Abstract
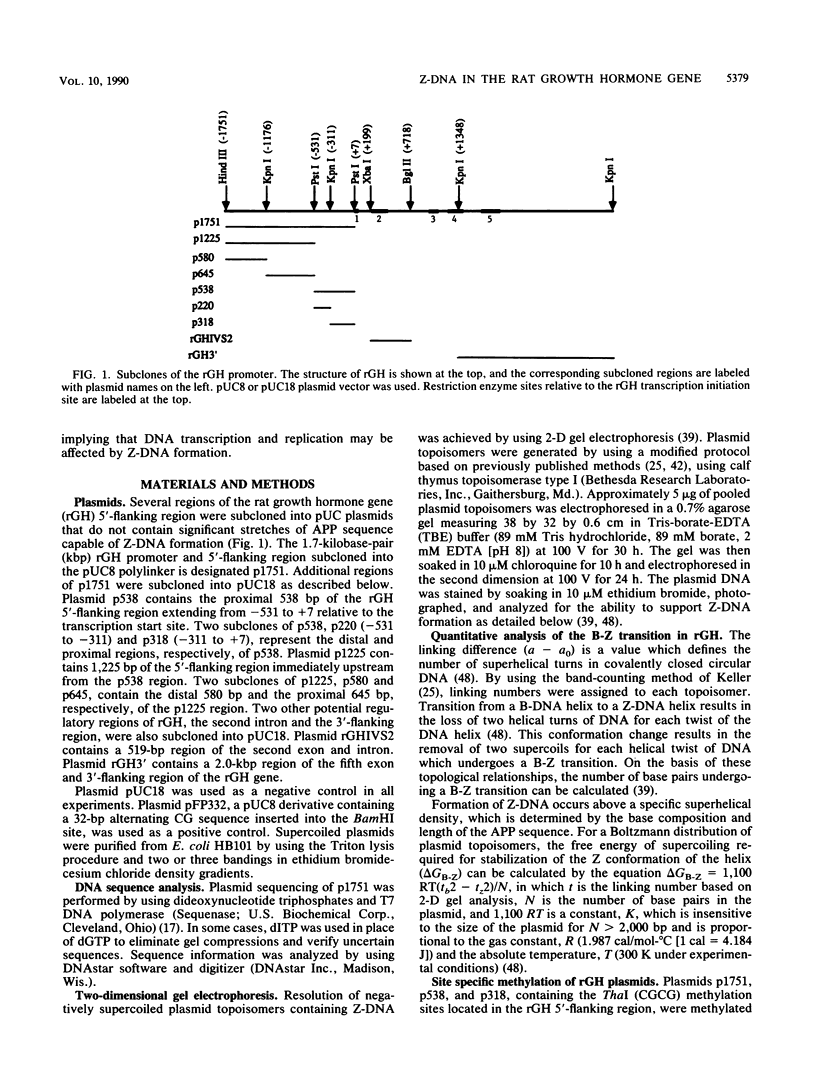
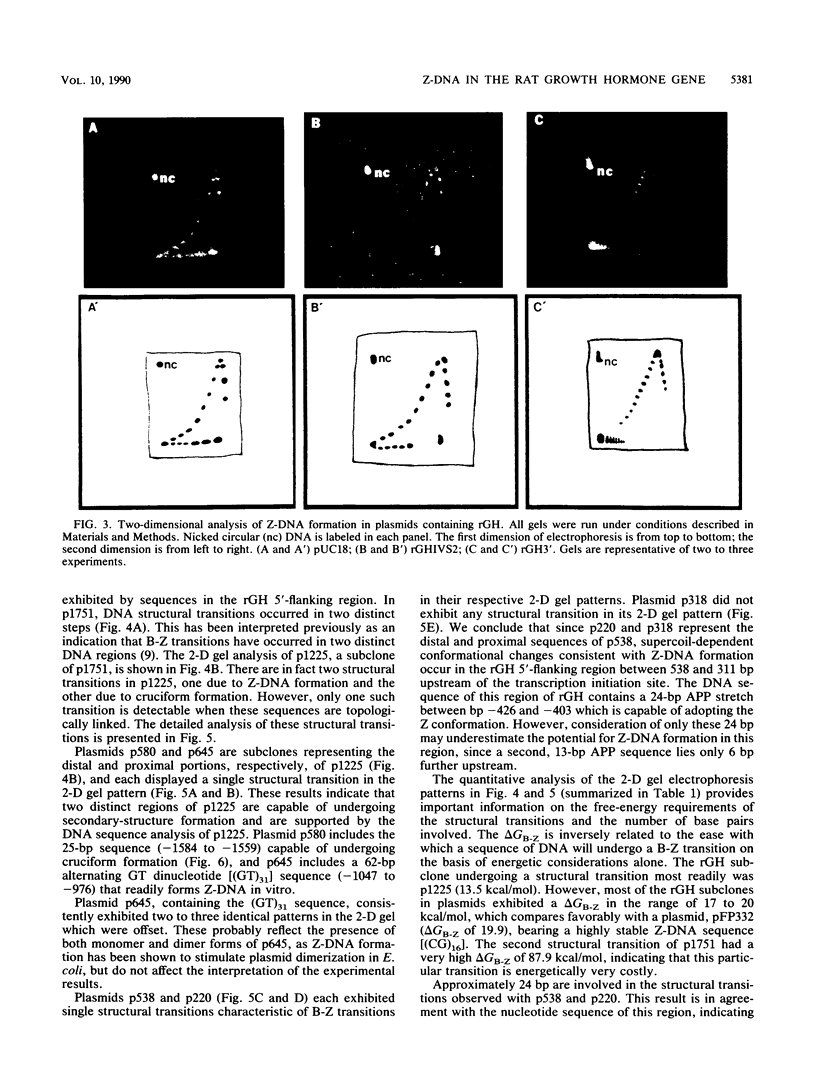
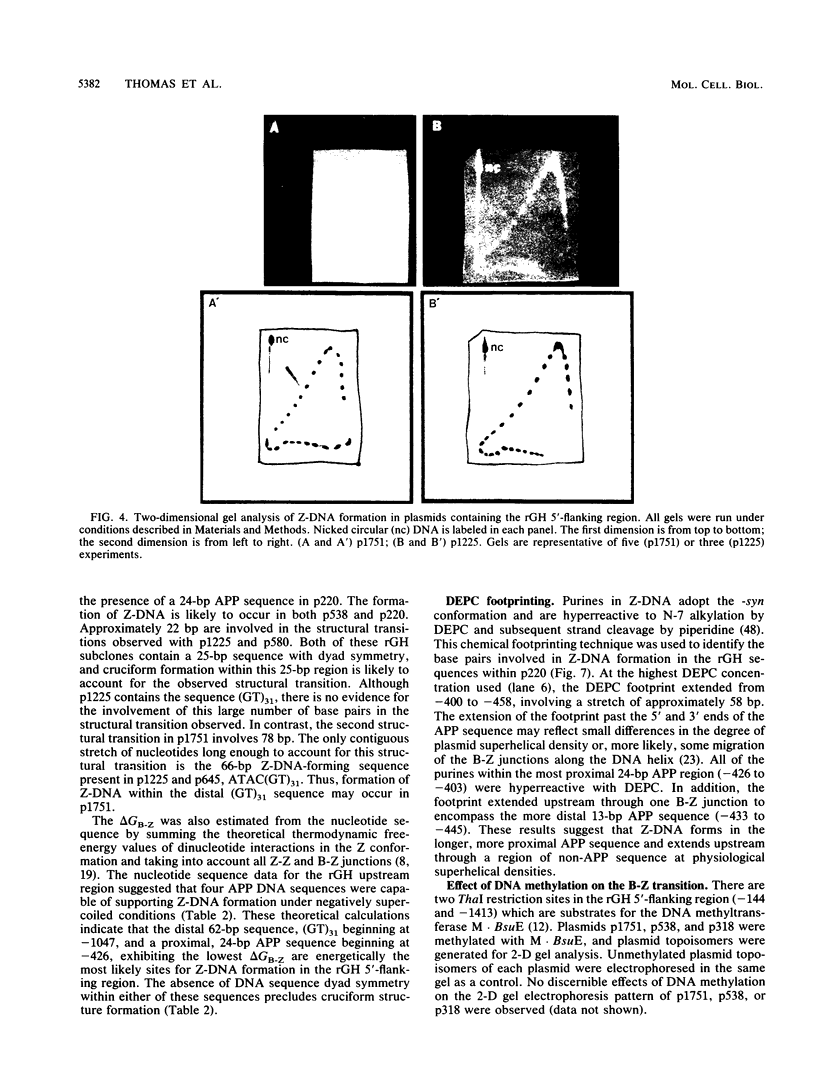
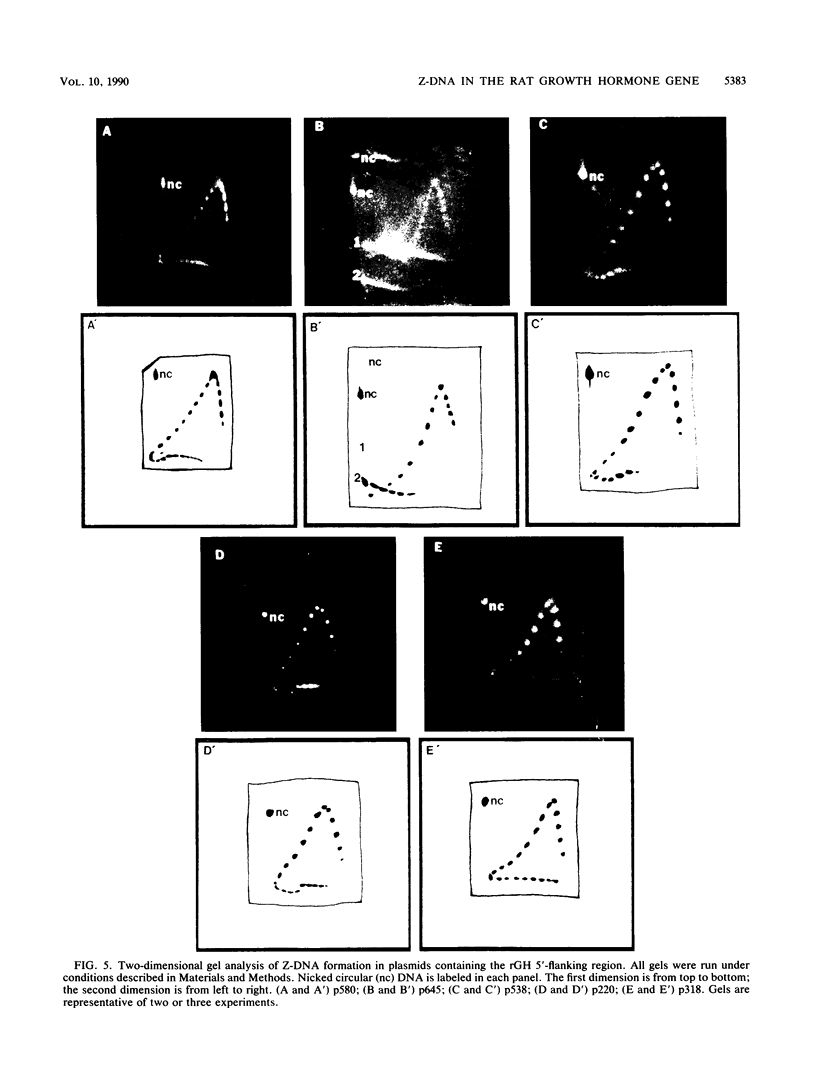
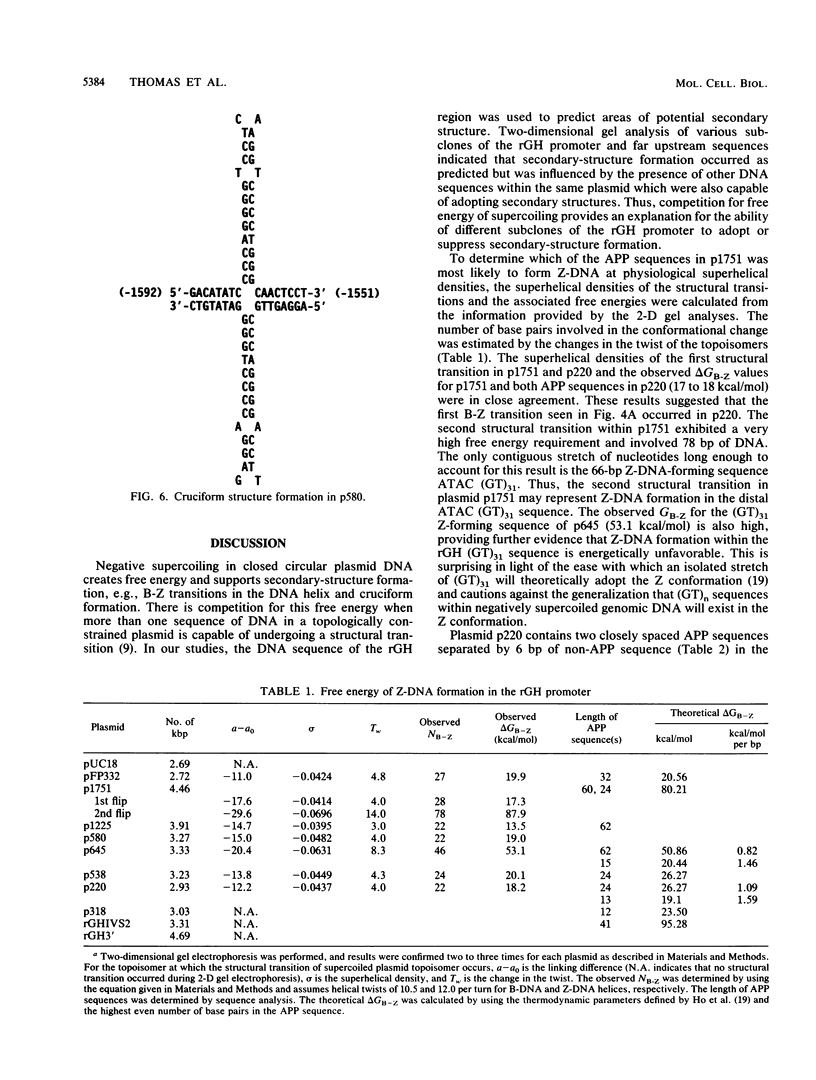
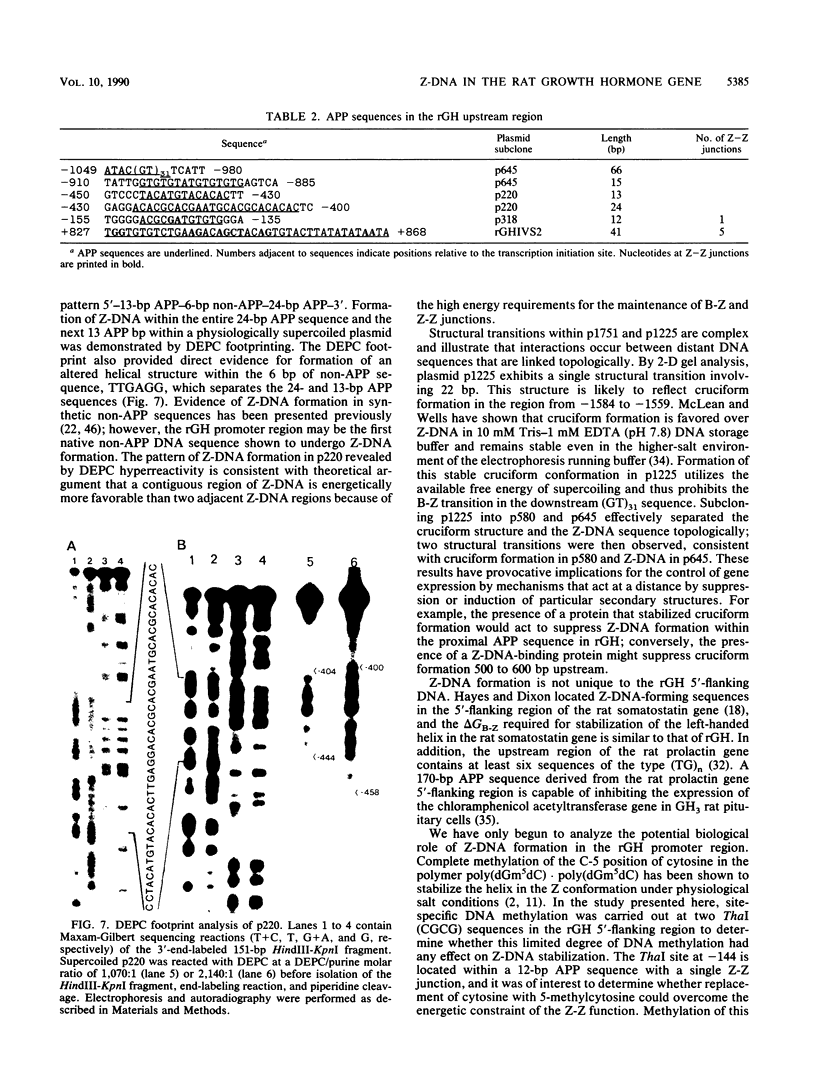
The complete DNA sequence of the 1.7 kilobase pairs (kbp) 5' of the rat growth hormone gene (rGH) has been determined and analyzed for Z-DNA-forming potential. Regions of alternating purine-pyrimidine (APP) sequences located between -1047 and -986 [(GT)31], between -445 and -433 bp, and between -426 and -403 bp relative to the rGH RNA transcription initiation site were identified and shown to form Z-DNA in negatively supercoiled plasmids by two-dimensional gel electrophoresis. Free-energy calculations indicated that Z-DNA forms most readily in the proximal Z-DNA regions. Diethyl pyrocarbonate footprinting of physiologically supercoiled plasmid DNA confirmed the presence of Z-DNA from -444 to -404 bp spanning the two most proximal APP sequences and a short non-APP sequence in between. DNA sequence analysis also predicted a region of DNA curvature near this proximal Z-DNA region. Formation of Z-DNA in the distal Z-DNA region consisting of a (GT)31 repeat was constrained at physiological plasmid superhelical densities. This may be related to the presence of DNA sequences (-1584 to -1559) 512 bp upstream of (GT)31 that undergo cruciform formation and thereby utilize the available free energy. Removal of 580 bp containing the cruciform region resulted in Z-DNA formation within (GT)31, thus demonstrating that deletion mutations can exert topological changes at a distance within the rGH 5'-flanking region. Methylation of two specific cytosines in the rGH 5'-flanking DNA that have been associated with inhibition of rGH promoter activity had no effect on Z-DNA formation. No evidence for DNA secondary structure formation within the rGH second exon-intron or 3'-flanking region was observed. We conclude that the rGH 5'-flanking region undergoes secondary-structure formation at physiological superhelical densities, thus providing a potential mechanism(s) for modulating rGH activity.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barta A., Richards R. I., Baxter J. D., Shine J. Primary structure and evolution of rat growth hormone gene. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4867–4871. doi: 10.1073/pnas.78.8.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behe M., Felsenfeld G. Effects of methylation on a synthetic polynucleotide: the B--Z transition in poly(dG-m5dC).poly(dG-m5dC). Proc Natl Acad Sci U S A. 1981 Mar;78(3):1619–1623. doi: 10.1073/pnas.78.3.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahms S., Vergne J., Brahms J. G., Di Capua E., Bucher P., Koller T. Natural DNA sequences can form left-handed helices in low salt solution under conditions of topological constraint. J Mol Biol. 1982 Dec 5;162(2):473–493. doi: 10.1016/0022-2836(82)90539-3. [DOI] [PubMed] [Google Scholar]
- Bullock P., Miller J., Botchan M. Effects of poly[d(pGpT).d(pApC)] and poly[d(pCpG).d(pCpG)] repeats on homologous recombination in somatic cells. Mol Cell Biol. 1986 Nov;6(11):3948–3953. doi: 10.1128/mcb.6.11.3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butzow J. J., Shin Y. A., Eichhorn G. L. Effect of template conversion from the B to the Z conformation on RNA polymerase activity. Biochemistry. 1984 Oct 9;23(21):4837–4843. doi: 10.1021/bi00316a004. [DOI] [PubMed] [Google Scholar]
- Delseny M., Laroche M., Penon P. Detection of sequences with Z-DNA forming potential in higher plants. Biochem Biophys Res Commun. 1983 Oct 14;116(1):113–120. doi: 10.1016/0006-291x(83)90388-1. [DOI] [PubMed] [Google Scholar]
- Ellison M. J., Feigon J., Kelleher R. J., 3rd, Wang A. H., Habener J. F., Rich A. An assessment of the Z-DNA forming potential of alternating dA-dT stretches in supercoiled plasmids. Biochemistry. 1986 Jun 17;25(12):3648–3655. doi: 10.1021/bi00360a026. [DOI] [PubMed] [Google Scholar]
- Ellison M. J., Fenton M. J., Ho P. S., Rich A. Long-range interactions of multiple DNA structural transitions within a common topological domain. EMBO J. 1987 May;6(5):1513–1522. doi: 10.1002/j.1460-2075.1987.tb02394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison M. J., Kelleher R. J., 3rd, Wang A. H., Habener J. F., Rich A. Sequence-dependent energetics of the B-Z transition in supercoiled DNA containing nonalternating purine-pyrimidine sequences. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8320–8324. doi: 10.1073/pnas.82.24.8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerstein B. G., Marton L. J., Keniry M. A., Wade D. L., Shafer R. H. New DNA polymorphism: evidence for a low salt, left-handed form of poly(dG-m5dC). Nucleic Acids Res. 1985 Jun 11;13(11):4133–4141. doi: 10.1093/nar/13.11.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaido M. L., Prostko C. R., Strobl J. S. Isolation and characterization of BsuE methyltransferase, a CGCG specific DNA methyltransferase from Bacillus subtilis. J Biol Chem. 1988 Apr 5;263(10):4832–4836. [PubMed] [Google Scholar]
- Gaido M. L., Strobl J. S. Inhibition of rat growth hormone promoter activity by site-specific DNA methylation. Biochim Biophys Acta. 1989 Jul 7;1008(2):234–242. doi: 10.1016/0167-4781(80)90014-7. [DOI] [PubMed] [Google Scholar]
- Gilmour R. S., Spandidos D. A., Vass J. K., Gow J. W., Paul J. A negative regulatory sequence near the mouse beta-maj globin gene associated with a region of potential Z-DNA. EMBO J. 1984 Jun;3(6):1263–1272. doi: 10.1002/j.1460-2075.1984.tb01961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H., Petrino M. G., Kakunaga T., Seidman M., Stollar B. D. Characterization of genomic poly(dT-dG).poly(dC-dA) sequences: structure, organization, and conformation. Mol Cell Biol. 1984 Dec;4(12):2610–2621. doi: 10.1128/mcb.4.12.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H., Seidman M., Howard B. H., Gorman C. M. Enhanced gene expression by the poly(dT-dG).poly(dC-dA) sequence. Mol Cell Biol. 1984 Dec;4(12):2622–2630. doi: 10.1128/mcb.4.12.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Hayes T. E., Dixon J. E. Z-DNA in the rat somatostatin gene. J Biol Chem. 1985 Jul 5;260(13):8145–8156. [PubMed] [Google Scholar]
- Ho P. S., Ellison M. J., Quigley G. J., Rich A. A computer aided thermodynamic approach for predicting the formation of Z-DNA in naturally occurring sequences. EMBO J. 1986 Oct;5(10):2737–2744. doi: 10.1002/j.1460-2075.1986.tb04558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoheisel J. D., Pohl F. M. Searching for potential Z-DNA in genomic Escherichia coli DNA. J Mol Biol. 1987 Feb 5;193(3):447–464. doi: 10.1016/0022-2836(87)90259-2. [DOI] [PubMed] [Google Scholar]
- Jaworski A., Hsieh W. T., Blaho J. A., Larson J. E., Wells R. D. Left-handed DNA in vivo. Science. 1987 Nov 6;238(4828):773–777. doi: 10.1126/science.3313728. [DOI] [PubMed] [Google Scholar]
- Jayasena V. K., Behe M. J. The B-Z transition in supercoiled DNA depends on sequence beyond nearest-neighbors. Nucleic Acids Res. 1989 Aug 25;17(16):6523–6529. doi: 10.1093/nar/17.16.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston B. H., Ohara W., Rich A. Stochastic distribution of a short region of Z-DNA within a long repeated sequence in negatively supercoiled plasmids. J Biol Chem. 1988 Apr 5;263(10):4512–4515. [PubMed] [Google Scholar]
- Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. I., Heuser J., Cox M. M. Enhanced recA protein binding to Z DNA represents a kinetic perturbation of a general duplex DNA binding pathway. J Biol Chem. 1989 Dec 25;264(36):21848–21856. [PubMed] [Google Scholar]
- Kmiec E. B., Holloman W. K. Homologous pairing of DNA molecules by Ustilago rec1 protein is promoted by sequences of Z-DNA. Cell. 1986 Feb 28;44(4):545–554. doi: 10.1016/0092-8674(86)90264-3. [DOI] [PubMed] [Google Scholar]
- Krishna P., Kennedy B. P., van de Sande J. H., McGhee J. D. Yolk proteins from nematodes, chickens, and frogs bind strongly and preferentially to left-handed Z-DNA. J Biol Chem. 1988 Dec 15;263(35):19066–19070. [PubMed] [Google Scholar]
- Lafer E. M., Möller A., Nordheim A., Stollar B. D., Rich A. Antibodies specific for left-handed Z-DNA. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3546–3550. doi: 10.1073/pnas.78.6.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafer E. M., Sousa R. J., Rich A. Z-DNA-binding proteins in Escherichia coli purification, generation of monoclonal antibodies and gene isolation. J Mol Biol. 1988 Sep 20;203(2):511–516. doi: 10.1016/0022-2836(88)90017-4. [DOI] [PubMed] [Google Scholar]
- Lafer E. M., Sousa R., Rosen B., Hsu A., Rich A. Isolation and characterization of Z-DNA binding proteins from wheat germ. Biochemistry. 1985 Sep 10;24(19):5070–5076. doi: 10.1021/bi00340a017. [DOI] [PubMed] [Google Scholar]
- Leith I. R., Hay R. T., Russell W. C. Detection of Z DNA binding proteins in tissue culture cells. Nucleic Acids Res. 1988 Sep 12;16(17):8277–8289. doi: 10.1093/nar/16.17.8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane D., Farrance I., Hall I., Morris J., Ivarie R. The rat prolactin gene contains at least six poly(dT-dG).poly(dC-dA) repeats. Nucleic Acids Res. 1986 Oct 10;14(19):7805–7805. doi: 10.1093/nar/14.19.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean M. J., Lee J. W., Wells R. D. Characteristics of Z-DNA helices formed by imperfect (purine-pyrimidine) sequences in plasmids. J Biol Chem. 1988 May 25;263(15):7378–7385. [PubMed] [Google Scholar]
- McLean M. J., Wells R. D. The role of DNA sequence in the formation of Z-DNA versus cruciforms in plasmids. J Biol Chem. 1988 May 25;263(15):7370–7377. [PubMed] [Google Scholar]
- Naylor L. H., Clark E. M. d(TG)n.d(CA)n sequences upstream of the rat prolactin gene form Z-DNA and inhibit gene transcription. Nucleic Acids Res. 1990 Mar 25;18(6):1595–1601. doi: 10.1093/nar/18.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordheim A., Meese K. Topoisomer gel retardation: detection of anti-Z-DNA antibodies bound to Z-DNA within supercoiled DNA minicircles. Nucleic Acids Res. 1988 Jan 11;16(1):21–37. doi: 10.1093/nar/16.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordheim A., Tesser P., Azorin F., Kwon Y. H., Möller A., Rich A. Isolation of Drosophila proteins that bind selectively to left-handed Z-DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7729–7733. doi: 10.1073/pnas.79.24.7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ní Bhriain N. A., Dorman C. J. A novel genetic locus determines in vivo B-Z DNA structural transitions in Escherichia coli. Trends Biochem Sci. 1988 Apr;13(4):130–130. doi: 10.1016/0968-0004(88)90068-0. [DOI] [PubMed] [Google Scholar]
- Pardoll D. M., Vogelstein B., Coffey D. S. A fixed site of DNA replication in eucaryotic cells. Cell. 1980 Feb;19(2):527–536. doi: 10.1016/0092-8674(80)90527-9. [DOI] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Energetics of B-to-Z transition in DNA. Proc Natl Acad Sci U S A. 1983 Oct;80(20):6206–6210. doi: 10.1073/pnas.80.20.6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh N., Shouche Y. S., Brahmachari S. K. Recognition of B and Z forms of DNA by Escherichia coli DNA polymerase I. J Mol Biol. 1986 Aug 20;190(4):635–638. doi: 10.1016/0022-2836(86)90248-2. [DOI] [PubMed] [Google Scholar]
- Rich A., Nordheim A., Wang A. H. The chemistry and biology of left-handed Z-DNA. Annu Rev Biochem. 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- Singleton C. K., Wells R. D. The facile generation of covalently closed, circular DNAs with defined negative superhelical densities. Anal Biochem. 1982 May 15;122(2):253–257. doi: 10.1016/0003-2697(82)90277-9. [DOI] [PubMed] [Google Scholar]
- Strobl J. S., Dannies P. S., Thompson E. B. Rat growth hormone gene expression is correlated with an unmethylated CGCG sequence near the transcription initiation site. Biochemistry. 1986 Jun 17;25(12):3640–3648. doi: 10.1021/bi00360a025. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N., Konopka A. K., Jovin T. M. Unusual frequencies of certain alternating purine-pyrimidine runs in natural DNA sequences: relation to Z-DNA. FEBS Lett. 1985 Jun 3;185(1):197–202. doi: 10.1016/0014-5793(85)80769-9. [DOI] [PubMed] [Google Scholar]
- Ulanovsky L., Bodner M., Trifonov E. N., Choder M. Curved DNA: design, synthesis, and circularization. Proc Natl Acad Sci U S A. 1986 Feb;83(4):862–866. doi: 10.1073/pnas.83.4.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. H., Gessner R. V., van der Marel G. A., van Boom J. H., Rich A. Crystal structure of Z-DNA without an alternating purine-pyrimidine sequence. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3611–3615. doi: 10.1073/pnas.82.11.3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Peck L. J., Becherer K. DNA supercoiling and its effects on DNA structure and function. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):85–91. doi: 10.1101/sqb.1983.047.01.011. [DOI] [PubMed] [Google Scholar]
- Wittig B., Dorbic T., Rich A. The level of Z-DNA in metabolically active, permeabilized mammalian cell nuclei is regulated by torsional strain. J Cell Biol. 1989 Mar;108(3):755–764. doi: 10.1083/jcb.108.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlrab F., Wells R. D. Enzymatic probes for left-handed Z-DNA. Gene Amplif Anal. 1987;5:247–256. [PubMed] [Google Scholar]