Summary
Molecular replacement (MR) is widely used for addressing the phase problem in X-ray crystallography. Historically, crystallographers have had limited success using NMR structures as MR search models. Here we report a comprehensive investigation of the utility of protein NMR ensembles as MR search models, using data for 25 pairs of X-ray and NMR structures solved and refined using modern NMR methods. Starting from NMR ensembles prepared by an improved protocol, FindCore, correct MR solutions were obtained for 22 targets. Based on these solutions, automatic model rebuilding could be done successfully. Rosetta refinement of NMR structures provided MR solutions for another two proteins. We also demonstrate that such properly prepared NMR ensembles and X-ray crystal structures have similar performance when used as MR search models for homologous structures, particularly for targets with sequence identity > 40%.
Introduction
One of the most critical stages in the process of determining the crystal structure of a protein involves estimating the phases of X-ray diffraction data. There are several ways to address this phase problem, including direct methods (Woolfson, 1971), multi-wavelength or single-wavelength anomalous diffraction (MAD or SAD) (Pahler, et al., 1990; Hendrickson, 1991), multiple or single isomorphous replacement (MIR or SIR) (Green et al., 1954; Perutz 1956; Blow and Rossmann, 1961), molecular replacement (MR) (Rossmann, 1972; Rossmann & Arnold, 1993), and/or a combination of these methods. Molecular replacement, first described by Rossmann and Blow (Rossmann and Blow, 1962), involves estimating the initial phases of diffraction data based on a known similar structure. In comparison to the experimental phase determination techniques, molecular replacement has the advantage of not requiring preparation of heavy atom derivatives, hence can be cost and time effective. In recent years, around 70 percent of deposited macromolecular structures have been solved by molecular replacement (Evans and McCoy, 2008). Additionally, both the number of structures deposited in PDB and the coverage of structure space are increasing rapidly (Liu, et al., 2007; Burley, et al., 2008; Nair, et al., 2009). These data, in combination with advances in homology modeling (Chivian et al., 2003; Eswar et al., 2006; Zhang, 2007; Schwede et al., 2009) and MR programs, make molecular replacement an increasingly important approach to the phase problem in protein X-ray crystallography.
In principle, given an accurate search model for a target protein structure, MR is quite straightforward. However, it can sometimes be very difficult to get a correct MR solution due to the enormous search space. Therefore, for successful MR phasing, it is critical to effectively prepare the initial search model so as to maximize its signal/noise ratio, and to enhance the signal detection capabilities of MR algorithms by finding an optimal target function and effective search strategy that can identify correct solutions. Significant efforts have been made to develop and improve both of these aspects in the last two decades.
A number of protocols to prepare MR search model have been proposed. These are generally designed to exclude structurally-disordered regions (e.g. by truncating long flexible side chains) or to incorporate structural flexibility information into search models by using a composite search model (Kleywegt et al., 1994; Leahy et al., 1992; Muller et al., 1995) or pseudo B-factors (Anderson et al., 1996; Baldwin et al., 1991; Wilmanns and Nilges, 1996). Armed with more accurate target functions, more advanced mathematical models and more effective search strategies, a number of software packages have been developed which have greatly improved the effectiveness of the MR approach, such as COMO (Jogl et al., 2001), XPLOR/CNS (Brunger et al., 1998), AMoRe (Navaza, 2001), MOLREP (Vagin and Teplyakov, 2000), EPMR (Kissinger et al., 1999), Queen of Spades (Glykos and Kokkinidis, 2000), SoMoRe (Jamrog et al., 2003), MrBUMP (Keegan and Winn, 2008), Phaser (McCoy et al., 2007), and others.
Nuclear magnetic resonance (NMR) is a powerful tool to determine protein structures in solution and in the solid state. Solution NMR methods have contributed a substantial fraction of the structures deposited in the Protein Data Bank (PDB). In 1987, Brunger et al. showed that solution NMR structures could be employed as search models for MR (Brunger et al., 1987). Since this early work, quite a few successful cases using NMR structures for MR have been published (for a useful review of this progress, see Chen et al., 2000). However, a common notion in the structural biology community is that the quality of NMR structure is often not good enough for MR, even when the sequence of the search model is identical to the target X-ray structure. There are various explanations for this observation. Some NMR structures, or parts of the structure, may be under-constrained due to insufficient data; in other cases, there may be genuine differences between structures in solution and in the crystal. Chen et. al have demonstrated, based on a few individual successful cases reported in previous literature, that success rate of using NMR structures in MR can be significantly improved by carefully preparing the initial search models (Chen and Clore, 2000; Chen et al., 2000; Chen, 2001). However, in most studies only the successful examples are reported and, to date, there have been no systematic studies to evaluate the general utility of NMR structures as initial search models for MR.
Over the last 10 years, there have been significant improvements in both phasing algorithms and the NMR structure determination process, particularly in structural genomics projects where state-of-the-art refinement and quality assessment tools are employed. These advances beg the question: given modern technologies for NMR structure determination and refinement, can NMR structures be used routinely as initial search models for molecular replacement? If that is the case, can we define an optimal protocol to prepare NMR structure ensembles as MR search models in order to maximize their phasing power in MR?
The Northeast Structural Genomics Consortium (NESG; www.nesg.org) is one of the large-scale structure production centers of the Protein Structure Initiative (PSI). NESG has contributed more than 400 NMR structures, as well as some 600 X-ray crystal structures, to the Protein Data bank (PDB) over the past ten years, representing a large fraction of the NMR structures deposited into the PDB by the PSI. The NESG Consortium, involving several NMR groups, has focused efforts on improving the efficiency and accuracy of its NMR structure determination pipeline, and has implemented strict quality control measures to ensure the production of high quality structures (Kim and Szyperski, 2003; Huang et al., 2007; Bhattacharya et al., 2007). Although most NESG structures have been solved by either NMR or X-ray crystallography, as of December 2009 the NESG consortium had solved 27 pairs of protein structures for identical construct sequences using both X-ray crystallography and NMR methods. These 3D structures of proteins with identical sequences, together with the raw NMR and crystallography data available in the BioMagRes and PDB, are an extremely valuable composite dataset for understanding structural variations between solution and crystal states, providing insights into protein dynamics and the effects of lattice packing in selecting conformations from solution, and for new methods development.
Model preparation is a cornerstone of many successful molecular replacement trials, given the fact that every atom in the search model contributes in MR analysis. In particular, it is critical to estimate structural variability in order to decide which portion of structure should be kept in the search model. There are alternative ways to assess the precision of a NMR structural ensemble, including RMSD (the root-mean-square-deviations from the average model), dihedral angle circular variance or order parameters (Hyberts et al., 1992), and inter-atomic variance matrices (Kelley et al., 1997; Snyder and Montelione, 2005). RMSD statistics depend on details of how the structural ensemble is superimposed. Dihedral angle order parameters are good estimators of local structural uncertainty, but generally do not provide a good measure of global consistency. Methods based on the inter-atomic variance matrix can identify one or more sets of “core atoms” whose positions are well defined with respect to one another. The FindCore algorithm (Snyder and Montelione, 2005) uses the inter-atomic variance matrix to define an “order parameter” for each atom, then identifies sets of “core atoms” using hierarchical clustering methods with an empirically-motivated stopping rule based on Chauvenet’s criterion for outlier detection. In some cases it partitions the protein structure into “multiple cores”, each of which is well-defined internally but exhibit structural variation between “cores”. The FindCore algorithm thus allows identification of the well-defined regions (i.e. groups of atoms) of the protein structure from the ensemble of NMR structures without the assumptions involved in generating a molecular superimposition.
We have used 25 NESG NMR/X-ray crystal structure pairs in a systematic investigation of the utility of NMR structures as initial search models for molecular replacement. Staring from NMR ensembles prepared by an improved protocol, FindCore, we obtained correct MR solutions for 22 of 25 targets. The NMR ensembles for two (2) additional proteins could also be used successfully for MR following Rosetta refinement. Based on these solutions, automatic model rebuilding could also be successfully done with high sequence completeness and model accuracy. We also demonstrate that these NMR structure ensembles can be used successfully as MR search models for homologous target X-ray structures, given sequence coverage and sequence identity of NMR structures to X-ray structures no less than 70% and 40% respectively. These studies indicate the high quality of the NMR structures that are being generated by structural genomics projects using routine modern NMR methods, and demonstrate that the FindCore protocol generally provides high success rates using NMR ensembles for phasing by MR.
Results
22 of 25 NESG NMR structures successfully provide MR solutions
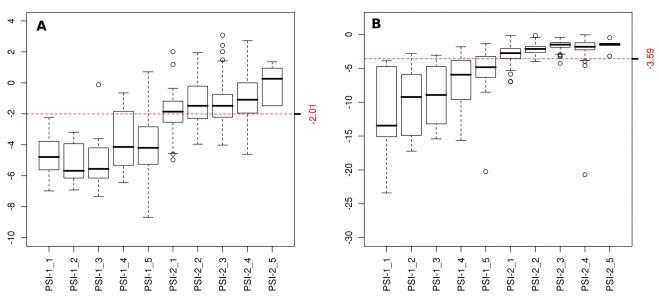
The NESG project uses the Protein Structure Validation Suite (PSVS) (Bhattacharya et al., 2007; http://psvs.nesg.org/) to monitor the quality of structures. Based on a set of 252 high resolution X-ray structures, PSVS provides Z scores for a variety of widely adopted structural quality measures, such as Procheck G-factor (Laskowski et al., 1996), Molprobity clashscore (Lovell et al., 2003), and other structure quality assessment metrics. The analysis aims to provide a multi-criteria estimate of protein structure quality. A time course study of the evolution of various PSVS Z scores for NESG NMR structures indicates that the quality of NESG structures has steadily improved over time. For example, significant improvements of knowledge-based stereochemical, geometric, and interatomic packing properties of protein NMR structures over the past few years are illustrated in Fig. 1. Most of the NMR structures used in this study were solved since 2006 (Table 1).
Fig. 1. Knowledge-based structure quality scores for NESG NMR structures have consistently improved as NMR methods have matured over the past several years.
Panel A and B show boxplots of the distribution of Z scores (y-axis) of Procheck ‘all-dihedral-angle’ G-factor and Molprobity clashscores, respectively, for all NMR structures solved by the NESG consortium in each PSI fiscal year (x-axis). The red dashed lines represent the average Z scores. One PSI fiscal year is a 12 month time period generally spanning July 1st through June 30th of the following year. The Procheck all-dihedral-angle G-factor is determined by the stereochemical quality of both backbone and side chain dihedral angles of proteins, and Molprobity clashscore is a measure to reflect the number of high-energy contacts in a structure calculated by the program probe. PSVS Z scores are calculated based on a calibrated dataset of 252 high quality X-ray crystal structures from the PDB with resolution ≤ 1.80 Å, R-factor ≤ 0.25, and R-free ≤ 0.28 (Bhattacharya et al. 2007).
Table 1.
NMR/X-ray Crystal Structure and Structural Statistics for MR Studies.
| Target | X-ray structure
|
NMR Structure
|
GDT-TS2
|
Phaser Solution3
|
ARP/wARP or Phenix model
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PDB id | Resolution | Space Group | Length1 | PDB id | Year | Mean | Max | LLG | TFZ | R7 | R-Free7 | Docked7 | Matched4 | GDT-TS7 | |
| BeR31 | 3cpk | 2.50 | P43212 | 150 | 2k2e | 2008 | 0.85 | 0.88 | 111 | 13.6 | 0.27 (0.26) | 0.43 (0.34) | 115 (115) | 89 (107) | 0.87 (0.95) |
| CcR55 | 2o0q | 1.80 | C222 | 115 | 2jqn | 2007 | 0.79 | 0.84 | 154 | 13.1 | 0.18 | 0.23 | 112 | 110 | 0.98 |
| CsR4 | 2ota | 2.20 | P212121 | 76 (2) | 2jr2 | 2007 | 0.95 | 0.97 | 388 | 27.7 | 0.23 | 0.30 | 123 | 116 | 0.96 |
| CtR107 | 3e0h | 1.81 | P212121 | 158 | 2kcu | 2009 | 0.72 | 0.77 | 54 | 8.7 | 0.23 | 0.29 | 136 | 120 | 0.88 |
| CtR148A | 3ibw | 1.93 | P43212 | 88 (2) | 2ko1 | 2009 | 0.94 | 0.96 | 219 | 15.7 | 0.20 | 0.24 | 154 | 149 | 0.99 |
| GmR137 | 3cwi | 1.90 | P43212 | 78 | 2k5p | 2008 | 0.79 | 0.84 | 64 | 8.8 | 0.23 | 0.26 | 67 | 65 | 0.97 |
| HR1958 | 1tvg | 1.60 | C121 | 153 | 1xpw | 2004 | 0.78 | 0.81 | 150 | 9.5 | 0.22 | 0.26 | 134 | 102 | 0.87 |
| HR3646E | 3fia | 1.45 | C121 | 121 | 2khn | 2009 | 0.75 | 0.78 | 26 | 3.6 | 0.20 | 0.26 | 93 | 90 | 0.97 |
| MbR242E | 3gw2 | 2.10 | P6422 | 108 | 2kko | 2009 | 0.88 | 0.93 | 178 | 18.1 | 0.23 | 0.26 | 89 | 84 | 0.95 |
| MrR110B | 3e0e | 1.60 | P212121 | 97 | 2k5v | 2008 | 0.93 | 0.96 | 136 | 12.7 | 0.20 | 0.25 | 94 | 91 | 0.98 |
| OR8C | 2rhk | 1.95 | P41 | 140 (2), 72 (2) | 2kkz | 2009 | 0.92 | 0.94 | 352 | 19.5 | 0.22 | 0.27 | 344 | 327 | 0.98 |
| PfR193A | 3idu | 1.70 | P1211 | 127 (2) | 2kl6 | 2009 | 0.87 | 0.88 | 262 | 17.1 | 0.23 | 0.27 | 209 | 188 | 0.9 |
| SgR42 | 3c4s | 1.70 | P32 | 66 (2) | 2jz2 | 2008 | 0.94 | 0.96 | 210 | 21 | 0.16 | 0.20 | 107 | 102 | 0.95 |
| SoR77 | 2qti | 2.30 | P43212 | 80 | 2juw | 2007 | 0.93 | 0.97 | 173 | 16 | 0.23 | 0.30 | 64 | 61 | 0.96 |
| SR213 | 2im8 | 2.00 | P212121 | 131 (2) | 2hfi | 2006 | 0.82 | 0.86 | 234 | 14.1 | 0.25 (0.29) | 0.47 (0.39) | 201 (218) | 183 (214) | 0.92 (0.89) |
| SR384 | 3bhp | 2.01 | C121 | 60 (3) | 2jvd | 2007 | 0.8 | 0.83 | 188 | 16.1 | 0.19 | 0.31 | 135 | 124 | 0.96 |
| SsR10 | 2q00 | 2.40 | I4122 | 129 (2) | 2jpu | 2007 | 0.84 | 0.88 | 454 | 24.6 | 0.27 | 0.33 | 218 | 155 | 0.84 |
| StR655 | 2es9 | 2.00 | I213 | 115 | 2jn8 | 2007 | 0.82 | 0.86 | 38 | 5.8 | 0.24 (0.30) | 0.39 (0.35) | 77 (88) | 51 (78) | 0.71 (0.86) |
| XcR50 | 1ttz | 2.11 | P65 | 87 | 1xpv | 2004 | 0.9 | 0.94 | 81 | 10.9 | 0.19 | 0.24 | 72 | 69 | 0.96 |
| HR41 | 3evx | 2.54 | P1 | 175 (4) | 2k07 | 2008 | 0.82 | 0.85 | 445 | 16.7 | 0.29 (0.24) | 0.62 (0.30) | 46 (616) | NA (596) | NA (0.96) |
| PsR293 | 3h9x | 2.51 | P1 | 125 (4) | 2kfp | 2009 | 0.81 | 0.85 | 227 | 12 | 0.28 (0.18) | 0.57 (0.23) | 10 (472) | NA (472) | NA (1.00) |
| StR70 | 2es7 | 2.80 | P1211 | 142 (4) | 2izt | 2008 | 0.76 | 0.82 | 927 | 28.6 | 0.40 (0.37) | 0.58 (0.44) | 0 (420) | NA (292) | NA (0.82) |
| DrR147D | 3ggn | 2.00 | P1211 | 155 (2) | 2kcz | 2009 | 0.48 | 0.52 | 48 | 4.7 | 0.53 | 0.57 | 0 | NA | NA |
| SR478 | 2gsv | 1.90 | P121 | 80 (2) | 2js1 | 2007 | 0.74 | 0.78 | 51 | 4.8 | 0.47 | 0.54 | 0 | NA | NA |
| ZR18 | 2ffm | 2.51 | P41212 | 91 | 1pqx6 | 2004 | 0.78 | 0.8 | 23 | 4.5 | 0.37 | 0.66 | 0 | NA | NA |
The number of subunits in the asymmetric unit is indicated in parentheses.
TM-score program is used to calculate GDT-TS between X-ray structure and NMR models.
TFZ and LLG values are extracted from MR solution with the highest TFZ score given LLG > 0
The number of residues with Cα-rmsd < 1Å between ARP/wARP model and X-ray structure
rms=1.8 is used in MR_AUTO mode of Phaser.
A11 NMR structures contain 20 models except for ZR18 (10 models).
R,R-free and GDT-TS values of Phenix models are in the parentheses
See also Tables S1, S3, and S4 and Figures S1 and S2.
Of 27 NESG NMR/X-ray crystal structure pairs available at the time this study was initiated, two were excluded from this investigation due to the following facts: One target (GR4) was reported as only a single structure, rather than as an ensemble. The NMR structure of target ER382A (PDB id: 2jn0) was solved as a monomer without a ligand, while its crystal structure counterpart (PDB id:3fif) has eight subunits in the asymmetric unit and was solved in complex with a heptapeptide ligand and appears to have a distinct structure; i.e. the Cα-rmsd between the NMR structure and chain A of crystal structure is 2.44 Å.
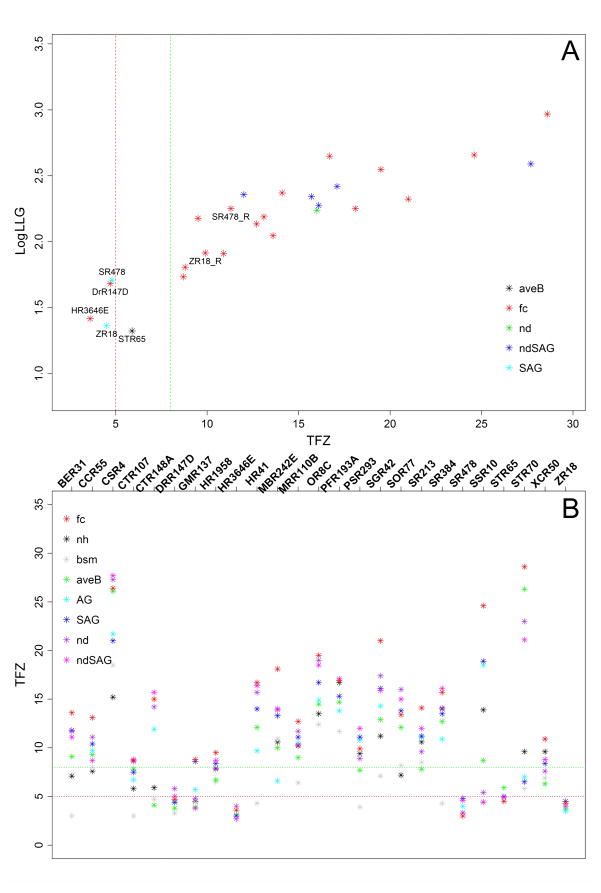
For each of the remaining 25 structures, MR search models were prepared from the NMR structure ensemble, using eight different methods to define the search models. We obtained definite MR solutions with Phaser, which have positive log likelihood gain (LLG) scores and translation function Z-score (TFZ) scores greater than 8, for 20 of 25 targets. For two additional targets, HR3646E and StR65, although their TFZ scores were relatively low (3.6 and 5.8 respectively), using the MR solutions with the highest TFZ scores, more than half of the residues could be accurately traced by ARP/wARP program; this indicates that the MR solutions were actually correct even though the TFZ scores were lower than 8 (see more details below). All together, useful phase information for 22 of 25 X-ray structures could be determined by Phaser based on their corresponding NMR structure ensembles (Fig. 2A, Table 1). In addition, for most targets with correct MR solutions and resolution better than 2.5 Å, highly accurate ARP/wARP models could be built with great sequence completeness. However, for five targets with definite Phaser solutions (TFZ >8), ARP/wARP either failed to build any legitimate model (HR41, StR70, PsR293) or eventually generated models with free R value worse than 0.4 (BeR31, SR213). To address these cases, we used phenix.autobuild (Terwilliger et al., 2007) for automatic model rebuilding, which was less sensitive to low resolution X-ray diffraction data. For all five of the targets that failed model building using ARP/wARP, we could build models using phenix.autobuild with free R factors better than 0.45. The free R factors of some models (HR41, PsR293) were even comparable with the free R factors of the corresponding crystal structures deposited in PDB (Supplementary Table S3). These results are particularly impressive since no manual intervention was used in these analyses. From this study, we conclude that good quality NMR structures, like those solved by the NESG consortium using standard modern NMR methods, are generally of sufficient accuracy to be routinely used as search models in MR.
Fig. 2. Using the fc method, Phaser phasing scores obtained using NMR structure ensembles as templates are generally sufficient to provide good MR solutions.
(A) LLG-TFZ scatter plot. LLG (log-likelihood gain) and TFZ (translation function Z-score) scores are calculated by Phaser, and log10(LLG) and TFZ scores are plotted on y-axis and x-axis respectively. The red vertical dash line delimits (TFZ=5), the typical cut-off of an invalid Phaser solution, while the green vertical dash line (TFZ=8) delimits the typical cut-off of a definite Phaser solution according to the Phaser manual. For each individual target, only the model with the highest TFZ score solution is plotted. Colors are coded by different model preparation methods. SR478_R and ZR18_R denote the two models following Rosetta refinement. (B) Comparisons of TFZ scores from different MR models prepared by the eight model preparation methods. Models are color coded by their respective preparation method. TFZ scores calculated by Phaser are plotted on y-axis, while each NESG target is plotted on x-axis in alphabetical order. The red horizontal dash line (at TFZ=5) delimits the typical cut-off of an invalid Phaser solution while the green horizontal dash line (at TFZ=8) delimits the typical cut-off of a definite Phaser solution, according to the Phaser Manual. See also Tables S2 and S5.
Structure similarity limit of search models to X-ray structures
A rule of thumb in MR is that a correct MR solution requires a Cα-RMSD between search model and target structure no greater than 1.5 Å over a large fraction of the molecule. In 2005, Giorgetti et. al (Giorgetti et al., 2005) demonstrated that the Global Distance Test (GDT) algorithm (Zemla et al, 2003) provides an even more robust measure to assess the usefulness of protein search model for MR than Cα-RMSD. They concluded that a GDT-TS higher than 0.84 is generally sufficient to guarantee the success of MR procedure, while a GDT-TS lower than 0.80 is essentially never successful in MR trials; GDT-TS values between 0.80 and 0.84 are in the “twilight zone” of mixed success rates. Our analysis confirms the first part of this conclusion. However, for two cases (NESG targets CtR107 and HR3646E), we obtained correct MR solutions using initial search structures with GDT-TS values lower than 0.8. In addition, we had almost perfect success rate of MR trials for targets in the “twilight zone” (Table 1).
We are in a better position today to push the limits of the application of MR than five years ago. In particular, recent advances in MR programs such as Phaser offer more powerful signal detection and more effective search strategies. In addition, improvements in NMR data analysis and structure refinement methods provide more accurate NMR models, and model uncertainty is better described by the reported NMR structure ensembles.
The FindCore protocol provides better search models for MR
The basic problem of preparing NMR search models for MR can be reduced to determining which subset of atoms have highest probability to contribute to signal instead of noise, and assigning appropriate weight to each atom proportional to its S/N ratio. Since it is impossible to know the X-ray structure beforehand without phase information, there is no direct criteria to assess the S/N level of each atom; i.e., the consistency of its relative position between solution and crystal states. However, structurally-ordered regions of the protein, such as atoms buried in the hydrophobic cores, generally have better “phasing power” than disordered residues, such as atoms in large surface side chains. This conclusion is supported by the work of Chen et al. (Chen and Clore, 2000; Chen et al., 2000; Chen, 2001) which demonstrated that phasing power of NMR structure ensemble can be significantly improved by removing structurally-disordered regions and by truncating long side chains to their common bases (Cβ or Cγ). Ensemble-derived pseudo-B factors or composite models can also improve the phasing power of NMR ensembles as search models (Wilmanns and Nilges, 1996).
The “dihedral angle order parameter” (S), a measure of dihedral angle circular variance, is one of the most commonly used measures to calculate the ordered region of a protein (Hyberts et al., 1992). In our study, the PSVS server (Bhattacharya et al., 2007) was used to identify ordered residues with S(phi) + S(psi) ≥ 1.8. Then, the areaimol program in the CCP4 software package (Lee et al.,1971; Saff et al., 1997) was used to identify surface exposed residues. As described in methods section and in Supplementary Table S2, eight search models were prepared for each target in order to compare their relative performance in MR experiments based on both Phaser solutions and ARP/wARP model building results. Most of these methods utilize the ensemble of NMR structures, trimmed in various ways, as the search model. We plotted TFZ scores against model preparation protocols for all the targets (Fig. 2B). TFZ scores of Phaser solutions derived using the whole ensemble model (nh) or single (best) NMR conformer (bsm) as the search model were among the lowest. Better TFZ scores could be attained by removing disordered residues (nd, aveB) or by truncating long side chain residues to common base (AG, SAG), but the level of improvement was case specific, and these protocols failed to find optimal MR solutions for some targets. A combination of removing disordered residues and truncating long surface side chains (ndSAG) showed no further significant improvement. TFZ scores of Phaser solutions using NMR ensembles trimmed to “core atom sets”, defined by the FindCore program (fc) which allows a robust estimate of model uncertainty at an atomic level, were always the highest or among the highest. Starting from these ‘fc’ MR solutions, more than half of the residues could be accurately built (Cα - rmsd < 1 Å) using ARP/wARP for 18 of 19 targets (i.e. except for StR65) which had both correct MR solutions and X-ray diffraction data resolution better than 2.5 Å (Supplementary Table S4). For target StR65, we only obtained a relatively weak solution using the ‘fc’ search model ensemble (TFZ = 5.8), and the quality of ARP/wARP model for this target was less satisfying (R-free=0.39 and GDT-TS=0.71). For targets BeR31 and SR213, although their ARP/wARP models were close to target X-ray structures, the free R values were relatively poor (> 0.4). In addition, for targets HR41, StR70 and PsR293 with resolution of X-ray diffraction data > 2.50 Å, no legitimate ARP/wARP models could be built from the ‘fc’ MR solutions (Table 1).
To validate the correctness of ‘fc’ MR solutions for targets that could not be modeled automatically with ARP/wARP, phenix.autobuild (Terwilliger et al., 2007) was used as an alternative automatic model rebuilding method. Models built by phenix.autobuild were generally of high quality (except for target StR70), with free R factors < 0.4, map correlation coefficient better than 0.75. and GDT-TS score to target X-ray structures > 0.85. For target StR70, although the quality of phenix.autobuild model was relatively poor with free R factor of 0.44 and map correlation coefficient of 0.62, it was still acceptable given the resolution of X-ray diffraction data is 2.80 Å (Supplementary Table S3); the R and Rfree values of the PDB deposited X-ray structure are 0.29 and 0.34 respectively. In conclusion, correct MR solutions were obtained and automatic model building of the crystal structure was done successfully for 22 of 25 of these NESG NMR/X-ray pairs, using the ‘fc’-trimmed NMR ensemble coordinates deposited in the PDB, Phaser, and either ARP/wARP or Phenix.
NMR structures can also be used as partial search models in solving complexes by MR
X-ray structure of NESG target OR8C, the “effector domain” of the influenza A virus non-structural protein 1 (NS1A), was determined as a tetrameric complex bound to the F2F3 Zn-finger fragment of human cellular polyadenylation and specificity factor 30 (CPSF30) (Das et al., 2008). In this complex, the asymmetric unit has four chains, two for OR8C and two for F2F3. The solution NMR structure of target OR8C is a monomer (Aramini et al., 2009). NMR search model ensembles trimmed using “core atom sets” determined by FindCore provide an unambiguous Phaser solution for the two OR8C chains, with final TFZ=19.5 and LLG=352. Starting from this MR solution from Phaser and using the 1.95 Å resolution X-ray data, ARP/wARP could build the structure of the entire complex automatically with high accuracy and almost complete sequence coverage. More specifically, for the ARP/wARP model, the R factor is 0.22, R-free is 0.27, and 344 of 361 residues were traced successfully. The Cα-rmsd between X-ray structure of the complex and the automated ARP/wARP model is less than 0.3 Å (Supplementary Fig. S2:A, Supplementary Fig. S2:B). These results demonstrate that NMR structures can also be used as partial search models for MR experiments, and can be used to solve the structures of protein-protein complexes when there are minimal structural rearrangements upon complex formation.
NMR structures that fail to provide good MR models can be improved by Rosetta refinement
Three NMR structures in our MR experiments failed to generate correct MR solutions with the methods described above. For NESG target DrR147D, the GDT-TS between NMR structure (PDB id: 2kcz) and X-ray structure (PDB ID: 3ggn) is quite low (0.48), as a large portion of the NMR structure [46 residues (i.e. residues 24–69) out of 155 residues] is not well defined. The X-ray crystal structure of target SR478 is a dimer of three-helix bundle domains, and the orientation of two N-terminal helices is somewhat different between NMR and X-ray structure, which accounts for about 40 percent of the X-ray structure. For ZR18, the overall agreement between secondary structure elements of the X-ray structure and the NMR structure are acceptable, however, the relative orientation between helix α1 (residues 40–47) and helix α2 (residues 71–81) is different in the NMR and X-ray structures; viz, the angles between those two helices in X-ray structure and NMR structure ensemble are 155.7 degree and 160.5–166.6 degree respectively. In addition, there are only 10 models in the reported NMR ensemble, which may not be large enough to properly sample the conformation space, providing an inaccurate estimate of precision that precludes proper elimination of inaccurately-defined regions in the initial model.
It has been pointed out previously that the phasing power of NMR structures that fail to provide good MR solutions can be significantly improved by Rosetta refinement (Qian et al., 2007; Ramelot et al., 2009). Therefore we carried out Rosetta loop rebuilding and all-atom refinement for NMR structure ensembles of NESG targets SR478 and ZR18, respectively. Improved agreement was observed between the X-ray structure and Rosetta-refined NMR structure compared to the NMR structure deposited in the PDB. For example, the angles between helix α1 and helix α2 of some Rosetta decoys for target ZR18 were within one degree variance from their corresponding X-ray structure. Both average GDT-TS and best GDT-TS between Rosetta models and X-ray structures were much higher than their PDB-deposited counterparts for those two targets (Supplementary Table S5). Using these Rosetta-refined NMR models, search models were prepared the same way as was done for the NMR structure ensembles. In both cases, we were able to obtain definite Phaser solutions starting from fc models with TFZ > 8 (Fig. 2A). Specifically, we obtained a solution with TFZ=9.9 for target ZR18 (identified by ZR18_R) and a solution with TFZ=11.3 for target SR478 (identified by SR178_R), which are significantly higher than the values of TFZ=4.5 for target ZR18 and TFZ=4.8 for target SR478, respectively, before Rosetta loop rebuilding and all-atom refinement. These results confirm the high value of the Rosetta loop-rebuilding and refinement protocol when using NMR structures for MR.
NMR structures can be successfully used as MR search models for homologous X-ray structures
As indicated by previous results, NESG NMR structures which have 100% sequence identity with target X-ray structures generally can be utilized successfully as MR search models. To further explore the value of NMR structures as MR search models, we identified homologous proteins in the PDB for nine (9) of the NESG NMR/X-ray structure pairs. These homologous X-ray structures were selected using the following criteria: (i) sequence identity with template sequence ≥ 20%, (ii) sequence coverage of the target by the template ≥ 70%, (iii) better than 3-Å diffraction data, and (iv) no more than 4 copies of the molecule in the asymmetric unit. These data sets for 9 homologous proteins are summarized in Supplementary Table S6.
For each target, we aligned the sequence of homologous protein with the sequence of our NMR/X-ray structure pair using the align2D function of Modeller software (Eswar et al., 2006). Unaligned residues were deleted from template NMR/X-ray structures, and unmatched sidechains were stripped back to the CG/OG coordinates. Based on these pre-processed NMR structure ensembles or X-ray structure coordinates, search models were prepared using each of the eight protocols summarized in Supplementary Table S2. Phaser was used to find MR solutions, and ARP/wARP was used for automatic model rebuilding.
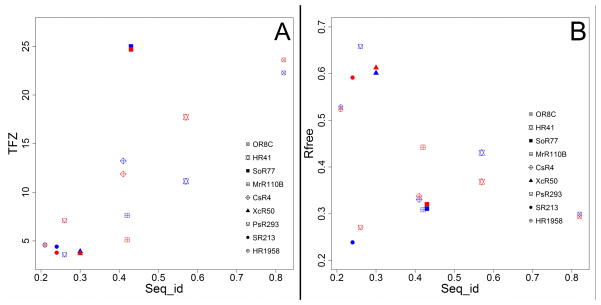
The results of this study can be divided into two subsets, distinguished by the sequence identity between the NMR/X-ray structure pair and the corresponding homologous X-ray crystal structures. For all five homologues with sequence identity > 40%, (i.e. for templates CsR4, HR41, MrR110B, OR8C and SoR77) correct MR solutions were found by Phaser, and a majority of residues could be successfully traced using ARP/wARP, with free R factors lower than 0.45 (Fig. 3B, Supplementary Table S7). On the other hand, for the four cases where the sequence identity between target X-ray sequence and template NMR/X-ray sequence is ≤ 30%, valid MR solutions were identified for only one case, SR213, with sequence identity of 24% and Phaser TFZ value of Z = 4.4. Subsequent model rebuilding demonstrates that this is indeed a correct solution, because the free R factor of the ARP/wARP model is only 0.24, and the GDT-TS value between the ARP/wARP model and target PDB structure is 0.94.
Fig. 3. NMR and X-ray structures are about equally useful as templates for obtaining MR solutions for homologous protein structures.
(A) Plot of TFZ scores of Phaser solutions vs. sequence identity (Seq_ID) between search model and target X-ray crystal structure. Solutions derived from X-ray crystal structure search models are colored red, and solutions derived from ‘fc’-trimmed NMR structure ensemble search models are colored blue. (B) Plot of free R factor values of final ARP/wARP models vs sequence identity between search models and target X-ray structures. Solutions derived from X-ray crystal structure search models are colored red, and solutions derived from ‘fc’-trimmed NMR structure ensemble search models are colored blue. See also Tables S6 and S7.
The same MR study was done using the corresponding NESG X-ray crystal structures, instead of the NMR structure ensembles, as MR templates. For all five targets with sequence identity greater than 40%, correct MR solutions could also be found using X-ray crystal structures as search models. Judged by TFZ scores of Phaser solutions and free R values of ARP/wARP models, for targets CsR4, OR8C and SoR77, the quality of MR solutions originating from either the NMR or X-ray search models was equally good. For target HR41, a better MR solution could be found using X-ray structure as a search model, while for target MrR110B a better MR solution was found using the ‘fc” trimmed NMR ensemble as the search model (Fig. 3, Supplementary Table S7). These results lead us to conclude that modern NMR structures can be as effective as X-ray crystal structures for MR of homologous protein structures, when the NMR coordinate ensemble is properly prepared.
Discussion
In this paper, we have shown that NESG NMR structures usually serve as excellent search models to estimate the phase information of their corresponding X-ray counterparts. Compared with X-ray crystallography, protein NMR structure determination is a relatively new field. The process of NMR structure determination is not as mature as the process of X-ray structure determination, and is still subject to intensive development. It is generally recognized that there is a gap between the quality of typical solution NMR structures and the best X-ray crystal structures (Bhattacharya et al., 2007). However, over the last decade protein NMR analysis of small (< 160-residue) proteins has become more routine, and the quality of protein NMR structures has improved significantly. NMR structures of such proteins generally have accuracies comparable to medium-resolution (2.0 – 2.5 Å) X-ray crystal structures (Bhattacharya et al., 2007). Moreover, as demonstrated in Fig. 1, the quality of NMR structures solved by structural genomics consortia, such as the NESG, has consistently improved over the past several years, as improved methods of data analysis and structure validation tools have been incorporated into the protein structure refinement process.
In this study, we failed to obtain MR solution for target DrR147D by all of the methods tested. Further investigation revealed that there are bona fide structural differences between these NMR and X-ray structures due to the fact they were solved at different pH values. Specifically, the solution NMR structure is a monomer solved at pH 4.5, while the crystal structure is a dimer solved at pH 6.0; most residues on the dimer interface observed in this crystal structure are disordered in the corresponding monomeric NMR structure (Supplementary Fig. S1), and this disorder to order transition is pH dependent (unpublished results).
In our 22 successful MR experiments, one case, NESG target HR3646E, is particularly interesting. Using the NMR ensemble to generate a ‘fc’-trimmed search model ensemble, we obtained one solution with TFZ=3.6 and LLG = 26, which was also the single solution reported by Phaser. Although we tried various model preparation methods and different Phaser parameters, this solution with low TFZ score was the best we could obtain; this was not unexpected since the best GDT-TS score between any individual NMR model and X-ray structure was only 0.77. None the less, a highly accurate model (GDT-TS relative to X-ray structure equals to 0.97) could be built by ARP/wARP using the initial MR solution, with 93 of 98 residues automatically-traced (Supplementary Fig S2:C). Although the resolution of the X-ray data is high (1.45 Å), ARP/wARP worked so well as to indicate that starting MR model produced by Phaser must be correct, even with a relatively low TFZ score of 3.6.
Recent developments in structural bioinformatics have further expanded the application of NMR data in molecular replacement. For example, for small proteins with less than 130 residues, CS-Rosetta models generated using only chemical shift data and energy calculations can be quite accurate (Shen et al., 2008), and have been used successfully as MR search models (Szymczyna, et al., 2009). In addition, as shown in Fig. 2A for NESG targets SR478 and ZR18, by focusing sampling on the most structurally variable regions, and then relaxing the whole NMR structure in the Rosetta all-atom energy field, Rosetta loop rebuilding protocol can be used to improve their agreement with X-ray structures to provide better phasing power (Qian et al., 2007; Ramelot et al., 2009). In this study, two NMR structures which did not initially provide MR solutions could be improved, both in phasing power and similarity with the crystal structure, by unconstrained Rosetta refinement. The generality of these results in using NMR structure ensembles as phasing models will be explored in future studies.
Methods
Data acquisition and preprocessing
The coordinates files of NMR structures and the structure factor files of X-ray structures were downloaded from PDB directly. The structure factor files, downloaded in mmCIF format, were converted to mtz format using the CCP4 program CIF2MTZ (Collaborative Computational Project, Number 4, 1994). Another CCP4 program uniquefy was used to standardize the mtz files and select reflections for free R calculation.
Search model preparation
For each NMR ensemble, eight different search models were prepared with various levels of simplification as detailed below. These methods are also summarized in Supplementary Table S2. For all those models, hydrogen atoms were deleted from NMR coordinates files.
nh model: A composite model including all the individual models in NMR ensemble and the coordinates of all the non-hydrogen atoms are kept.
bsm model: Single NMR model which has the highest structural similarity with X-ray structure.
aveB model: Average structure of NMR ensemble with distance based pseudo B-factor (Wilmanns and Nilges, 1996); coordinates of ‘not-well-defined’ residues calculated by the PSVS program based on dihedral order parameter values (Bhattacharya et al., 2007; Hyberts et al., 1992) are deleted.
AG model: Composite model including all the individual models in NMR ensemble residues with side chains longer than Ala are truncated to Ala. This model is based on the protocol as defined in the script multiprobe (ftp://X-ray.bmc.uu.se/pub/gerard/omac/multi_probe).
SAG model: Composite model including all the individual models in NMR ensemble and residues with side chains longer than Ser are truncated to Ser. This model is based on the protocol as defined in the script multiprobe (ftp://X-ray.bmc.uu.se/pub/gerard/omac/multi_probe).
nd model: Composite model including all the individual models in NMR ensemble for which coordinates of ‘not-well-defined’ residues calculated by PSVS program based on dihedral order parameter values (Bhattacharya et al., 2007; Hyberts et al., 1992) are deleted.
ndSAG model: Composite model including all the individual models in NMR ensemble. Coordinates of ‘not-well-defined’ residues calculated by PSVS program based on dihedral order parameter values (Bhattacharya et al., 2007; Hyberts et al., 1992) are removed, and surface residues with side chains longer than Ser are truncated to Ser.
fc model: Composite model with NMR ensemble trimmed by results of FindCore analysis. The atomic precision of the NMR structure ensemble was assessed by a pseudo B-factor, which was calculated from a variance distance matrix using the FindCore program (Snyder and Montelione, 2005). Each residue was treated as a tree data structure with backbone atoms (N, Cα, C, O) being defined as the root, and side chain heavy atoms were defined as child nodes and their precedence were determined by their relative distance to Cα; e.g., Cβ is the child node of Cα, and Cγ is the child node of Cβ. Any nodes together with their child nodes were removed from search model if their pseudo B-factors calculated by FindCore, were equal or greater than 60.
MR trials and automatic model building and refinement
The program Phaser (McCoy et al., 2007) (version 2.1) was used for molecular replacement. MR_AUTO mode was adopted with RMS being set to 1.5. Program ARP/wARP version 7.0 (Perrakis et al., 2001) was used for automatic model building starting from the Phaser MR solution. The ARP/wARP expert system mode was employed for automatic model building, and Refmac5 (Murshudov et al., 2003) was used in refinement, staring from the positioned search model and a maximum of 10 building cycles were allowed. Phenix.autobuild (Terwilliger et al., 2007) was employed for automatic model rebuilding if ARP/wARP failed to generate good quality models. No manual model building was applied to any case, to allow a fair comparison of each MR trials.
We developed a pipeline using Perl script language to run Phaser and ARP/wARP jobs on a cluster of 128 CPUs in a highly automated manner. TFZ and LLG values were extracted from Phaser solutions to assess the quality of MR solutions. The quality of models automatically built by ARP/wARP was judged by R, R-free, and the completeness of auto-tracing. In addition, structural similarity between ARP/wARP models and corresponding X-ray structures were evaluated by GDT-TS score (Zemla et al., 2003).
Coot (Emsley and Cowtan, 2004) was used to check the models and electron density maps, after molecular replacement, and after model building in ARP/wARP. The TM-score program (Zhang and Skolnick, 2004) was used to perform structural alignment and GDT-TS calculation (Zemla et al., 2003).
Rosetta loop rebuilding and all atom refinement
The Robetta fragment server (http://robetta.bakerlab.org/fragmentsubmit.jsp) (Chivian et al., 2003; Kim et al., 2004) was used to generate fragment library, based on sequence and chemical shift data of each target protein. Then loop rebuilding and all atom refinement (Misura et al., 2005; Bradley et al., 2005) was done by Rosetta cyclic coordinate descent (CCD) and kinematic closure (KIC) loop modeling application (Version 3.0), ‘fastrelax’ mode was used to allow the whole structure to relax in Rosetta all-atom force field, and could be 5–10 times faster than normal relaxation mode. For each target protein, loop regions were defined by the consensus of secondary structure, “not-well-defined” residues were identified by the PSVS program based on dihedral order parameter values (Bhattacharya et al., 2007; Hyberts et al., 1992), and non-core residues defined by FindCore program (Snyder and Montelione, 2005). 1000 decoys were generated from each individual model of NMR structure ensemble, and the overall top 20 decoys with the lowest Rosetta energy were selected and combined as a composite model to be used in molecular replacement the same way as their NMR counterpart.
Supplementary Material
Highlights.
Modern protein NMR structures are generally accurate enough for MR applications.
Variance matrix methods allow NMR structures to be used for MR applications.
Rosetta refinement can sometimes improve the phasing power of NMR structures.
Acknowledgments
We thank Drs. L. Tong and F. Faroud for helpful comments on this manuscript. This work was supported by the National Institutes of General Medical Science Protein Structure Initiative program, grants U54 GM074958 and U54 GM094597. PDB and BMRB codes for the NMR NOESY peak list and chemical shift data, as well as crystallographic structure factor, data for the 25 proteins used in this study are summarized in Supplementary Table S1, and available on line at http://psvs-1_4-dev.nesg.org/MR/dataset.html and as a link from the BioMagResDB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson DH, Weiss MS, Eisenberg D. A challenging case for protein crystal structure determination: the mating pheromone Er-1 from Euplotes raikovi. Acta Crystallogr D Biol Crystallogr. 1996;52:469–480. doi: 10.1107/S0907444995014235. [DOI] [PubMed] [Google Scholar]
- Aramini JM, Ma L, Lee H, Zhao L, Cunningham K, Ciccosanti C, Janjua H, Fang Y, Xiao R, Krug RM, Montelione GT. Solution NMR structure of the monomeric W187R mutant of A/Udorn NS1 effector domain. Northeast Structural Genomics target OR8C[W187R] journal. 2009 page numbers, etc. [Google Scholar]
- Baldwin ET, Weber IT, St Charles R, Xuan JC, Appella E, Yamada M, Matsushima K, Edwards BF, Clore GM, Gronenborn AM, et al. Crystal structure of interleukin 8: symbiosis of NMR and crystallography. Proc Natl Acad Sci U S A. 1991;88:502–506. doi: 10.1073/pnas.88.2.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley SK, Joachimiak A, Montelione GT, Wilson IA, et al. Contributions to the NIH Protein Structure Initiative from the four large-scale production centers. Structure. 2008;16:5–11. doi: 10.1016/j.str.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Tejero R, Montelione GT. Evaluating protein structures determined by structural genomics consortia. Proteins. 2007;66:778–795. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- Blow DM, Rossmann MG. the single isomorphous replacement method. Acta Cryst. 1961;14:1195–1202. [Google Scholar]
- Bradley P, Malmstrom L, Qian B, Schonbrun J, Chivian D, Kim DE, Meiler J, Misura KM, Baker D. Free modeling with Rosetta in CASP6. Proteins. 2005;61(Suppl 7):128–134. doi: 10.1002/prot.20729. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Campbell RL, Clore GM, Gronenborn AM, Karplus M, Petsko GA, Teeter MM. Solution of a Protein Crystal-Structure with a Model Obtained from Nmr Interproton Distance Restraints. Science. 1987;235:1049–1053. doi: 10.1126/science.235.4792.1049. [DOI] [PubMed] [Google Scholar]
- Chen YW. Solution solution: using NMR models for molecular replacement. Acta Crystallogr D Biol Crystallogr. 2001;57:1457–1461. doi: 10.1107/s0907444901010824. [DOI] [PubMed] [Google Scholar]
- Chen YW, Clore GM. A systematic case study on using NMR models for molecular replacement: p53 tetramerization domain revisited. Acta Crystallogr D Biol Crystallogr. 2000;56:1535–1540. doi: 10.1107/s0907444900012002. Standard journal name. [DOI] [PubMed] [Google Scholar]
- Chen YW, Dodson EJ, Kleywegt GJ. Does NMR mean “not for molecular replacement”? Using NMR-based search models to solve protein crystal structures. Structure. 2000;8:R213–220. doi: 10.1016/s0969-2126(00)00524-4. [DOI] [PubMed] [Google Scholar]
- Chivian D, Kim DE, Malmstrom L, Bradley P, Robertson T, Murphy P, Strauss CE, Bonneau R, Rohl CA, Baker D. Automated prediction of CASP-5 structures using the Robetta server. Proteins. 2003;53(Suppl 6):524–533. doi: 10.1002/prot.10529. [DOI] [PubMed] [Google Scholar]
- Cohen SX, Ben Jelloul M, Long F, Vagin A, Knipscheer P, Lebbink J, Sixma TK, Lamzin VS, Murshudov GN, Perrakis A. ARP/wARP and molecular replacement: the next generation. Acta Crystallogr D Biol Crystallogr. 2008;64:49–60. doi: 10.1107/S0907444907047580. Standard journal name. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K, Ma LC, Xiao R, Radvansky B, Aramini J, Zhao L, Marklund J, Kuo RL, Twu KY, Arnold E, et al. Structural basis for suppression of a host antiviral response by influenza A virus. Proc Natl Acad Sci U S A. 2008;105:13093–13098. doi: 10.1073/pnas.0805213105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. Standard journal name. [DOI] [PubMed] [Google Scholar]
- Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics. 2006;Chapter 5(Unit 5):6. doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P, McCoy A. An introduction to molecular replacement. Acta Crystallogr D Biol Crystallogr. 2008;64:1–10. doi: 10.1107/S0907444907051554. Standard journal name. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgetti A, Raimondo D, Miele AE, Tramontano A. Evaluating the usefulness of protein structure models for molecular replacement. Bioinformatics. 2005;21(Suppl 2):ii, 72–76. doi: 10.1093/bioinformatics/bti1112. [DOI] [PubMed] [Google Scholar]
- Glykos NM, Kokkinidis M. A stochastic approach to molecular replacement. Acta Crystallogr D Biol Crystallogr. 2000;56:169–174. doi: 10.1107/s0907444999015322. [DOI] [PubMed] [Google Scholar]
- Green DW, Jngram VM, Perutz MF. The structure of haemoglobin IV. Sign determination by the isomorphous replacement method. Proc Roy Soc. 1954;A225(1954):287–307. [Google Scholar]
- Hendrickson WA. Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science. 1991;254:51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- Huang YJ, Powers R, Montelione GT. Protein NMR recall, precision, and F-measure scores (RPF scores): structure quality assessment measures based on information retrieval statistics. J Am Chem Soc. 2005;127:1665–1674. doi: 10.1021/ja047109h. [DOI] [PubMed] [Google Scholar]
- Hyberts SG, Goldberg MS, Havel TF, Wagner G. The solution structure of eglin c based on measurements of many NOEs and coupling constants and its comparison with X-ray structures. Protein Sci. 1992;1:736–751. doi: 10.1002/pro.5560010606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamrog DC, Zhang Y, Phillips GN., Jr SOMoRe: a multidimensional search and optimization approach to molecular replacement. Acta Crystallogr D Biol Crystallogr. 2003;59:304–314. doi: 10.1107/s0907444902021935. [DOI] [PubMed] [Google Scholar]
- Jogl G, Tao X, Xu Y, Tong L. COMO: a program for combined molecular replacement. Acta Crystallogr D Biol Crystallogr. 2001;57:1127–1134. doi: 10.1107/s0907444901006783. [DOI] [PubMed] [Google Scholar]
- Keegan RM, Winn MD. MrBUMP: an automated pipeline for molecular replacement. Acta Crystallogr D Biol Crystallogr. 2008;64:119–124. doi: 10.1107/S0907444907037195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Gardner SP, Sutcliffe MJ. An automated approach for defining core atoms and domains in an ensemble of NMR-derived protein structures. Protein Eng. 1997;10:737–741. doi: 10.1093/protein/10.6.737. [DOI] [PubMed] [Google Scholar]
- Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32:W526–531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Szyperski T. GFT NMR, a new approach to rapidly obtain precise high-dimensional NMR spectral information. J Am Chem Soc. 2003;125:1385–1393. doi: 10.1021/ja028197d. [DOI] [PubMed] [Google Scholar]
- Kissinger CR, Gehlhaar DK, Fogel DB. Rapid automated molecular replacement by evolutionary search. Acta Crystallogr D Biol Crystallogr. 1999;55:484–491. doi: 10.1107/s0907444998012517. [DOI] [PubMed] [Google Scholar]
- Kleywegt GJ, Bergfors T, Senn H, Le Motte P, Gsell B, Shudo K, Jones TA. Crystal structures of cellular retinoic acid binding proteins I and II in complex with all-trans-retinoic acid and a synthetic retinoid. Structure. 1994;2:1241–1258. doi: 10.1016/s0969-2126(94)00125-1. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Leahy DJ, Axel R, Hendrickson WA. Crystal structure of a soluble form of the human T cell coreceptor CD8 at 2.6 A resolution. Cell. 1992;68:1145–1162. doi: 10.1016/0092-8674(92)90085-q. [DOI] [PubMed] [Google Scholar]
- Lee B, Richards FM. JMolBiol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Liu G, Li Z, Chiang Y, Acton T, Montelione GT, Murray D, Szyperski T. High-quality homology models derived from NMR and X-ray structures of E. coli proteins YgdK and Suf E suggest that all members of the YgdK/Suf E protein family are enhancers of cysteine desulfurases. Protein Sci. 2005;14:1597–1608. doi: 10.1110/ps.041322705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Montelione GT, Rost B. Novel leverage of structural genomics. Nat Biotechnol. 2007;25:849–851. doi: 10.1038/nbt0807-849. [DOI] [PubMed] [Google Scholar]
- Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- Mccoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misura KM, Baker D. Progress and challenges in high-resolution refinement of protein structure models. Proteins. 2005;59:15–29. doi: 10.1002/prot.20376. [DOI] [PubMed] [Google Scholar]
- Muller T, Oehlenschlager F, Buehner M. Human interleukin-4 and variant R88Q: phasing X-ray diffraction data by molecular replacement using X-ray and nuclear magnetic resonance models. J Mol Biol. 1995;247:360–372. doi: 10.1006/jmbi.1994.0144. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nair R, Liu J, Soong TT, Acton TB, Everett JK, Kouranov A, Fiser A, Godzik A, Jaroszewski L, Orengo C, et al. Structural genomics is the largest contributor of novel structural leverage. J Struct Funct Genomics. 2009;10:181–191. doi: 10.1007/s10969-008-9055-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaza J. Implementation of molecular replacement in AMoRe. Acta Crystallogr D Biol Crystallogr. 2001;57:1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- Pahler A, Smith JL, Hendrickson WA. A probability representation for phase information from multiwavelength anomalous dispersion. Acta Crystallogr A. 1990;46(Pt 7):537–540. doi: 10.1107/s0108767390002379. [DOI] [PubMed] [Google Scholar]
- Pannu NS, Read RJ. The application of multivariate statistical techniques improves single-wavelength anomalous diffraction phasing. Acta Crystallogr D Biol Crystallogr. 2004;60:22–27. doi: 10.1107/s0907444903020808. [DOI] [PubMed] [Google Scholar]
- Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS. ARP/wARP and molecular replacement. Acta Crystallogr D Biol Crystallogr. 2001;57:1445–1450. doi: 10.1107/s0907444901014007. [DOI] [PubMed] [Google Scholar]
- Perutz MF. Isomorphous replacement and phase determination in noncentrosymmetric space groups. Acta Cryst. 1956;9(1956):867–873. [Google Scholar]
- Qian B, Raman S, Das R, Bradley P, McCoy AJ, Read RJ, Baker D. High-resolution structure prediction and the crystallographic phase problem. Nature. 2007;450:259–264. doi: 10.1038/nature06249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramelot TA, Raman S, Kuzin AP, Xiao R, Ma LC, Acton TB, Hunt JF, Montelione GT, Baker D, Kennedy MA. Improving NMR protein structure quality by Rosetta refinement: a molecular replacement study. Proteins. 2009;75:147–167. doi: 10.1002/prot.22229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D Biol Crystallogr. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- Rossmann MG. The Molecular Replacement Method. Godon & Breach; New York: 1972. [Google Scholar]
- Rossmann MG, Arnold E. Patterson and molecular-replacement techniques. International Tables for Crystallography. 1993;B:230–263. [Google Scholar]
- Rossmann MG, Blow DM. The detection of sub-units within the crystallographic asymmetric unit. Acta Cryst. 1962;15:24–31. [Google Scholar]
- Saff EB, Kuijlaars ABJ. The Mathematical Intelligencer. 1997;19:5–11. [Google Scholar]
- Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu G, Eletsky A, Wu Y, Singarapu KK, Lemak A, et al. Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci U S A. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder DA, Montelione GT. Clustering algorithms for identifying core atom sets and for assessing the precision of protein structure ensembles. Proteins. 2005;59:673–686. doi: 10.1002/prot.20402. [DOI] [PubMed] [Google Scholar]
- Szymczyna BR, Taurog RE, Young MJ, Snyder JC, Johnson JE, Williamson JR. Synergy of NMR, computation, and X-ray crystallography for structural biology. Structure. 2009;17:499–507. doi: 10.1016/j.str.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede T, Sali A, Honig B, Levitt M, Berman HM, Jones D, Brenner SE, Burley SK, Das R, Dokholyan NV, et al. Outcome of a workshop on applications of protein models in biomedical research. Structure. 2009;17:151–159. doi: 10.1016/j.str.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, Adams PD. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr D Biol Crystallogr. 2000;56:1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- Wilmanns M, Nilges M. Molecular replacement with NMR models using distance-derived pseudo B factors. Acta Crystallogr D Biol Crystallogr. 1996;52:973–982. doi: 10.1107/S0907444996003393. [DOI] [PubMed] [Google Scholar]
- Woolfson MM. Direct methods in crystallography. Rep Prog Phys. 1971:369–434. [Google Scholar]
- Zemla A. LGA: A method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003;31:3370–3374. doi: 10.1093/nar/gkg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins. 2007;69(Suppl 8):108–117. doi: 10.1002/prot.21702. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.