Abstract
Dietary intake of macronutrients (carbohydrate, protein, and fat) has been associated with risk of chronic conditions such as obesity and diabetes. Family studies have reported a moderate contribution of genetics to variation in macronutrient intake. In a genome-wide meta-analysis of a population-based discovery cohort (n = 33 533), rs838133 in FGF21 (19q13.33), rs197273 near TRAF family member-associated NF-kappa-B activator (TANK) (2p24.2), and rs10163409 in FTO (16q12.2) were among the top associations (P < 10−5) for percentage of total caloric intake from protein and carbohydrate. rs838133 was replicated in silico in an independent sample from the Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium (CHARGE) Nutrition Working Group (n = 38 360) and attained genome-wide significance in combined analysis (Pjoint = 7.9 × 10−9). A cytokine involved in cellular metabolism, FGF21 is a potential susceptibility gene for obesity and type 2 diabetes. Our results highlight the potential of genetic variation for determining dietary macronutrient intake.
INTRODUCTION
Dietary intake is tightly related to human health and excess intake of energy has been associated with obesity, diabetes and cardiovascular disease, as well as many other chronic diseases in epidemiology studies (1). Total energy intake can be split into energy derived from three primary macronutrients: fat, protein, and carbohydrate. In spite of wide variety in food ingredients and preparation according to culture and geography, mean proportions of dietary macronutrient intake are relatively constant across many populations from industrialized nations providing ∼40–50, 10–20 and 25–40% of total caloric intake from carbohydrate, protein and fat, respectively (1). Inter-individual variation in macronutrient intake remains substantial however, and is heritable as judged by family and twin studies, although the range of heritability estimates is broad at 11–65% (2). Prior approaches to discovering genetic influences on macronutrient intake have focused on candidate hypotheses related to metabolic function (3) and taste (4), and have suggested some genetic links between dietary habits, obesity and diabetes. However, there have been no genome-wide association studies (GWASs) investigating macronutrient intake to date. To gain additional insights into genetic control of macronutrient intake, we conducted a GWAS by performing a genome-wide meta-analysis among population-based cohorts of European ancestry.
RESULTS
We performed a genome-wide meta-analysis of 33 355 male and female participants of European ancestry in DietGen, a consortium consisting of three large population-based studies investigating the genetics of dietary intake and nutrition (the Women's Genome Health Study: WGHS, n = 22 691; the Health Professionals Follow-up Study: HPFS, n = 4077 among three nested case–control studies and the Nurses’ Health Study: NHS, n = 6765 among four nested case–control studies; Supplementary Material, Table S1). The proportion of total energy derived from each of the three macronutrients as a percentage was estimated from similarly designed, self-administered, semi-quantitative and highly validated food frequency questionnaires (FFQs) (5) in all the three cohorts. Discovery-stage genome-wide association scans and meta-analyses across the three cohorts were performed for all three macronutrient phenotypes adjusted for age, location, sub-population stratification, with and without adjustment for body mass index (BMI). BMI adjustments were performed to decrease variance of the macronutrient phenotypes and to account for genetic effects mediated through body composition.
A genome-wide significant (P < 5 × 10−8) association was identified for SNPs in an intron of the FTO gene at 16q12.2 with percentage of total caloric intake from carbohydrate in models with or without BMI adjustment (index SNP rs10163409, P = 3.4 × 10−8 and P = 7.4 × 10−9, respectively; Table 1), although this association appeared to be driven by the WGHS cohort (P = 5.7 × 10−9 and P = 1.2 × 10−9 with and without adjustment for BMI; see Supplementary Material, Figure S1). A total of 22 independent SNPs (see Methods) were associated with macronutrient intake with or without BMI adjustment at sub-genome-wide significance (5 × 10−8 < P < 1 × 10−5); 13 for percentage carbohydrate intake, 7 for percentage protein intake, and 2 for percentage fat intake (Table 1, Supplementary Material, Table S2 and Figure S2).
Table 1.
Genome-wide association results from meta-analysis for percentage of total caloric intake from macronutrients (carbohydrate and protein) in the discovery (DietGen), replication (CHARGE) and combined (Joint) cohort analyses
| Model | SNP | Gene | Chr | BP | EA/ nonEA | EAF | DietGen |
CHARGE |
Joint Analysis |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SE | P(2GC) | Beta | SE | P(2GC) | one-sided P | Beta | SE | P | N | |||||||
| Carbohydrate | |||||||||||||||||
| Model1a | rs10163409 | FTO | 16 | 52 696 381 | A/T | 0.69 | 0.44 | 0.08 | 7.4E−09 | −0.03 | 0.07 | 6.9E−01 | 3.4E−01 | 0.19 | 0.05 | 2.2E−04 | 71 885 |
| Model2b | rs197273 | TANK | 2 | 161 602 909 | A/G | 0.48 | 0.31 | 0.06 | 2.0E−06 | 0.17 | 0.06 | 5.7E−03 | 2.9E−03 | 0.23 | 0.04 | 9.6E−08 | 71 328 |
| rs10163409 | FTO | 16 | 52 696 381 | A/T | 0.69 | 0.42 | 0.07 | 3.4E−08 | −0.05 | 0.07 | 4.6E−01 | 2.3E−01 | 0.17 | 0.05 | 1.1E−03 | 71 326 | |
| Protein | |||||||||||||||||
| Model2b | rs838133 | FGF21 | 19 | 53 951 341 | A/G | 0.45 | −0.12 | 0.03 | 2.3E−06 | −0.10 | 0.03 | 1.5E−03 | 7.3E−04 | −0.11 | 0.02 | 7.9E−09 | 68 968 |
CHARGE, Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium; SNP, single-nucleotide polymorphism; Chr, chromosome; BP, base pair position—build 36; EA, effect allele; nonEA, non-effect allele; EAF, effect allele frequency; beta, beta-coefficient; SE, standard error; P(2GC), P-value corrected for genomic control; one-sided P, one-sided P-value; P, P-value; N, sample size
aModel 1 adjusts for age, sex (in CHARGE), location and sub-population stratification.
bModel 2 adjusts for model 1 covariates in addition to BMI (kg/m2).
The most significantly associated variant at each locus was further evaluated through in silico replication from a parallel genome-wide meta-analysis of 38 360 samples among 12 cohorts from the Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium (CHARGE) Nutrition Working Group (6). The sub-genome-wide association between rs838133 in the FGF21 gene at 19q13.33 and percentage protein intake adjusted for BMI from DietGen was replicated in a model adjusted for BMI after correcting for multiple hypothesis testing (Pone-sided = 7.2 × 10−4; αprotein = 7.1 × 10−3 = 0.05/7 protein loci), and attained a genome-wide significant P-value in a joint inverse-variance weighted meta-analysis combining the discovery cohorts with the replication cohorts (Pjoint = 7.9 × 10−9). In the replication cohorts, each copy of the minor allele was associated with a decrease of 0.098% (SE 0.030%, P = 7.3 × 10−4) protein intake (Table 1). The sub-genome-wide association of percentage carbohydrate intake adjusted for BMI with rs197273 ∼100 kb to the 5′ end of the TANK gene on chromosome 2q24 was replicated in a model adjusting for BMI after correction for multiple hypothesis testing (Pone-sided = 2.9 × 10−3; αcarbohydrate = 3.8 × 10−3 = 0.05/13 carbohydrate loci), but did not attain genome-wide significance in the joint analysis (Pjoint = 9.6 × 10−8). The genome-wide significant association between the FTO variant rs10163409 and percentage carbohydrate intake was not replicated by CHARGE either with or without BMI adjustment (P = 0.34 and PBMI adjusted = 0.23, respectively) (Table 1). For this SNP, there was evidence of moderate heterogeneity in CHARGE (I2 = 34.8 and 32.3%, unadjusted and adjusted for BMI, respectively, Supplementary Material, Table S2).
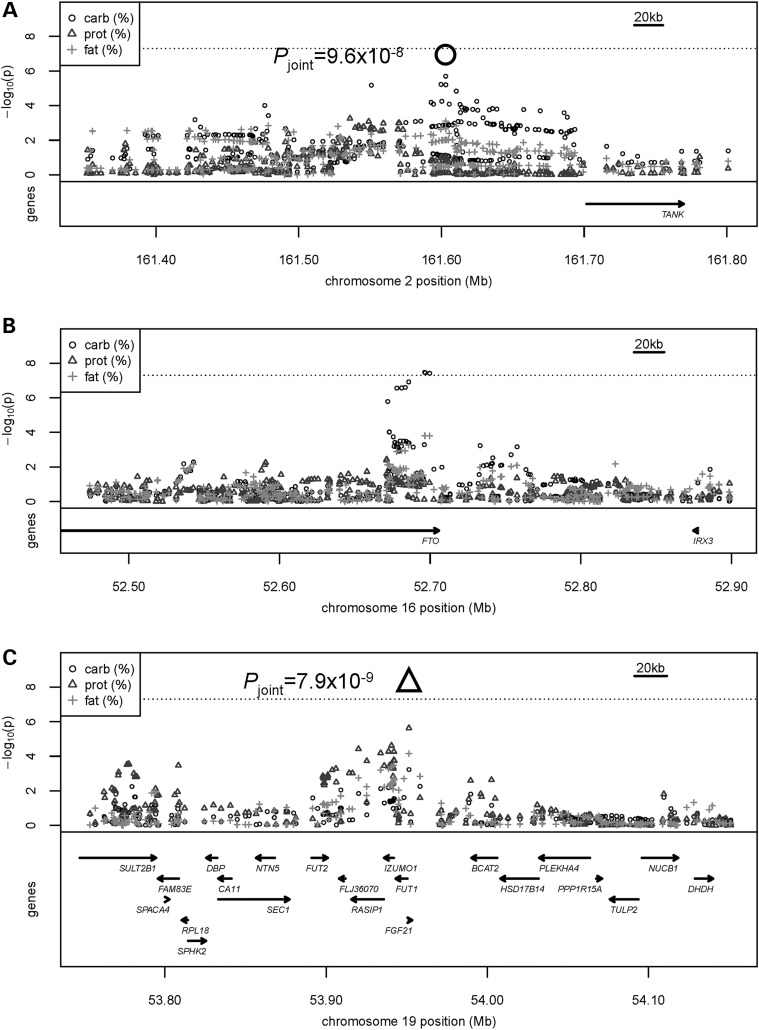
Together, the three macronutrients account for the majority of caloric intake, raising the potential for simultaneous associations between the lead SNPs and more than one macronutrient. In discovery, the variant rs838133 at the FGF21 locus, which was identified in association with decreased protein intake, was also associated with increased carbohydrate intake (beta[se]: 0.23% [0.07%], P = 5.9 × 10−4) and decreased fat intake (beta[se]: −0.21% [0.05%], P = 7.1 × 10−5) in models adjusted for BMI (Fig. 1). The variant rs10163409 in FTO, which was discovered in association with increased percentage carbohydrate intake in models adjusted for BMI, was associated with decreased fat intake (beta[se]: −0.22%[0.05%], P = 1.6 × 10−4), but not associated with protein intake (beta[se]: −0.05%[0.03%], P = 0.08; Fig. 1). Variant rs197273 near the TANK gene, in addition to a sub-genome-wide association with increased carbohydrate intake, was significantly associated with decreased fat intake (beta[se]: −0.17%[0.05%], P = 0.7.8 × 10−4) and marginally associated with decreased protein intake (beta[se]: −0.05%[0.02%], P = 0.068) in models adjusted for BMI. Associations at all three loci were similar in the absence of BMI adjustment (Supplementary Material, Table S3).
Figure 1.
Regional association plots showing −log10 P-values for percentage of total energy intake from macronutrients. Closest genes and -log10 P-values for SNPs at (A) chromosome 2q24, (B) chromosome16q12 and (C) chromosome 19q13 in association with percentage of total energy intake from carbohydrate (circles), protein (triangles) and fat (crosses) adjusted for BMI in the discovery cohort. The P-values from joint meta-analysis of DietGen and CHARGE Nutrition are indicated for association between (A) rs197273 and carbohydrate intake by the bolded circle and (C) rs838133 and protein intake by the bolded triangle. The threshold for genome-wide significance (P < 5 × 10−8) is indicated by the horizontal dotted line.
To find evidence for additional sub-genome-wide associations, we estimated the proportion of variance in macronutrient intake in the WGHS using GCTA (7) (see Methods). This procedure found that 6.6% (SE 2.1%, P = 9.1 × 10−5), 8.0% (SE 2.2%, P = 2.7 × 10−5) and 7.3% (SE 2.2%, P = 3.8 × 10−5) of the variance in total energy from carbohydrate, protein and fat, respectively, could be explained by the common WGHS tag-SNPs. By comparison, SNP rs838133 in FGF21 explained 0.052, 0.062, and 0.057% of the variance in carbohydrate, protein, and fat intake, respectively, in the WGHS. Adjustment for age, location and population substructure as measured by 10 eigenvectors did not substantially affect the estimated genetic contribution to the variance, nor did additional adjustment for BMI (Supplementary Material, Table S4). Partitioning the contribution to the variance according to MAF suggested that the estimates were mostly due to the SNPs with MAF > 5% compared with less common SNPs with MAF between 1 and 5% (Supplementary Material, Table S4).
DISCUSSION
We performed a two-stage genome-wide meta-analysis of percentage of total energy intake from carbohydrate, protein and fat among 33 533 participants with independent replication in a sample of equivalent size. The replication yielded a locus at 19q33 (rs838133) associated with protein intake at genome-wide significance, and provided nominal support for a second locus at 2q24 (rs197273) associated with carbohydrate intake. In addition, heritability analyses in a subset of the discovery cohort showed that common SNPs explained a modest but statistically significant proportion of genetic variance for carbohydrate, protein and fat intake (6–8%).
Variant rs838133 maps to exon 1 of the FGF21 gene, but also maps in the vicinity of a gene-dense region on chromosome 19 that includes other genes FUT1, FUT2, IZUMO1 and RASIP1—all within 100 kb. While we cannot exclude the possibility this variant could be influencing other genes in this region, FGF21 is a strong candidate due to its biology and location. Additional potential SNP associations in the region were fully attenuated by conditional meta-analysis of rs838133, reinforcing choice of FGF21 as the candidate gene. rs838133 is a synonymous SNP located in the first exon of the FGF21 gene encoding fibroblast growth factor 21. Secreted by hepatocytes and adipocytes, FGF21 promotes glucose uptake in adipocytes and has other functions in cellular metabolism, such as regulation of carbohydrate and lipid metabolism (8). FGF21 expression can be induced through fasting or feeding in animal models (9). Serum levels of FGF21 have been associated with increased odds of type 2 diabetes (10,11) and obesity(12), and there is suggestive evidence that human FGF21 infusion into rodents may regulate food intake(13,14), although the effects were not consistently observed (15). And FGF21 protein is being investigated as a potential agent for pharmacologic intervention targeting these conditions (15).
A different variant at the FGF21 locus was identified from the CHARGE Nutrition Working Group meta-analysis (6) in association with carbohydrate and fat intake at genome-wide significance, whereas the FGF21 locus was associated with protein intake in the DietGen analysis. The FGF21 variants identified in the DietGen and CHARGE analyses are in moderate linkage disequilibrium (r2 = 0.7, 1000 Genomes) and likely represent the same signal. Because macronutrient intake is expressed as a proportion of the total caloric intake, a SNP association with one macronutrient could imply association with one or both of the others. The FGF21 variant identified in DietGen in association with percentage protein intake was also associated with carbohydrate and fat intake at a lower significance level in our analyses. Which macronutrient is identified with the greatest significance in one study or another may reflect differences in instruments used for nutritional assessment, true differences in the dietary habits of the underlying populations, or genetic influence on the composition of macronutrient intake rather than individual macronutrient intake, among other explanations. All of these possibilities still support the association of the FGF21 variant with macronutrient intake.
However, there are some published findings that may be inconsistent with the identification of FGF21 as the causal gene. For example, rs838133 was mapped as an eQTL for FUT2, RASIP1 and NTN5 (P < 3.8 × 10−5) in skin tissue, but not adipose tissue or lymphoblasts (16, 17). RASIP1 and NTN5 have not been associated with dietary intake, anthropometric measures or metabolic disorders, but variants in FUT2 have been associated with plasma vitamin B-12 levels (18), and the FUT2 polymorphism rs601338 (r2 = 0.39 with rs838133) is known to determine glycoprotein and glycolipid secretor phenotype (19). Taken together the evidence suggests that there remains some ambiguity to the causality of FGF21, and indicates that while FGF21 is a top candidate for macronutrient intake at this locus, other genes in the region cannot be entirely excluded.
It is unlikely that the association between the FGF21 locus and protein intake in our meta-analysis could be mediated or confounded by BMI, or other related characteristics. We did not observe any association between rs838133 and BMI within the individual DietGen cohorts, and the variant is only nominally associated with BMI in the publically available summary statistics from the most recent GIANT meta-analysis (P = 0.027, n = 123 702) (20). Currently available GIANT results do not allow investigation of whether this weak effect is mediated by dietary intake. In addition, no FGF21 candidate variants reached genome-wide significance in the recent DIAGRAM+ meta-analysis of 34 840 type 2 diabetes cases and 114 981 controls (21). Furthermore, the DietGen meta-analysis results for FGF21 were stronger with the inclusion of BMI than without (beta[se]: −0.124[0.026], P = 2.3 × 10−6; beta[se]: −0.115[0.026], P = 1.1 × 10−5, respectively). Sensitivity analyses from the WGHS demonstrated that this association was robust to adjustment for BMI, baseline diabetes status, physical activity, current smoking and daily alcohol intake (data not shown). Residual confounding, if any, is unlikely expected to be mediated through clinical characteristics typically thought to influence the diet.
TANK, the nearest gene to rs197273 on chromosome 2 encodes a signaling protein that inhibits tumor necrosis factor receptor-associated factor (TRAF) function. The TRAF family of proteins plays a role in inflammation and immune response, and has not been previously linked to regulation of dietary macronutrient intake.
Although the FTO locus at 16q12 is strongly and consistently associated with BMI and adiposity, and there is evidence that it influences dietary intake and energy balance, the genome-wide significant association of rs10163409 with percentage carbohydrate was not replicated by the independent meta-analysis from the CHARGE Nutrition Working Group. Heterogeneity for rs10163409 was estimated to be qualitatively larger among the CHARGE cohorts (I2 = 34.8% and I2BMI adjusted = 32.3%) compared with the DietGen cohorts (I2 = 8.7%, and I2BMI adjusted = 19.9%). It is possible that this heterogeneity may have partially influenced the lack of replication at this locus. Furthermore, rs10163409 is not in LD with SNPs at FTO that are strongly associated with BMI (rs1558902, r2 = 0.07; 1000 Genomes), including in the DietGen cohorts (20), nor is it associated with BMI in the individual DietGen cohorts (data not shown). These results suggest that there may be additional effects of FTO on dietary composition and intake unrelated to its effect on the body size and composition.
Unhealthy habitual dietary behaviors especially recent changes in the proportion of macronutrients consumed underlie the current epidemic of obesity and its associated risk of metabolic diseases, including cardiovascular disease and type 2 diabetes (22). The present work, together with the study by Tanaka et al. (6) on behalf of the CHARGE Nutrition Working Group, represents the first and largest genetic investigations of macronutrient intake on a genome-wide scale. Our results demonstrate associations with macronutrient intake at two loci, one at genome-wide significance, while using heritability analysis to infer that additional genetic associations with macronutrient intake remain to be discovered. The proportion of variance explained in the GCTA analysis likely underestimates the actual heritability due to residual imprecision in measuring macronutrient intake from FFQs and to intrinsic limitations in using tag rather than causal SNPs to assess the correlation between relatedness and macronutrient intake (23). In sum, our results provide a compelling impetus for learning more about the biological function of FGF21 and for performing additional analysis to further delineate the genetic determinants of macronutrient intake.
MATERIALS AND METHODS
Study populations
Discovery cohorts
DietGen consortium
DietGen is composed of three US population-based cohorts: HPFS, NHS and WGHS investigating the genetics of dietary intake and nutrition.
Health professionals follow-up study (HPFS)
The study population was derived from the HPFS, a prospective cohort of 51 529 male health care professionals between the ages of 40–75 years at enrollment in 1986 (24). Since the inception of the study, data on lifestyle and medical history have been ascertained through a self-administered questionnaire, including a semi-quantitative FFQ every 4 years. For this analysis, we included participants who completed the 1986 FFQ. Participants of validated self-reported European ancestry from three sets of the GWAS from nested case-control studies were included in this analysis: type 2 diabetes (25) (HPFS T2D, n = 2316), coronary heart disease (26) (HPFS CHD, n = 1212), and kidney stones (27) (HPFS KS, n = 549). The study was approved by the Institutional Review Board (IRB) of the Harvard School of Public Health.
Nurses’ health study (NHS)
The study population was derived from the NHS, a prospective cohort of 121 700 female registered nurses between the ages of 30–55 years at enrollment in 1976 (28). Since the inception of the NHS, data on lifestyle and medical history have been ascertained through a self-administered questionnaire, including a semi-quantitative FFQ every 2–4 years. For this analysis, we included participants who responded to the 1986 FFQ. Participants of validated self-reported European ancestry from four sets of the GWAS from nested case–control studies were included in this analysis: type 2 diabetes (25) (NHS T2D, n = 3219), coronary heart disease (26) (NHS CHD, n = 1013), kidney stones (27) (NHS KS, n = 490) and breast cancer (29) (NHS BC, n = 2043). The study was approved by the IRB of the Harvard School of Public Health.
Women's genome health study (WGHS)
The study population was derived from the WGHS (30), a prospective cohort of women from the Women's Health Study aged 45 years or older with no history of cardiovascular disease, cancer or major chronic illness, and who provided blood samples at baseline from which genomic DNA was extracted. Individuals of confirmed self-reported European ancestry and had genotyping information available were included in this analysis (n = 23 294). Among these participants, 22 691 provided FFQ data at baseline. The study was approved by the IRB of Brigham and Women's Hospital.
Replication cohorts
Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium (CHARGE) (31). A subset of CHARGE, the CHARGE Nutrition Working Group, is composed of 12 cohorts from the United States and Europe with a total sample size of 38 360 (6) and includes the Atherosclerosis Risk in Communities Study (ARIC), Cardiovascular Health Study (CHS), The European Prospective Investigation of Cancer-Norfolk (EPIC-Norfolk), Family Heart Study, Fenland study, Framingham Heart Study, Genetics of Lipid Lowering Drugs and Diet Network study (GOLDN), Health Aging and Body Composition study (Health ABC), the InCHIANTI study, Multi-Ethnic Study of Atherosclerosis (MESA), the Rotterdam study and the Young Finns Study.
Assessment of percentage of caloric intake from macronutrients
In the NHS and HPFS, dietary intake was assessed every 2–4 years using the Willett semi-quantitative food frequency questionnaire containing 118-food items (32,33). The FFQ from 1986 was used for this analysis. In the WGHS, dietary intake was assessed at baseline between 1992 and 1994 using the Willett semi-quantitative FFQ derived from the NHS containing 131-food items (34). For all cohorts, participants reported an average frequency of consumption and portion sizes of all food items listed in the FFQ during the previous year. Average daily intake per food item was calculated by multiplying the frequency of consumption by portion size (1). Absolute macronutrient intake was calculated by multiplying the average daily intake by the nutrient content derived from food composition tables from the Harvard School of Public Health.
In the replication cohorts, dietary intake was assessed using similar but slightly varied FFQs. Average daily intake of food and nutrients was calculated using the frequency of consumption, portion sizes and study-specific nutrient and food composition tables.
Genotyping
Genotyping for each of the discovery cohort studies was performed using either the Illumina HumanHap 300 Duo+ (WGHS), Illumina 550 K (NHS BC), Illumina 610Q (NHS/HPFS KS cohort), or the Affymetrix 6.0 (NHS/HPFS T2D and CHD) array; each panel was imputed up to ∼2.6 million SNPs using MaCH software (reference panel—HapMap release 22/NCBI build 36)(35,36). Imputed genotypes were expressed as allelic dosage values between 0 and 2.
Similarly, the replication cohorts were genotyped with either Affymetrix- or Illumina-based genotyping arrays and imputed to HapMap release 21 or 22/NCBI build 35 or 36.
Statistical analysis
Three primary outcomes were examined in this study: percentage of total energy intake from (i) carbohydrate, (ii) protein, and (iii) fat. Percentage of total energy intake from each of the three macronutrients was calculated by multiplying the absolute macronutrient intake in grams by the caloric content of each type of macronutrient (4 kcal/g for carbohydrate and protein, 9 kcal/g for fat) (1), and dividing by the total energy intake. Absolute macronutrient intake and total energy intake were computed by multiplying the relative frequency of consumption by the nutrient content using United States Department of Agriculture food composition sources.
Study-specific genome-wide association analyses of percentage macronutrient intake were performed using linear regression in ProbABEL (37) for ∼2.6 million SNPs, adjusted for age, location, measures of population stratification, with and without adjustment for BMI. Distributions of the residuals from each of the macronutrient intake analyses are provided in the Supplementary material, Figure S3. If the genomic inflation factor was >1.0, the analysis was corrected before the meta-analysis.
We conducted a two-stage genome-wide meta-analysis for macronutrient intake. In the initial stage, meta-analyses for each phenotype–model combination were performed in METAL (38). Fixed-effects meta-analyses using the inverse-variance weighting scheme were performed for each macronutrient and model combination (total of six meta-analyses). The meta-analysis results were corrected if the genomic inflation factor was >1.0. Heterogeneity was estimated using the I2 statistic. Meta-analyses were limited to 2.1 million SNPs with minor allele frequencies (MAF) ≥ 0.05 and imputation quality scores (R2) ≥ 0.30. The genome-wide significance threshold was set at P < 5 × 10−8, and the sub-genome-wide significance threshold was set at 1 × 10−5 > P > 5 × 10−8. Lead SNPs were deemed independent if they had the most significant association at a locus, and were >500 kb from a lead SNP at an adjacent locus. Conditional meta-analysis in GCTA(7) on the lead SNP at each locus did not reveal any remaining SNP associations with P < 1 × 10−5 at the candidate loci.
In the second stage, we approached the CHARGE consortium and requested to investigate the association of our index genome-wide and sub-genome-wide SNPs in their parallel genetic analysis of macronutrient intake (6). The Bonferroni method was used to correct for multiple hypothesis testing by dividing the α(=0.05) by the number of loci brought forward for replication per macronutrient phenotype. The P-values from one-tailed z-distributions were used to determine the significance in replication. Genome-wide significance was declared if the P-value from the joint analysis was <5 × 10−8.
GCTA estimation of heritability
We randomly selected 15 000 participants (out of 23 294) from the WGHS to estimate heritability of macronutrient intake using the GCTA software (7); we were unable to include all WGHS participants in the heritability estimate due to computational restraints. We restricted the analysis to 339 596 genotyped SNPs with 98.5% complete genotyping and MAF > 0.01.
SUPPLEMENTARY MATERIAL
FUNDING
The HPFS and NHS were supported by grants DK091718, HL71981, HL073168, CA87969, CA49449, HL34594, HL088521, U01HG004399, DK080140, DK58845 and DK46200 from the National Institutes of Health. The WGHS is supported by HL043851, HL080467 and CA058988 from the National Institutes of Health with collaborative scientific support and funding for genotyping provided by Amgen. L.Q. is a recipient of the American Heart Association Scientist Development Award (0730094N).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the participants and the staff of the HPFS, NHS and the WGHS for their continued contributions.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Willett W. Nutritional Epidemiology. New York: Oxford University Press; 1998. [Google Scholar]
- 2.Rankinen T., Bouchard C. Genetics of food intake and eating behavior phenotypes in humans. Annu. Rev. Nutr. 2006;26:413–434. doi: 10.1146/annurev.nutr.26.061505.111218. [DOI] [PubMed] [Google Scholar]
- 3.Qi L., Kraft P., Hunter D.J., Hu F.B. The common obesity variant near MC4R gene is associated with higher intakes of total energy and dietary fat, weight change and diabetes risk in women. Hum. Mol. Genet. 2008;17:3502–3508. doi: 10.1093/hmg/ddn242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reed D.R., Bachmanov A.A., Beauchamp G.K., Tordoff M.G., Price R.A. Heritable variation in food preferences and their contribution to obesity. Behav. Genet. 1997;27:373–387. doi: 10.1023/a:1025692031673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Willett W.C., Reynolds R.D., Cottrell-Hoehner S., Sampson L., Browne M.L. Validation of a semi-quantitative food frequency questionnaire: comparison with a 1-year diet record. J. Am. Diet Assoc. 1987;87:43–47. [PubMed] [Google Scholar]
- 6.Tanaka T., Ngwa J.S., van Rooij F.J.A., Zillikens M.C., Wojczynski M.K., Frazier-Wood A.C., Houston D.K., Kanoni S., Lemaitre R.N., Luan J., et al. Genome-wide meta-analysis of observational studies reveals common genetic variants associated with macronutrient intake. Am. J. Clin. Nutr. 2013 doi: 10.3945/ajcn.112.052183. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berglund E.D., Li C.Y., Bina H.A., Lynes S.E., Michael M.D., Shanafelt A.B., Kharitonenkov A., Wasserman D.H. Fibroblast growth factor 21 controls glycemia via regulation of hepatic glucose flux and insulin sensitivity. Endocrinology. 2009;150:4084–4093. doi: 10.1210/en.2009-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uebanso T., Taketani Y., Yamamoto H., Amo K., Ominami H., Arai H., Takei Y., Masuda M., Tanimura A., Harada N., et al. Paradoxical regulation of human FGF21 by both fasting and feeding signals: is FGF21 a nutritional adaptation factor? PLoS. One. 2011;6:e22976. doi: 10.1371/journal.pone.0022976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen W.W., Li L., Yang G.Y., Li K., Qi X.Y., Zhu W., Tang Y., Liu H., Boden G. Circulating FGF-21 levels in normal subjects and in newly diagnose patients with Type 2 diabetes mellitus. Exp. Clin. Endocrinol Diabetes. 2008;116:65–68. doi: 10.1055/s-2007-985148. [DOI] [PubMed] [Google Scholar]
- 11.Bobbert T., Schwarz F., Fischer-Rosinsky A., Pfeiffer A.F., Mohlig M., Mai K., Spranger J. Fibroblast growth factor 21 predicts the metabolic syndrome and type 2 diabetes in Caucasians. Diabetes Care. 2013;36:145–149. doi: 10.2337/dc12-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X., Yeung D.C., Karpisek M., Stejskal D., Zhou Z.G., Liu F., Wong R.L., Chow W.S., Tso A.W., Lam K.S., et al. Serum FGF21 levels are increased in obesity and are independently associated with the metabolic syndrome in humans. Diabetes. 2008;57:1246–1253. doi: 10.2337/db07-1476. [DOI] [PubMed] [Google Scholar]
- 13.Coskun T., Bina H.A., Schneider M.A., Dunbar J.D., Hu C.C., Chen Y., Moller D.E., Kharitonenkov A. Fibroblast growth factor 21 corrects obesity in mice. Endocrinology. 2008;149:6018–6027. doi: 10.1210/en.2008-0816. [DOI] [PubMed] [Google Scholar]
- 14.Sarruf D.A., Thaler J.P., Morton G.J., German J., Fischer J.D., Ogimoto K., Schwartz M.W. Fibroblast growth factor 21 action in the brain increases energy expenditure and insulin sensitivity in obese rats. Diabetes. 2010;59:1817–1824. doi: 10.2337/db09-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dutchak P.A., Katafuchi T., Bookout A.L., Choi J.H., Yu R.T., Mangelsdorf D.J., Kliewer S.A. Fibroblast growth factor-21 regulates PPARgamma activity and the antidiabetic actions of thiazolidinediones. Cell. 2012;148:556–567. doi: 10.1016/j.cell.2011.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang T.P., Beazley C., Montgomery S.B., Dimas A.S., Gutierrez-Arcelus M., Stranger B.E., Deloukas P., Dermitzakis E.T. Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics. 2010;26:2474–2476. doi: 10.1093/bioinformatics/btq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grundberg E., Small K.S., Hedman A.K., Nica A.C., Buil A., Keildson S., Bell J.T., Yang T.P., Meduri E., Barrett A., et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 2012;44:1084–1089. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hazra A., Kraft P., Selhub J., Giovannucci E.L., Thomas G., Hoover R.N., Chanock S.J., Hunter D.J. Common variants of FUT2 are associated with plasma vitamin B12 levels. Nat. Genet. 2008;40:1160–1162. doi: 10.1038/ng.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rouquier S., Lowe J.B., Kelly R.J., Fertitta A.L., Lennon G.G., Giorgi D. Molecular cloning of a human genomic region containing the H blood group alpha(1,2)fucosyltransferase gene and two H locus-related DNA restriction fragments. Isolation of a candidate for the human secretor blood group locus. J. Biol. Chem. 1995;270:4632–4639. doi: 10.1074/jbc.270.9.4632. [DOI] [PubMed] [Google Scholar]
- 20.Speliotes E.K., Willer C.J., Berndt S.I., Monda K.L., Thorleifsson G., Jackson A.U., Allen H.L., Lindgren C.M., Luan J., Magi R., et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morris A.P., Voight B.F., Teslovich T.M., Ferreira T., Segre A.V., Steinthorsdottir V., Strawbridge R.J., Khan H., Grallert H., Mahajan A., et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Popkin B.M., Adair L.S., Ng S.W. Global nutrition transition and the pandemic of obesity in developing countries. Nutr. Rev. 2012;70:3–21. doi: 10.1111/j.1753-4887.2011.00456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Visscher P.M., Yang J., Goddard M.E. A commentary on ‘common SNPs explain a large proportion of the heritability for human height’ by Yang et al. (2010) Twin Res. Hum. Genet. 2011;13:517–524. doi: 10.1375/twin.13.6.517. [DOI] [PubMed] [Google Scholar]
- 24.Rimm E.B., Giovannucci E.L., Willett W.C., Colditz G.A., Ascherio A., Rosner B., Stampfer M.J. Prospective study of alcohol consumption and risk of coronary disease in men. Lancet. 1991;338:464–468. doi: 10.1016/0140-6736(91)90542-w. [DOI] [PubMed] [Google Scholar]
- 25.Qi L., Cornelis M.C., Kraft P., Stanya K.J., Linda Kao W.H., Pankow J.S., Dupuis J., Florez J.C., Fox C.S., Pare G., et al. Genetic variants at 2q24 are associated with susceptibility to type 2 diabetes. Hum. Mol. Genet. 2010;19:2706–2715. doi: 10.1093/hmg/ddq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jensen M.K., Pers T.H., Dworzynski P., Girman C.J., Brunak S., Rimm E.B. Protein interaction-based genome-wide analysis of incident coronary heart disease. Circ. Cardiovasc. Genet. 2011;4:549–556. doi: 10.1161/CIRCGENETICS.111.960393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Curhan G.C., Taylor E.N. 24-h uric acid excretion and the risk of kidney stones. Kidney Int. 2008;73:489–496. doi: 10.1038/sj.ki.5002708. [DOI] [PubMed] [Google Scholar]
- 28.Colditz G.A., Hankinson S.E. The Nurses’ Health Study: lifestyle and health among women. Nat. Rev. Cancer. 2005;5:388–396. doi: 10.1038/nrc1608. [DOI] [PubMed] [Google Scholar]
- 29.Hunter D.J., Kraft P., Jacobs K.B., Cox D.G., Yeager M., Hankinson S.E., Wacholder S., Wang Z., Welch R., Hutchinson A., et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ridker P.M., Chasman D.I., Zee R.Y., Parker A., Rose L., Cook N.R., Buring J.E. Rationale, design, and methodology of the Women's Genome Health Study: a genome-wide association study of more than 25,000 initially healthy American women. Clin. Chem. 2008;54:249–255. doi: 10.1373/clinchem.2007.099366. [DOI] [PubMed] [Google Scholar]
- 31.Psaty B.M., O'Donnell C.J., Gudnason V., Lunetta K.L., Folsom A.R., Rotter J.I., Uitterlinden A.G., Harris T.B., Witteman J.C., Boerwinkle E. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ. Cardiovasc. Genet. 2009;2:73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willett W.C., Sampson L., Stampfer M.J., Rosner B., Bain C., Witschi J., Hennekens C.H., Speizer F.E. Reproducibility and validity of a semiquantitative food frequency questionnaire. Am. J. Epidemiol. 1985;122:51–65. doi: 10.1093/oxfordjournals.aje.a114086. [DOI] [PubMed] [Google Scholar]
- 33.Rimm E.B., Giovannucci E.L., Stampfer M.J., Colditz G.A., Litin L.B., Willett W.C. Reproducibility and validity of an expanded self-administered semiquantitative food frequency questionnaire among male health professionals. Am. J. Epidemiol. 1992;135:1114–1126. doi: 10.1093/oxfordjournals.aje.a116211. discussion 1127–1136. [DOI] [PubMed] [Google Scholar]
- 34.Liu S., Manson J.E., Lee I.M., Cole S.R., Hennekens C.H., Willett W.C., Buring J.E. Fruit and vegetable intake and risk of cardiovascular disease: the Women's Health Study. Am. J. Clin. Nutr. 2000;72:922–928. doi: 10.1093/ajcn/72.4.922. [DOI] [PubMed] [Google Scholar]
- 35.Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aulchenko Y.S., Struchalin M.V., van Duijn C.M. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genome-wide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.