Abstract
The Nephrotic Syndrome Study Network (NEPTUNE) is a North American multi-center collaborative consortium established to develop a translational research infrastructure for Nephrotic Syndrome. This includes a longitudinal observational cohort study, a pilot and ancillary studies program, a training program, and a patient contact registry. NEPTUNE will enroll 450 adults and children with minimal change disease, focal segmental glomerulosclerosis and membranous nephropathy for detailed clinical, histopathologic, and molecular phenotyping at the time of clinically-indicated renal biopsy. Initial visits will include an extensive clinical history, physical examination, collection of urine, blood and renal tissue samples, and assessments of quality of life and patient-reported outcomes. Follow-up history, physical measures, urine and blood samples, and questionnaires will be obtained every 4 months in the first year and bi-annually, thereafter. Molecular profiles and gene expression data will be linked to phenotypic, genetic, and digitalized histologic data for comprehensive analyses using systems biology approaches. Analytical strategies were designed to transform descriptive information to mechanistic disease classification for Nephrotic Syndrome and to identify clinical, histological, and genomic disease predictors. Thus, understanding the complexity of the disease pathogenesis will guide further investigation for targeted therapeutic strategies.
Introduction
Primary non-inflammatory glomerular diseases that include minimal change disease (MCD), focal and segmental glomerulosclerosis (FSGS) and membranous nephropathy (MN) are rare diseases that cause serious morbidity and high mortality accounting for approximately 15% of prevalent ESRD cases (2008) at an annual cost in the USA of more than $3 billion.1, 2 Our limited knowledge of underlying glomerular disease mechanisms is a major barrier to successful therapeutic intervention. To date, this void has not been overcome by surrogate clinical and histopathology-based classification methods, which are of limited value in reliably predicting disease natural history or responsiveness to therapy. Hence, there is a need for basic and translational investigation to identify underlying disease mechanisms, specific biomarkers, and therapeutic targets for further testing in clinical trials.
Integration of large-scale multi-dimensional molecular and clinical data sets obtained from a well-characterized patient population could enhance our ability to understand the spectrum of molecular disease entities that present as a common histological phenotype, elucidate pathogenesis, and expedite scientific innovation toward safe and effective treatments for glomerular diseases3.
To facilitate studies of this type, the Nephrotic Syndrome Study Network (NEPTUNE), supported by the National Institutes of Diabetes and Digestive and Kidney Disease (NIDDK), and the Office for Rare Diseases Research (ORDR) at the National Institutes of Health (NIH), and The NephCure and Halpin Foundations, was designed to conduct multi-disciplinary innovative research to address the “translational gap,” which is a barrier to successful management of adults and children with FSGS, MCD and MN.
The overall objectives of the NEPTUNE network (Table 1) are incorporated into the NEPTUNE observational cohort study that is designed to address the following general and disease-specific hypotheses:
After accounting for types, duration, and frequency of immunomodulating therapy, the probability of remission of FSGS, MCD and MN, defined as change in urinary protein excretion and/or renal function, will be predicted by baseline demographics, clinical, genetic, and histopathological characteristics at the time of presentation.
Genome-wide molecular profiles obtained from renal biopsy tissue at the time of diagnosis of FSGS, MCD and MN will independently predict the proportion of subjects entering remission.
Individual genetic causes of FSGS and MCD activate specific transcriptional pathways in renal tissue and the molecular characterization of these transcriptional pathways will identify novel therapeutic targets yielding more complete and sustained disease remission.
The use of non-immunomodulating therapy (including anti-hypertensive, lipid-lowering, and antithrombotic agents and dietary manipulation) will have an independent effect on the probability of remission of nephrotic syndrome.
The composite rates of major medical complications (vascular thrombosis, complications of the kidney biopsy procedure, infection, and death) will vary according to the baseline characteristics and types of treatment in patients.
Nephrotic syndrome, its treatment and clinical outcomes, will have a varied measurable effect on quality of life of patients.
Table 1.
Objectives of the Nephrotic Syndrome Study Network (NEPTUNE)
| Objectives of the Nephrotic Syndrome Study Network |
|---|
| Establish a collaborative, integrated, cost-effective investigational infrastructure to conduct clinical and translational research in FSGS, MCD, and MN |
| Perform a longitudinal observational cohort study on patients with incipient biopsy proven NS |
| Establish Pilot and Ancillary Projects Programs using the unique resources, clinical data, or specimens collected by NEPTUNE |
| Establish a Training Program for post-doctoral/junior faculty trainees to prepare for clinical and translational research in glomerular disease |
| Collaborate with the ORDR Data Management and Coordinating Center and the NephCure Foundation to establish a web-based exchange platform for lay people, physicians, and scientists. |
Results
Overview of Study Design and Organization
The NEPTUNE study is a prospective observational study that will enroll 450 children and adults with FSGS, MCD and MN at the time of first clinically-indicated renal biopsy for diagnosis of proteinuria. Enrollment from 18 clinical centers in the U.S. and Canada (Table 2) will occur over 30 months with a minimum follow-up of 30 months. The two primary study outcomes are change in urinary protein excretion and change in renal function as indicated in Table 3. Secondary study outcomes are quality of life assessment, development of new-onset diabetes, malignancies, infections, thromboembolic events, hospitalization, acute kidney injury, and death.
Table 2.
Nephrotic Syndrome Study Network Clinical Centers
| NEPTUNE Clinical Centers* | Recruitment Population |
|---|---|
| Case Western Medical Center, Cleveland, OH | Adult and Pediatric |
| Cohen’s Children’s Medical Center, Manhasset, NY | Pediatric |
| Columbia University, New York, NY | Adult |
| Emory University, Atlanta, GA | Pediatric |
| Harbor Biomedical Research Institute, Torrance, CA | Adult |
| Johns Hopkins Medical Center, Baltimore, MD | Pediatric |
| Montefiore Medical Center, Bronx, NY | Adult and Pediatric |
| The Mayo Clinic, Rochester, MN | Adult and Pediatric |
| National Institute of Diabetes and Digestive and Kidney Diseases, NIH, Bethesda, MD | Adult |
| New York University School of Medicine, New York, NY | Adult and Pediatric |
| Temple University Health Systems, Philadelphia, PA | Adult |
| University Health Network, Toronto, Canada | Adult |
| University of Michigan Health Systems, Ann Arbor, MI | Adult and Pediatric |
| University of Miami Medical Center, Miami, FL | Adult and Pediatric |
| University of North Carolina, Chapel Hill, NC | Adult and Pediatric |
| University of Pennsylvania, Philadelphia, PA | Adult and Pediatric |
| University of Southern California Children’s Hospital, Los Angeles, CA | Pediatric |
| University of Washington, Seattle, WA | Adult and Pediatric |
For full list of sites enrolling NEPTUNE participants, see Appendix
Table 3.
NEPTUNE Primary Outcome Variables and Definitions
| Rate of Change in Urinary Protein Excretion from Baseline (UP:C ratio) | Rate of Change in Renal Function from Baseline |
||
|---|---|---|---|
| Definitions | Standard | Cohort Study | Cohort Study |
| Reduction in UP:C ratio of ≤0.3 Reduction in UP:C ratio > 50% and final U:C ratio ≥0.3 and ≤3.5 |
Complete Remission Partial Remission |
Complete/Partial Remission |
25 ml/min/1.73m2 reduction in eGFR* |
| Reduction in UP:C ratio > 50% and final >3.5 Reduction in UP:C ratio < 50% (includes increases < 50%) Urine P:C ratio increases by >50% |
Limited Response Non-Response Progressive Proteinuria |
No Response | 50% decline in eGFR from baseline |
| New development of UP:C ratio >3.5 after complete or partial remission | Relapse | Relapse | ESRD (initiation of dialysis, kidney transplantation or eGFR ≤ 10 ml/min/1.73m2 |
“Abbreviations: UP:C = urine protein: creatinine (g/g); eGFR = estimated glomerular filtration rate; ESRD = end stage renal disease”
The NEPTUNE consortium infrastructure consists of a Steering Committee, a Central Digitalized Histopathological Archive at the NIH, a Central Biorepository, Biochemical Laboratory and Data Analysis and Coordinating Center (DACC) at the University of Michigan, an Observational Study Monitoring Board at the NIDDK, and an external Scientific Advisory Board.
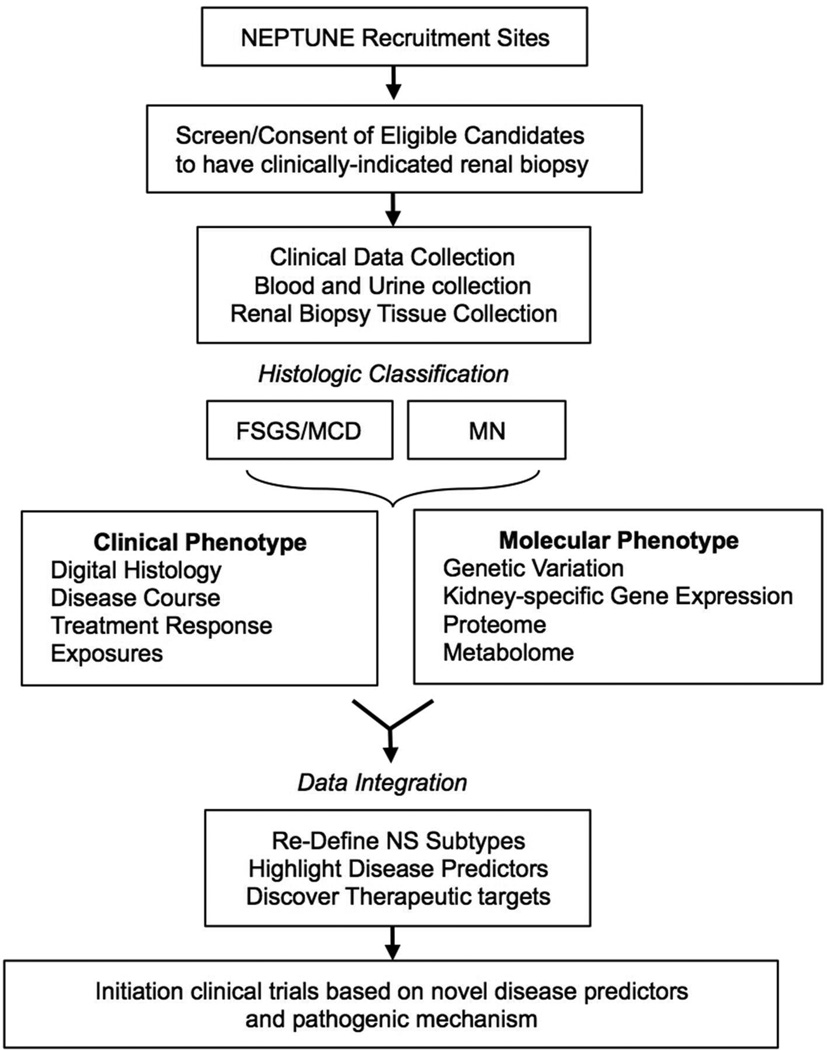
The overall design of the NEPTUNE cohort study is depicted in Figure 1 and described in detail below, NEPTUNE is registered at clinicaltrials.gov under NCT01209000.
Figure 1. NEPTUNE Cohort Study Design.
NEPTUNE Cohort Participants
The NEPTUNE study will enroll a diverse population reflective of the disease burden in North America. Eligible candidates for the NEPTUNE study are individuals of any age scheduled for a clinically-indicated renal biopsy with proteinuria ≥ 500 mg/day estimated from a 24-hour or spot (protein to creatinine ratio) urine collection. Patients with sub-nephrotic proteinuria are included to capture the broad spectrum of clinical presentations4, 5. Exclusion criteria include patients with clinical, serological, or histological evidence of other renal diseases (e.g. diabetic nephropathy, systemic lupus erythematosus, etc), prior solid organ transplant, life expectancy < 6 months, and those who are unwilling or unable to consent. Cohort assignment is based on detailed renal histopathological analysis. Participants whose histopathology is not determined to be consistent with FSGS, MCD and MN are assigned to an “Other Glomerulopathies” cohort and retained in the study to serve as a comparison group and for future studies addressing these histopathologically-distinct entities. Eligible candidates over age 18 are enrolled after providing written, informed consent.
NEPTUNE Pediatric Population
Children are an important population affected by nephrotic syndrome (NS) and are enrolled in the NEPTUNE study at many clinical centers as indicated in Table 2. The NEPTUNE study will enroll eligible children after parental written consent (and child assent when appropriate) prior to the diagnostic renal biopsy. Information on medications prescribed for NS prior to enrollment will be collected in all participants including children who may have a longer exposure to pre-biopsy therapies consistent with standard pediatric nephrotic syndrome management6. Additional funding to enroll children from initial nephrotic syndrome presentation is currently being pursued.
Study Enrollment and Follow-Up Visits
The enrollment process includes a screening visit to assess eligibility and willingness to participate, a baseline visit for collection of clinical and demographic information, and a renal biopsy visit to obtain renal tissue samples. Renal biopsy material will be used to confirm assignment into the specific study cohort and for research purposes. The baseline visit includes collection of a detailed medical history, sociodemographic information, physical measures, and blood and urine samples (see https://rarediseasesnetwork.epi.usf.edu/NEPTUNE/professional/studynetwork.htm# for details). In addition to age-appropriate questionnaires to assess quality of life, self-reported health will be evaluated using the Patient Reported Outcomes Measurement Information System (PROMIS, www.nihpromis.org). After enrollment, NEPTUNE participants are evaluated at 4-month intervals during the first year and every 6 months thereafter with similar data collection as performed in the baseline visit.
NEPTUNE Pathology Digital Imaging and Scoring System
Pathology material including glass slides, immunofluorescence images, and electron-micrographs, will be scanned into high resolution digital images and stored in the NEPTUNE pathology database along with copies of the local pathology reports. This database can be accessed remotely by the NEPTUNE Histopathology committee for review, scoring, and cohort assignment by disease classification.
The NEPTUNE Histopathology Committee has developed a novel strategy for scoring renal biopsies designed to standardize morphologic analysis7,8. The pathology protocol is designed to provide 1) multi-level annotation of glomeruli for multi-dimensional reconstruction to allow simultaneous review and scoring of the same glomerulus on each digital slide, and 2) identification and scoring of specific pre-defined morphologic “descriptors”. This approach for kidney tissue analysis has the potential to redefine histopathologic classification and link these novel criteria to clinical and molecular phenotypes. In addition, the NEPTUNE pathology database will serve as a major resource available to investigators for future pilot and ancillary studies.
Overview of Applied Systems Biology Analysis Strategy
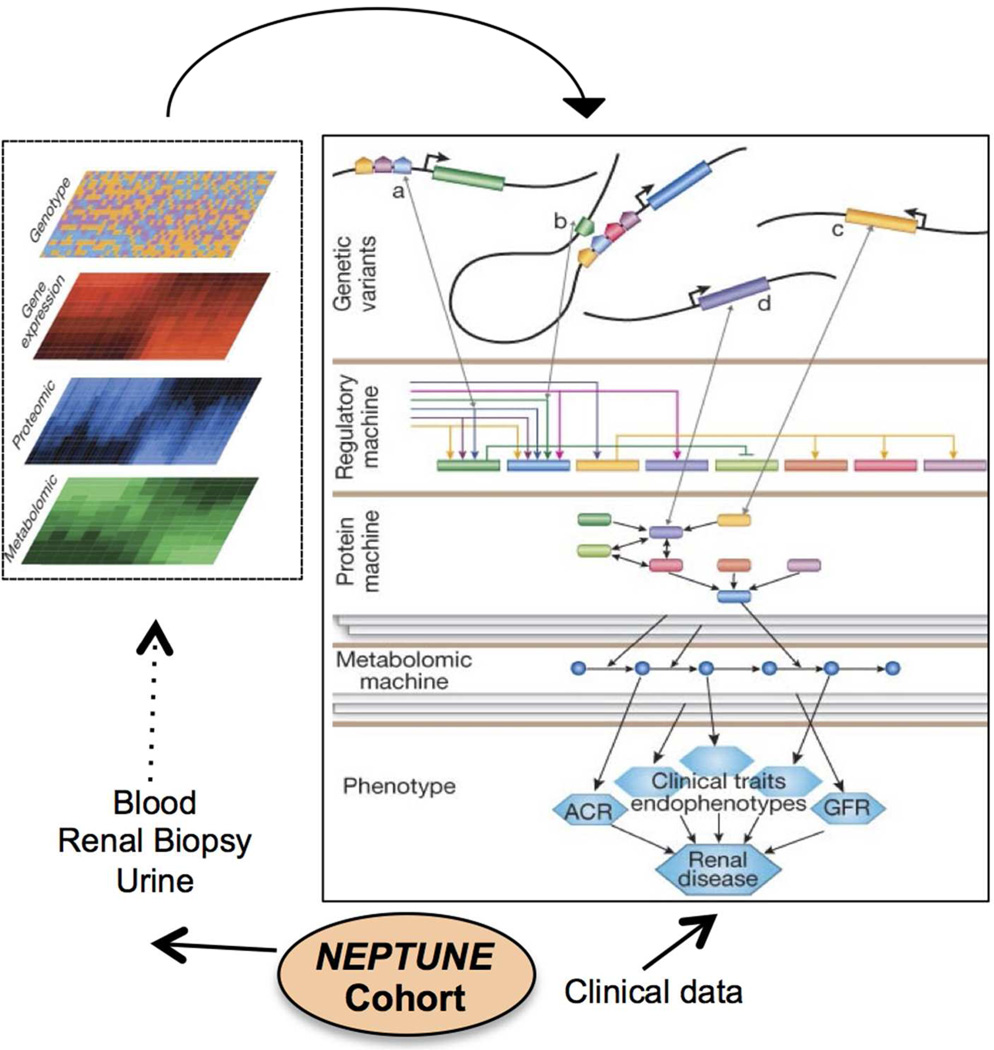
Systems biology is an emerging scientific approach that aims to address the ‘pluralism of cause and effects in biological networks’9 that defines the steady state and its disturbances in health and disease10,11,12,13, 14. The NEPTUNE study aims to integrate comprehensive clinical and pathologic phenotypes with extensive analysis of the molecular regulatory networks active in glomerular diseases. As depicted in figure 2, NEPTUNE will prospectively procure human samples for analyses focused on genome-wide profiling of genetic variation, epigenetic markers and transcriptomic, proteomic, and metabolomic profiles. Emerging systems biology approaches will be used to define associations between these multi-phasic and inter-related genome scale profiles and the detailed clinical and structural phenotypes.
Figure 2. Overview of Multilevel Data Integration in NEPTUNE.
“The integrative analytical approach in NEPTUNE relies on obtaining comprehensive molecular and clinical information from each subject. For molecular analysis, the initial step is to generate large-scale datasets of genome-wide genetic variation and targeted gene sequencing, compartment specific gene expression data on renal biopsy tissue, leukocytes and urine, and proteomic/metabolomic data from both urine and blood samples. Each molecular dataset will be interrogated alone and in relationship with the others in order to understand multidimensional molecular interactions. In addition, the molecular data will be analyzed with the comprehensive clinical information to determine associations between molecular events and clinical outcomes. The overall goal is to develop a framework for studies to define the molecular heterogeneity of NS for disease stratification, biomarker identification and molecular target definition. (Figure modified from Keller et al10).”
The NEPTUNE study will establish a core molecular data set containing genetic profiles and renal tissue, compartment-specific, genome wide mRNA expression data to facilitate multi- and trans-disciplinary research exploration along the genotype – phenotype continuum10,11,12,13.The NEPTUNE core data sets will provide a foundation to link mechanistic information with clinical disease presentation, and link molecular information from model systems with human nephrotic syndrome data sets. Patient subgroups with activation of specific molecular mechanisms can be used to assess the efficacy of novel therapeutics in a highly targeted manner. Defining patients by the underlying disease mechanism will reduce the heterogeneity inherent to the current histology-based glomerular disease classification and, as a result, will focus the analysis and interpretation of clinical trials.
The core and ancillary data generated in NEPTUNE can be a significant resource to the renal research community. NEPTUNE will utilize Nephromine (www.Nephromine.org), a renal-specific systems biology search engine, to enable large-scale data analysis by nephrology investigators with and without systems biology expertise.
Statistical Considerations
The research aims will be explored separately in FSGS, MCD cohort and MN cohort. The primary objective of the statistical analysis plan is to build a predictive model that utilizes multiple sources of data, including molecular biomarkers and auxiliary predictors (e.g. race/ethnicity, edema), to classify patients according to the proteinuria outcome (remission) as the primary endpoint. Secondary study endpoints include longitudinal measures of renal function. The primary analysis plan involves deriving a prediction model to rigorously evaluate and identify potential predictors for study endpoints. Subsequently, the overall prediction model will be used to assess relative risk and association via odds ratios for targeted sub-group analysis. In analysis of data from the network, random-effects models will be used to account for potential heterogeneity and unstructured interventions across and within the clinical centers. Also, the analysis is tailored to handle dropouts, incomplete longitudinal data, and other types of missing data15. For further details on statistical methodology, see Supplement.
For biomarker discovery and validation, the sample will be split into the training and the validation sets at the ratio of 16, 17. With regard to the primary endpoint, selection of biomarkers (e.g. gene expression levels or serum or urine proteomic markers) will proceed in two steps: first, univariate variable screening will identify a small pool of promising biomarkers and, subsequently, joint screening to narrow identified candidate biomarkers to those with the optimal predictive power. Using selected biomarkers, a prediction model will be developed that is further validated via the test cohort by the criteria of ROC curves and net reclassification index18, 19. In the analysis of secondary endpoints, similar strategies will be applied to build prediction models to classify longitudinal trajectories (e.g. estimated GFR).
Sample Size Consideration
The NEPTUNE study will have adequate power to identify important molecular biomarkers predicting the primary endpoints. Power calculations were based on the probability of correct classification (PCC)20. The expected probability of PCC is a weighted average of sensitivity (probability o f remission) and specificity (probability of non-remission). An estimate 20,000 genes (probes) and 75 auxiliary predictors will be screened to identify true gene expression markers and true auxiliary predictor for the defined endpoints (Table 4). The PCCs are 0.874, 0.942 and 0.966 for the sample sizes of 100, 150 and 200, respectively, based on the identification of 50 true gene expression markers and 10 true auxillary markers. The NEPTUNE study enrollment goal is to enroll 450 subjects.
Table 4.
Sample size and Classification Power for the NEPTUNE Study
| Prevalence = 0.6 | Prevalence = 0.3 | |||||||
|---|---|---|---|---|---|---|---|---|
| rm = 5 | rC = 10 | rm = 5 | rC = 10 | |||||
| δm/σm | PCC* | PCC | PCC* | PCC | ||||
| n=100 | n=150 | n=200 | n=100 | n=150 | n=200 | |||
| 0.2 | 0.788 | 0.638 | 0.667 | 0.687 | 0.812 | 0.670 | 0.675 | 0.690 |
| 0.3 | 0.881 | 0.776 | 0.803 | 0.818 | 0.892 | 0.779 | 0.807 | 0.822 |
| rm = 50 | rC = 10 | rm = 50 | rC = 10 | |||||
| δm/σm | PCC* | PCC | PCC* | PCC | ||||
| n=100 | n=150 | n=200 | n=100 | n=150 | n=200 | |||
| 0.2 | 0.941 | 0.639 | 0.695 | 0.753 | 0.948 | 0.694 | 0.697 | 0.754 |
| 0.3 | 0.990 | 0.866 | 0.938 | 0.964 | 0.991 | 0.867 | 0.939 | 0.964 |
“Sample size and power determination are based on a classification model for the primary endpoints with significant predictors. PCC* is the maximal classification power that would be obtained under rm genetic predictors (of 20,000 genes) and rc clinical predictors (of 75 clinical parameters) with respect to the standardized effect-sizes, δm/σm, where δm and σm are the effect size and standard deviation of an important biomarker, respectively. In parallel, PCC is the achievable power in the actual study with n patients enrolled in the FSGS, MCD, or MN subcohorts in NEPTUNE.”
NEPTUNE Training, Ancillary Study and Outreach Program
In the spirit of accelerating NS research, NEPTUNE has organized a pilot and ancillary studies program to facilitate high-quality investigation within the research community utilizing the resources provided by the NEPTUNE infrastructure. The ancillary studies mechanism is open to investigators regardless of institutional affiliation. Details on the available data, specimens and process required to gain access to these resources are provided at https://rarediseasesnetwork.epi.usf.edu/NEPTUNE/professional/studynetwork.htm#. Further, NEPTUNE supports a training program for post-doctoral fellows and junior faculty to promote career development in clinical and translational research in glomerular disease (https://rarediseasesnetwork.epi.usf.edu/NEPTUNE/professional/fellowship/). Lastly, NEPTUNE has organized a contact registry within the Rare Diseases Clinical Research Network (www.neptune-study.org) for patients with NS. This registry contains self-reported information, including diagnostic, demographic, and contact information that can be used for patient education and to facilitate enrollment of patients into clinical trials and other research studies. At the time of this publication, over 1300 patients are actively enrolled in the NEPTUNE contact registry and a substantial number of registrants have provided consent to be directly approached for participation in clinical studies.
Discussion
Primary NS is a debilitating clinical entity affecting individuals of all ages and associated with end-stage renal disease and premature death. We remain limited in our ability to effectively treat the specific diseases presenting as NS due to our lack of understanding of disease mechanisms and lack of predictors to identify clinical course and therapeutic responsiveness. The NEPTUNE cohort study is designed to address these gaps in knowledge in prominent NS disorders, FSGS, MCD and MN, by developing a platform for comprehensive longitudinal assessment of patients with incipient disease. This study represents strong commitment to bring together expertise from a broad range of disciplines focused on large-scale integration of clinical, histopathologic and molecular data. This state-of-the-art approach to identify pathogenic mechanisms and novel biomarkers has the greatest potential to advance medical management of these complex diseases.
Epidemiologic data demonstrates an increased incidence of primary NS due to FSGS over several decades in adults and children21. In young children, MCD remains the predominant etiology of primary NS. However, 10–15% of primary NS is caused by MCD in adults. FSGS and MCD are often considered related entities due to their similarities in clinical presentation, renal pathologic features as well as common genetic polymorphisms22, 23, but a systematic comparative analysis in sporadic patients is lacking. Although the clinical course can be highly variable, patients with FSGS are more likely resistant to glucocorticoid therapy and have greater risks for kidney failure. A variety of monogenetic mutations (both dominant and recessive patterns) with distinct functional consequences have been identified that present with indistinguishable FSGS-type lesions signifying heterogeneity in disease pathogenesis22, 24–29. Therefore, it is evident that the current classification and diagnostic tools are insufficient in guiding therapy. The paucity of conclusive clinical interventional studies for glomerular disease in general underscores the need to establish predictive biomarkers and therapeutic targets for future trials in molecularly-defined NS cohorts30.
Recent discovery of genetic risk loci in linkage disequilibrium with APOL1 and MYH931–33 among people of African descent with FSGS (and potentially other kidney disease) is a major step toward understanding racial/ethnic disparities in kidney disease. Recruitment of a diverse study population from sites located across North America provides opportunities to explore, in a longitudinal fashion, the clinical manifestations and therapeutic implications of these and other genetic variants.
MN is a common cause of primary NS in adults34, 35 and has a highly variable course from spontaneous remission to progression to end-stage renal disease. As with FSGS and MCD, renal tissue classification is descriptive and not associated with clinical course. The clinical course is highly variable and difficult to predict at presentation often leading to prolonged observation of patients by clinicians to determine prognosis and guide therapy36, 37,38. Recent evidence suggests that circulating phospholipase A2 receptor autoantibodies may be an immunologic marker for the majority of cases of primary MN39, 40. A major emphasis for the MN cohort in NEPTUNE is to understand the value of these new biomarkers in predicting disease progression and response to treatment.
In line with the major goals of the consortium, the NEPTUNE pathology protocol for scoring renal biopsies has the potential to re-define morphologic descriptors utilizing innovative digital methodology and serve as an extensive resource for future ancillary studies. This approach to renal biopsy analysis has potential applicability for risk stratification, disease classification and clinical trial design.
There is rapid expansion of information about monogenetic diseases and genetic loci associated with NS. However, discovering the functional links that bridge the gap from genetic risk to disease phenotype is one of the main challenges ahead. The NEPTUNE cohort study has been designed to bridge this genotype-phenotype gap using the integrated approaches of systems biology. These emerging innovative strategies will be the first steps towards identifying and testing rational interventions in primary NS. Overall, the NEPTUNE study and its complementary programs provide unparalleled opportunities to make fundamental observations using state-of-the-art research methodology, foster collaborative scientific activity, and facilitate research career development to stimulate progress in the management of glomerular diseases.
Methods
See supplement for a more detailed definition of statistical methodology.
Supplementary Material
Acknowledgments
Sources of support that require acknowledgement:
NEPTUNE is a part of NIH Rare Diseases Clinical Research Network (RDCRN). Funding and/or programmatic support for this project has been provided by U54 DK083912 from the Office of Rare Diseases Research (ORDR)/NCATS, NIDDK, the NephCure Foundation, and the University of Michigan. The views expressed in written materials or publications do not necessarily reflect the official policies of the Department of Health and Human Services; nor does mention by trade names, commercial practices, or organizations imply endorsement by the U.S. Government.
Footnotes
Disclosure:
The authors do not have any relationships with companies that may have a financial interest in the information contained in the manuscript.
REFERENCES
- 1.Maisonneuve P, Agodoa L, Gellert R, et al. Distribution of primary renal diseases leading to end-stage renal failure in the United States, Europe, and Australia/New Zealand: results from an international comparative study. Am J Kidney Dis. 2000;35:157–165. doi: 10.1016/S0272-6386(00)70316-7. [DOI] [PubMed] [Google Scholar]
- 2.U S Renal Data System, USRDS. 2011 Annual Data Report: Atlas of Chronic Kidney Disease and End-Stage Renal Disease in the United States. Bethesda MD: National Institute of Diabetes and Digestive and Kidney Diseases; 2011. 2011. [Google Scholar]
- 3.Kretzler M, Cohen CD. Integrative biology of renal disease: toward a holistic understanding of the kidney's function and failure. Semin Nephrol. 2010;30:439–442. doi: 10.1016/j.semnephrol.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hladunewich MA, Troyanov S, Calafati J, et al. The natural history of the non-nephrotic membranous nephropathy patient. Clinical journal of the American Society of Nephrology : CJASN. 2009;4:1417–1422. doi: 10.2215/CJN.01330209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwartz MM, Evans J, Bain R, et al. Focal segmental glomerulosclerosis: prognostic implications of the cellular lesion. Journal of the American Society of Nephrology : JASN. 1999;10:1900–1907. doi: 10.1681/ASN.V1091900. [DOI] [PubMed] [Google Scholar]
- 6.Gipson DS, Massengill SF, Yao L, et al. Management of childhood onset nephrotic syndrome. Pediatrics. 2009;124:747–757. doi: 10.1542/peds.2008-1559. [DOI] [PubMed] [Google Scholar]
- 7.Working Group of the International Ig ANN, the Renal Pathology S. Cattran DC, et al. The Oxford classification of IgA nephropathy: rationale, clinicopathological correlations, and classification. Kidney international. 2009;76:534–545. doi: 10.1038/ki.2009.243. [DOI] [PubMed] [Google Scholar]
- 8.Barisoni L, Jennette JC, Colvin R, Sitaraman S, Bragat A, Castelli J, Boudes P. A novel quantitative method to evaluate GL-3 inclusions in renal peritubular capillaries by virtual microscopy in patients with Fabry's disease. Archives of pathology & laboratory medicine. doi: 10.5858/arpa.2011-0350-OA. in press. [DOI] [PubMed] [Google Scholar]
- 9.Sauer U, Heinemann M, Zamboni N. Genetics. Getting closer to the whole picture. Science. 2007;316:550–551. doi: 10.1126/science.1142502. [DOI] [PubMed] [Google Scholar]
- 10.Keller BJ, Martini S, Sedor JR, et al. A systems view of genetics in chronic kidney disease. Kidney international. 2012;81:14–21. doi: 10.1038/ki.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Velic A, Macek B, Wagner CA. Towards quantitative proteomics of organ substructures: implications for renal physiology. Sem Nephrol. 2010 doi: 10.1016/j.semnephrol.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 12.Perco P, Oberbauer R. Integrative analysis of ‘-omics’ data and histological scoring in renal disease and transplantation: renal histogenomics. Sem Nephrol. 2010 doi: 10.1016/j.semnephrol.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ju W, Brosius FC., 3rd Understanding Kidney Disease: Toward the Integration of Regulatory Networks Across Species. Sem Nephrol. 2010 doi: 10.1016/j.semnephrol.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He JC, Chuang PY, Ma'ayan A, et al. Systems biology of kidney diseases. Kidney international. 2012;81:22–39. doi: 10.1038/ki.2011.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diggle P, Heagerty P, Liang K-Y, Zeger SL. Analysis of longitudinal data. 2nd edn. New York: Oxford University Press; 2002. [Google Scholar]
- 16.Dudoit SFJ, Speed TP. Comparison of discrimination methods for tumor classification based on microarray data. JASA. 2002;97:77–87. [Google Scholar]
- 17.Ma S, Song X, Huang J. Regularized binormal ROC method in disease classification using microarray data. BMC bioinformatics. 2006;7:253. doi: 10.1186/1471-2105-7-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou X, Obuchowski A, McClish DK. Statistical methods in diagnostic medicine. New York: Wiley; 2002. [Google Scholar]
- 19.Pencina MJ, D'Agostino RB, Sr, D'Agostino RB, Jr, et al. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Statistics in medicine. 2008;27:157–172. doi: 10.1002/sim.2929. discussion 207-112. [DOI] [PubMed] [Google Scholar]
- 20.Dobbin KKSR. Sample size planning for developing classifiers using high-dimensional DNA microarray data. Biostatistics. 2007:101–117. doi: 10.1093/biostatistics/kxj036. [DOI] [PubMed] [Google Scholar]
- 21.Srivastava T, Simon SD, Alon US. High incidence of focal segmental glomerulosclerosis in nephrotic syndrome of childhood. Pediatr Nephrol. 1999;13:13–18. doi: 10.1007/s004670050555. [DOI] [PubMed] [Google Scholar]
- 22.Weber S, Gribouval O, Esquivel EL, et al. NPHS2 mutation analysis shows genetic heterogeneity of steroid-resistant nephrotic syndrome and low post-transplant recurrence. Kidney international. 2004;66:571–579. doi: 10.1111/j.1523-1755.2004.00776.x. [DOI] [PubMed] [Google Scholar]
- 23.Vats A, Nayak A, Ellis D, et al. Familial nephrotic syndrome: clinical spectrum and linkage to chromosome 19q13. Kidney international. 2000;57:875–881. doi: 10.1046/j.1523-1755.2000.057003875.x. [DOI] [PubMed] [Google Scholar]
- 24.Barisoni L, Schnaper HW, Kopp JB. A proposed taxonomy for the podocytopathies: a reassessment of the primary nephrotic diseases. Clinical journal of the American Society of Nephrology : CJASN. 2007;2:529–542. doi: 10.2215/CJN.04121206. [DOI] [PubMed] [Google Scholar]
- 25.Albaqumi M, Barisoni L. Current views on collapsing glomerulopathy. Journal of the American Society of Nephrology : JASN. 2008;19:1276–1281. doi: 10.1681/ASN.2007080926. [DOI] [PubMed] [Google Scholar]
- 26.Winn MP, Conlon PJ, Lynn KL, et al. Clinical and genetic heterogeneity in familial focal segmental glomerulosclerosis. International Collaborative Group for the Study of Familial Focal Segmental Glomerulosclerosis. Kidney international. 1999;55:1241–1246. doi: 10.1046/j.1523-1755.1999.00384.x. [DOI] [PubMed] [Google Scholar]
- 27.Hinkes B, Wiggins RC, Gbadegesin R, et al. Positional cloning uncovers mutations in PLCE1 responsible for a nephrotic syndrome variant that may be reversible. Nat Genet. 2006;38:1397–1405. doi: 10.1038/ng1918. [DOI] [PubMed] [Google Scholar]
- 28.Diomedi-Camassei F, Di Giandomenico S, Santorelli FM, et al. COQ2 nephropathy: a newly described inherited mitochondriopathy with primary renal involvement. Journal of the American Society of Nephrology : JASN. 2007;18:2773–2780. doi: 10.1681/ASN.2006080833. [DOI] [PubMed] [Google Scholar]
- 29.McTaggart SJ, Algar E, Chow CW, et al. Clinical spectrum of Denys-Drash and Frasier syndrome. Pediatr Nephrol. 2001;16:335–339. doi: 10.1007/s004670000541. [DOI] [PubMed] [Google Scholar]
- 30.Gipson DS, Trachtman H, Kaskel FJ, et al. Clinical trial of focal segmental glomerulosclerosis in children and young adults. Kidney international. 2011;80:868–878. doi: 10.1038/ki.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kopp JB, Smith MW, Nelson GW, et al. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat Genet. 2008;40:1175–1184. doi: 10.1038/ng.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tzur S, Rosset S, Shemer R, et al. Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum Genet. 2010;128:345–350. doi: 10.1007/s00439-010-0861-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Genovese G, Tonna SJ, Knob AU, et al. A risk allele for focal segmental glomerulosclerosis in African Americans is located within a region containing APOL1 and MYH9. Kidney international. 2010;78:698–704. doi: 10.1038/ki.2010.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cameron JS. Membranous nephropathy and its treatment. Nephrol Dial Transplant. 1992;7(Suppl 1):72–79. [PubMed] [Google Scholar]
- 35.Haas M, Meehan SM, Karrison TG, et al. Changing etiologies of unexplained adult nephrotic syndrome: a comparison of renal biopsy findings from 1976–1979 and 1995–1997. Am J Kidney Dis. 1997;30:621–631. doi: 10.1016/s0272-6386(97)90485-6. [DOI] [PubMed] [Google Scholar]
- 36.Cattran DC, Pei Y, Greenwood CM, et al. Validation of a predictive model of idiopathic membranous nephropathy: its clinical and research implications. Kidney international. 1997;51:901–907. doi: 10.1038/ki.1997.127. [DOI] [PubMed] [Google Scholar]
- 37.Pei Y, Cattran D, Greenwood C. Predicting chronic renal insufficiency in idiopathic membranous glomerulonephritis. Kidney international. 1992;42:960–966. doi: 10.1038/ki.1992.374. [DOI] [PubMed] [Google Scholar]
- 38.Perna A, Schieppati A, Zamora J, et al. Immunosuppressive treatment for idiopathic membranous nephropathy: a systematic review. Am J Kidney Dis. 2004;44:385–401. [PubMed] [Google Scholar]
- 39.Stanescu HC, Arcos-Burgos M, Medlar A, et al. Risk HLA-DQA1 and PLA(2)R1 alleles in idiopathic membranous nephropathy. The New England journal of medicine. 2011;364:616–626. doi: 10.1056/NEJMoa1009742. [DOI] [PubMed] [Google Scholar]
- 40.Ronco P, Debiec H. Advances in membranous nephropathy: success stories of a long journey. Clin Exp Pharmacol Physiol. 2011;38:410–416. doi: 10.1111/j.1440-1681.2011.05506.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.