Abstract
Predicting the molecular complexity of a genomic sequencing library has emerged as a critical but difficult problem in modern applications of genome sequencing. Available methods to determine either how deeply to sequence, or predict the benefits of additional sequencing, are almost completely lacking. We introduce an empirical Bayesian method to implicitly model any source of bias and accurately characterize the molecular complexity of a DNA sample or library in almost any sequencing application.
Modern DNA sequencing experiments often interrogate hundreds of millions or even billions of reads, possibly to achieve deep coverage or to observe very rare molecules. Low complexity DNA sequencing libraries are problematic in such experiments: many sequenced reads will correspond to the same original molecules and deeper sequencing either provides redundant data that is discarded, or introduces biases in downstream analyses. When sequencing depth appears insufficient, investigators may be presented with the decision to sequence more deeply from an existing library or to generate another. Perhaps this situation has been anticipated during experimental design, and investigators can select from several libraries or samples for deep sequencing based on preliminary “shallow” surveys. The underlying question is how much new information will be gained from additional sequencing? The Lander-Waterman model1 was essential to understanding traditional sequencing experiments but does not account for the various biases typical in applications of high-throughput sequencing.
We present a new empirical Bayes method for understanding the molecular complexity of sequencing libraries or samples based on data from very shallow sequencing runs. We define complexity as the expected number of distinct molecules sequenced in a given set of reads produced in a sequencing experiment2. This function, which we call the complexity curve, efficiently summarizes new information to be obtained from additional sequencing and is generally robust to variation between sequencing runs (Supplementary Note). Importantly, our method also applies to understanding the complexity of molecular species in a sample (e.g. RNA from different isoforms) and since we require no specific assumptions about the sources of biases, out method is applicable in a surprising variety of contexts (Supplementary Note).
Consider a sequencing experiment as sampling at random from a DNA library. The distinct molecules in the library have different probabilities of being sequenced, and we assume those probabilities will change very little if the same library is sequenced again. Our goal is to accurately estimate the number of previously unsequenced molecules that would be observed if some amount of additional reads were generated.
We borrow methodology from capture-recapture statistics, which has dealt with analogous statistical questions of estimating the sizes of animal populations or the diversity of animal species3. The specific model we borrow is the classic Poisson non-parametric empirical Bayes model4. Based on the initial sequencing experiment, we identify unique molecules by some unique molecular identifier5 and obtain the frequency of each unique observation (e.g. each genomic position, transcript, allele, etc.). These frequencies are used to estimate the expected number of molecules that would be observed once, twice, and so on, in an experiment of the same size from the same library. The formula for the expected number of unique observations in a larger sequencing experiment then takes the form of an alternating power series with the estimated expectations as coefficients (full derivation provided in Online Methods).
The power series is extremely accurate for small extrapolations but major problems are encountered when attempting to extrapolate past twice the size of the initial experiment6. At that point the estimates show extreme variation depending on the number of terms included in the sum. Technically the series is said to diverge and therefore cannot be used directly to make inferences about properties of experiments more than twice as large as the initial experiment. Methods traditionally applied to help these series converge in practice, including Euler’s series transformation7, are not sufficient when data is on the scale produced in high-throughput sequencing experiments or for long range predictions.
We investigated a technique called rational function approximation, which is commonly used in theoretical physics8. Rational functions are ratios of polynomials and when used to approximate a power series, they often have a vastly increased radius of convergence. Algorithms to fit a rational function approximation essentially rearrange the information in the coefficients of the original power series, under the constraint that the resulting rational function closely approximates the power series. The convergence properties of rational function approximations are known to be especially good for a class of functions that includes the Good-Turing power series (discussion in Supplementary Note). By combining the Good-Turing power series with rational function approximations we developed an algorithm that can make optimal use of information from the initial sample and accurately predict the properties of sequencing data sets several orders of magnitude larger than the initial “shallow” sequencing run. We implemented our methods as a command line software package licensed under GPL and available from Supplementary Software or http://smithlab.usc.edu/software.
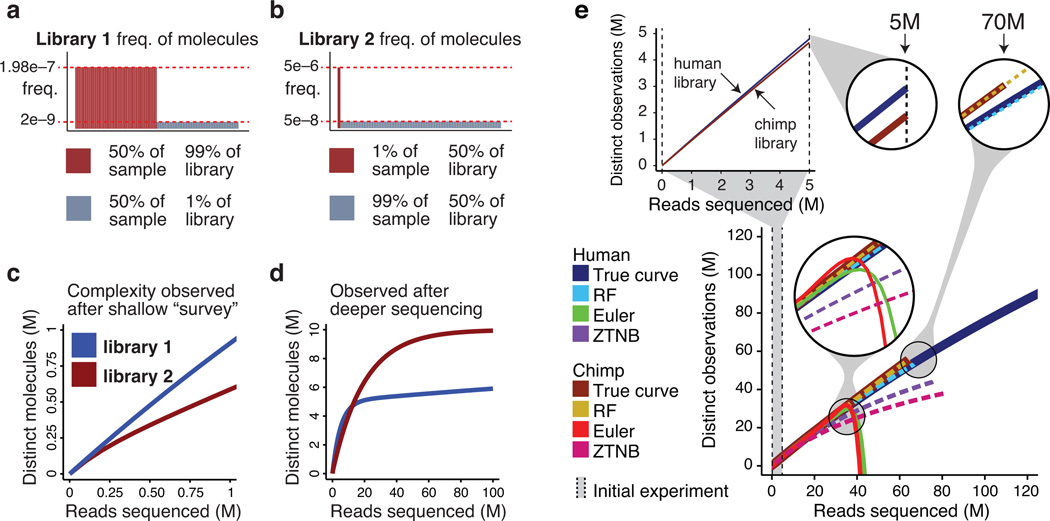
We illustrate the concepts of library complexity by means of a toy example, which also shows how naive analysis can lead to incorrect predictions (Fig. 1a-d). In the example two hypothetical libraries have complexity curves that initially appear linear (Fig. 1c), but eventually cross (Fig. 1d). Such extreme behavior can actually arise in practice. We used a small sample of reads (i.e. the initial sample) from human and chimp sperm BS-seq experiments9 (Supplementary Table 1) and produced complexity curves for the libraries (Fig. 1e). Both complexity curves appear linear through the initial experiment (5 million (M) reads) and the curve for the chimp library has a lower trajectory, and on that basis a naive analysis might predict this library to saturate first. However, the complexity curves cross after deeper sequencing (at 22 M reads), with the chimp library showing greater yield of distinct observations. Based on the initial sample of 5 M reads, we estimated the complexity of these two libraries using the rational function approximation (RF), as well as Euler’s transform (ET) and a zero-truncated negative binomial (ZTNB). The ZTNB is the natural next step when counts data are not Poisson, and the ET is the traditional method for improving convergence of the Good-Turing series. Initially the ET method gave accurate estimates, but these estimates diverge and are useless after 40 M reads. The ZTNB estimates show a substantial downward bias (more than 35% error for both libraries) and do not predict that the complexity curves cross, indicating that this distribution is not sufficiently flexible to account for the biases in the libraries. The RF method estimates the complexity of both libraries almost perfectly, and in the case of the chimp library this amounts to extrapolating 60x the size of the initial sample, incurring only 4% error for the human library and well under 1% error for the chimp library.
Figure 1.
Two hypothetical libraries containing 10 million (M) distinct molecules. (a) In library 1, half of the molecules (5 M) exist at the same level making up 99 % of the library. (b) In library 2, ten thousand molecule represents half the material in the library. (c) Based on a shallow sequencing run (1 M reads), library 1 appears to contain a greater diversity of molecules. (d) After additional sequencing, library 2 yields more distinct observations. (e) Such situations do occur in practice. Initial observed complexity from 5 M reads for two BS-seq libraries indicates the Human Sperm is the more complex library. Observed library complexity curves cross after additional sequencing, with the Chimp Sperm library yielding more distinct reads. Estimates using Rational Function (RF) and Euler’s transform (ET) fit to initial experiments predict crossing (though ET becomes unstable), while zero-truncated negative binomial (ZTNB) does not.
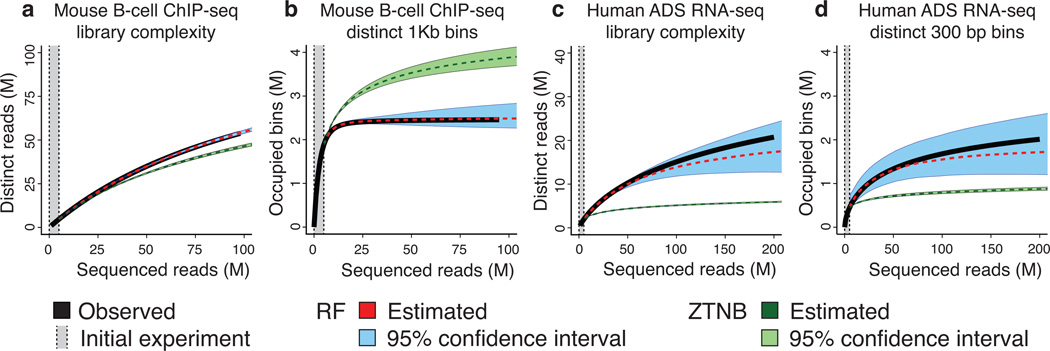
In sequencing applications to identify genomic intervals, for example protein binding sites in ChIP-seq or expressed exons in RNA-seq, the number of distinct molecules in the library might be secondary to the number of distinct genomic intervals identified through some post-processing of mapped reads. To demonstrate the broad applicability of our method (further discussed in Supplementary Note), we investigated how well our method could predict the number of non-overlapping genomic windows identified in a ChIP-seq experiment (1 kb) and an RNA-seq experiment (300 bp), in both cases using an initial experiment size of 5 M reads. These non-overlapping windows represent a proxy for some more sophisticated method of identifying binding sites or exons. For the ChIP-seq experiment (CTCF; mouse B-cells10), saturation of distinct reads was not reached even after sequencing 90 M (Fig. 2a), while the number of identified windows saturated after approximately 25 M reads (Fig. 2b). The RF predicted this saturation correctly (Fig. 2b) and estimated the complexity in terms of distinct reads with very high accuracy (Fig. 2a). The ZTNB over-estimated the saturation of identified windows at 4 M, more than possible in the mouse genome, while significantly under-estimating the yield of distinct reads. The RNA-seq experiment (Human adipose-derived mesenchymal stem cells11) did not saturate for either distinct reads (Fig. 2c) or identified windows (Fig. 2d), suggesting additional sequencing from this library would yield more information. Only the RF accurately predicted absence of saturation for both windows and reads, showing significantly lower relative error than the ZTNB at 200 M sequenced reads.
Figure 2.
Library complexity can be estimated both in terms of distinct molecules sequenced and in terms of distinct loci identified. (a) A ChIP-seq library (CTCF; mouse B-Cells) yields additional molecules after sequencing 100 million (M) reads; the RF remains accurate while the ZTNB loses accuracy. (b) In the same library, the number of mapped distinct genomic 1 kb windows saturates after 25 M reads. The rational function approximation (RF) is accurate and forecasts saturation, while the zero-truncated Negative Binomial (ZTNB) significantly overestimates. (c) An RNA-seq (Human adipose-derived mesenchymal stem (ADS) cells) library continues to yield additional molecules after 200 M reads; the RF remains accurate while the ZTNB predicts saturation. (d) In the same library, reads continued mapping to new 300 bp windows after 200 M reads. ZTNB incorrectly predicts saturation, while RF does not.
Sequencing data will always be subject to some amount of technical variation between sequencing instruments or even between runs on the same machine. We applied our method to data from a single library sequenced on different instruments (slightly differing sequencing technologies), and comparisons of the complexity estimates are within the range expected due to stochastic noise (Supplementary Fig. 1). For such run-to-run variation to impact the library complexity estimates, the variation must be dramatic and would likely be caused by detectable sequencing error at levels sufficient to warrant discarding the run.
As the cost, throughput, and read lengths of sequencing technologies improve, the usefulness of methods for understanding molecular complexity in a DNA sample will increase. The approach we have described, based on rational function approximation to the power series of Good & Toulmin, can be applied to an immense diversity of sequencing applications (Supplementary Note). As the age of clinical sequencing approaches, significant resources will be dedicated to refining quality control, protocol optimization and automation; methods for evaluating libraries will be essential to controlling costs and interpreting the results of sequencing that potentially could inform clinical decisions.
Supplementary Material
References
- 1.Lander E, Waterman M. Genomics. 1988;2:231–239. doi: 10.1016/0888-7543(88)90007-9. [DOI] [PubMed] [Google Scholar]
- 2.Chen Y, et al. Nat. Methods. 2012;9:609–614. doi: 10.1038/nmeth.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fisher RA, Corbet S, Williams CB. J. Anim. Ecol. 1943;12:42–58. [Google Scholar]
- 4.Good IJ. Biometrika. 1953;40:237–264. [Google Scholar]
- 5.Kivioja T, et al. Nat. Methods. 2012;9:72–74. doi: 10.1038/nmeth.1778. [DOI] [PubMed] [Google Scholar]
- 6.Good IJ, Toulmin GH. Biometrika. 1956;43:45–63. [Google Scholar]
- 7.Efron B, Thisted R. Biometrika. 1976;63:435–447. [Google Scholar]
- 8.Baker G, Graves-Morris P. Pade approximants. Cambridge, UK: Cambrige University Press; 1996. [Google Scholar]
- 9.Molaro A, et al. Cell. 2011;146:1029–1041. doi: 10.1016/j.cell.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Almeida CR, et al. Immunity. 2011;35:501–513. [Google Scholar]
- 11.Lister R, et al. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.