., Crechet J. B., De Vendittis E., Zahn R., Feger G., Vitelli A., Parmeggiani A. Yeast mutants temperature-sensitive for growth after random mutagenesis of the chromosomal RAS2 gene and deletion of the RAS1 gene. EMBO J. 1988 Nov;7(11):3375–3383. doi: 10.1002/j.1460-2075.1988.tb03210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field J., Broek D., Kataoka T., Wigler M. Guanine nucleotide activation of, and competition between, RAS proteins from Saccharomyces cerevisiae. Mol Cell Biol. 1987 Jun;7(6):2128–2133. doi: 10.1128/mcb.7.6.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frech M., John J., Pizon V., Chardin P., Tavitian A., Clark R., McCormick F., Wittinghofer A. Inhibition of GTPase activating protein stimulation of Ras-p21 GTPase by the Krev-1 gene product. Science. 1990 Jul 13;249(4965):169–171. doi: 10.1126/science.2164710. [DOI] [PubMed] [Google Scholar]
- Gibbs J. B., Marshall M. S., Scolnick E. M., Dixon R. A., Vogel U. S. Modulation of guanine nucleotides bound to Ras in NIH3T3 cells by oncogenes, growth factors, and the GTPase activating protein (GAP). J Biol Chem. 1990 Nov 25;265(33):20437–20442. [PubMed] [Google Scholar]
- Gibbs J. B., Marshall M. S. The ras oncogene--an important regulatory element in lower eucaryotic organisms. Microbiol Rev. 1989 Jun;53(2):171–185. doi: 10.1128/mr.53.2.171-185.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs J. B., Schaber M. D., Allard W. J., Sigal I. S., Scolnick E. M. Purification of ras GTPase activating protein from bovine brain. Proc Natl Acad Sci U S A. 1988 Jul;85(14):5026–5030. doi: 10.1073/pnas.85.14.5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs J. B., Sigal I. S., Poe M., Scolnick E. M. Intrinsic GTPase activity distinguishes normal and oncogenic ras p21 molecules. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5704–5708. doi: 10.1073/pnas.81.18.5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock J. F., Magee A. I., Childs J. E., Marshall C. J. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell. 1989 Jun 30;57(7):1167–1177. doi: 10.1016/0092-8674(89)90054-8. [DOI] [PubMed] [Google Scholar]
- Hata Y., Kikuchi A., Sasaki T., Schaber M. D., Gibbs J. B., Takai Y. Inhibition of the ras p21 GTPase-activating protein-stimulated GTPase activity of c-Ha-ras p21 by smg p21 having the same putative effector domain as ras p21s. J Biol Chem. 1990 May 5;265(13):7104–7107. [PubMed] [Google Scholar]
- Housey G. M., Johnson M. D., Hsiao W. L., O'Brian C. A., Murphy J. P., Kirschmeier P., Weinstein I. B. Overproduction of protein kinase C causes disordered growth control in rat fibroblasts. Cell. 1988 Feb 12;52(3):343–354. doi: 10.1016/s0092-8674(88)80027-8. [DOI] [PubMed] [Google Scholar]
- Kataoka T., Powers S., Cameron S., Fasano O., Goldfarb M., Broach J., Wigler M. Functional homology of mammalian and yeast RAS genes. Cell. 1985 Jan;40(1):19–26. doi: 10.1016/0092-8674(85)90304-6. [DOI] [PubMed] [Google Scholar]
- Kawata M., Farnsworth C. C., Yoshida Y., Gelb M. H., Glomset J. A., Takai Y. Posttranslationally processed structure of the human platelet protein smg p21B: evidence for geranylgeranylation and carboxyl methylation of the C-terminal cysteine. Proc Natl Acad Sci U S A. 1990 Nov;87(22):8960–8964. doi: 10.1073/pnas.87.22.8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawata M., Matsui Y., Kondo J., Hishida T., Teranishi Y., Takai Y. A novel small molecular weight GTP-binding protein with the same putative effector domain as the ras proteins in bovine brain membranes. Purification, determination of primary structure, and characterization. J Biol Chem. 1988 Dec 15;263(35):18965–18971. [PubMed] [Google Scholar]
- Kim S., Mizoguchi A., Kikuchi A., Takai Y. Tissue and subcellular distributions of the smg-21/rap1/Krev-1 proteins which are partly distinct from those of c-ras p21s. Mol Cell Biol. 1990 Jun;10(6):2645–2652. doi: 10.1128/mcb.10.6.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayama H., Matsuzaki T., Ikawa Y., Noda M. Genetic analysis of the Kirsten-ras-revertant 1 gene: potentiation of its tumor suppressor activity by specific point mutations. Proc Natl Acad Sci U S A. 1990 Jun;87(11):4284–4288. doi: 10.1073/pnas.87.11.4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayama H., Sugimoto Y., Matsuzaki T., Ikawa Y., Noda M. A ras-related gene with transformation suppressor activity. Cell. 1989 Jan 13;56(1):77–84. doi: 10.1016/0092-8674(89)90985-9. [DOI] [PubMed] [Google Scholar]
- Krengel U., Schlichting I., Scherer A., Schumann R., Frech M., John J., Kabsch W., Pai E. F., Wittinghofer A. Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules. Cell. 1990 Aug 10;62(3):539–548. doi: 10.1016/0092-8674(90)90018-a. [DOI] [PubMed] [Google Scholar]
- Marshall M. S., Gibbs J. B., Scolnick E. M., Sigal I. S. An adenylate cyclase from Saccharomyces cerevisiae that is stimulated by RAS proteins with effector mutations. Mol Cell Biol. 1988 Jan;8(1):52–61. doi: 10.1128/mcb.8.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall M. S., Hill W. S., Ng A. S., Vogel U. S., Schaber M. D., Scolnick E. M., Dixon R. A., Sigal I. S., Gibbs J. B. A C-terminal domain of GAP is sufficient to stimulate ras p21 GTPase activity. EMBO J. 1989 Apr;8(4):1105–1110. doi: 10.1002/j.1460-2075.1989.tb03480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay I. A., Marshall C. J., Calés C., Hall A. Transformation and stimulation of DNA synthesis in NIH-3T3 cells are a titratable function of normal p21N-ras expression. EMBO J. 1986 Oct;5(10):2617–2621. doi: 10.1002/j.1460-2075.1986.tb04542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy C. J., Bottaro D. P., Fleming T. P., Marshall M. S., Gibbs J. B., Aaronson S. A. PDGF induction of tyrosine phosphorylation of GTPase activating protein. Nature. 1989 Dec 7;342(6250):711–714. doi: 10.1038/342711a0. [DOI] [PubMed] [Google Scholar]
- Pai E. F., Kabsch W., Krengel U., Holmes K. C., John J., Wittinghofer A. Structure of the guanine-nucleotide-binding domain of the Ha-ras oncogene product p21 in the triphosphate conformation. Nature. 1989 Sep 21;341(6239):209–214. doi: 10.1038/341209a0. [DOI] [PubMed] [Google Scholar]
- Pizon V., Chardin P., Lerosey I., Olofsson B., Tavitian A. Human cDNAs rap1 and rap2 homologous to the Drosophila gene Dras3 encode proteins closely related to ras in the 'effector' region. Oncogene. 1988 Aug;3(2):201–204. [PubMed] [Google Scholar]
- Satoh T., Endo M., Nakafuku M., Nakamura S., Kaziro Y. Platelet-derived growth factor stimulates formation of active p21ras.GTP complex in Swiss mouse 3T3 cells. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5993–5997. doi: 10.1073/pnas.87.15.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaber M. D., Garsky V. M., Boylan D., Hill W. S., Scolnick E. M., Marshall M. S., Sigal I. S., Gibbs J. B. Ras interaction with the GTPase-activating protein (GAP). Proteins. 1989;6(3):306–315. doi: 10.1002/prot.340060313. [DOI] [PubMed] [Google Scholar]
- Sigal I. S., Gibbs J. B., D'Alonzo J. S., Scolnick E. M. Identification of effector residues and a neutralizing epitope of Ha-ras-encoded p21. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4725–4729. doi: 10.1073/pnas.83.13.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal I. S., Marshall M. S., Schaber M. D., Vogel U. S., Scolnick E. M., Gibbs J. B. Structure/function studies of the ras protein. Cold Spring Harb Symp Quant Biol. 1988;53(Pt 2):863–869. doi: 10.1101/sqb.1988.053.01.099. [DOI] [PubMed] [Google Scholar]
- Temeles G. L., Gibbs J. B., D'Alonzo J. S., Sigal I. S., Scolnick E. M. Yeast and mammalian ras proteins have conserved biochemical properties. Nature. 1985 Feb 21;313(6004):700–703. doi: 10.1038/313700a0. [DOI] [PubMed] [Google Scholar]
- Vogel U. S., Dixon R. A., Schaber M. D., Diehl R. E., Marshall M. S., Scolnick E. M., Sigal I. S., Gibbs J. B. Cloning of bovine GAP and its interaction with oncogenic ras p21. Nature. 1988 Sep 1;335(6185):90–93. doi: 10.1038/335090a0. [DOI] [PubMed] [Google Scholar]
- Willumsen B. M., Papageorge A. G., Kung H. F., Bekesi E., Robins T., Johnsen M., Vass W. C., Lowy D. R. Mutational analysis of a ras catalytic domain. Mol Cell Biol. 1986 Jul;6(7):2646–2654. doi: 10.1128/mcb.6.7.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G. F., Lin B., Tanaka K., Dunn D., Wood D., Gesteland R., White R., Weiss R., Tamanoi F. The catalytic domain of the neurofibromatosis type 1 gene product stimulates ras GTPase and complements ira mutants of S. cerevisiae. Cell. 1990 Nov 16;63(4):835–841. doi: 10.1016/0092-8674(90)90149-9. [DOI] [PubMed] [Google Scholar]
- Zhang K., Noda M., Vass W. C., Papageorge A. G., Lowy D. R. Identification of small clusters of divergent amino acids that mediate the opposing effects of ras and Krev-1. Science. 1990 Jul 13;249(4965):162–165. doi: 10.1126/science.2115210. [DOI] [PubMed] [Google Scholar]
- de Vos A. M., Tong L., Milburn M. V., Matias P. M., Jancarik J., Noguchi S., Nishimura S., Miura K., Ohtsuka E., Kim S. H. Three-dimensional structure of an oncogene protein: catalytic domain of human c-H-ras p21. Science. 1988 Feb 19;239(4842):888–893. doi: 10.1126/science.2448879. [DOI] [PubMed] [Google Scholar]