Abstract
Instances in which natural selection maintains genetic variation in a population over millions of years are thought to be extremely rare. We conducted a genome-wide scan for long-lived balancing selection by looking for combinations of SNPs shared between humans and chimpanzees. In addition to the major histocompatibility complex (MHC), we identified 125 regions in which the same haplotypes are segregating in the two species, all but two of which are non-coding. In six cases, there is evidence for an ancestral polymorphism that persisted to the present in humans and chimpanzees. Regions with shared haplotypes are significantly enriched for membrane glycoproteins, and a similar trend is seen among shared coding polymorphisms. These findings indicate that ancient balancing selection has shaped human variation and point to genes involved in host-pathogen interactions as common targets.
Introduction
Balancing selection is a mode of adaptation that leads to the persistence of variation in a population or species in the face of stochastic loss by genetic drift. In humans, examples include the sickle cell hemoglobin polymorphism, maintained by heterozygote advantage in environments in which Plasmodium falciparum is endemic, as well as other cases that likely arose recently in evolution in response to malaria (1). Beyond humans, examples of balancing selection are known in a wide range of organisms and often seem to arise from predator-prey or host-pathogen interactions (e.g., (2–8)). Most are not thought to be due to heterozygote advantage but to negative frequency dependent selection, as occurs at self-incompatibility loci in plants (5, 9), or to temporally or spatially varying selection, as seen, for example, at R genes in Arabidopsis (4). The genetic basis is known only in a small subset of cases, however, and the age-old question (10–12) of how much genetic variation is maintained by balancing selection remains largely open.
When balancing selection pressures result in the stable maintenance of genetic variation in the population for long periods of time, neutral diversity accumulates at nearby sites; in other words, ancient balancing selection leads to deep coalescence times to a common ancestor at the selected site(s) and closely linked ones (13). One approach to identify targets is therefore to scan the genome for regions of high diversity or other related features, such as intermediate allele frequencies (14). A challenge is that such patterns of diversity can occur by chance, because of the tremendous variance in coalescence times due to genetic drift alone (14). As an illustration, under a simple demographic model with no selection, the probability that two human lineages do not coalesce before the split with chimpanzee is on the order of 10−4 (15, 16). While this probability is small, the human genome is large and so many such regions could occur by chance. To circumvent this difficulty, we looked for cases where an ancestral polymorphism has persisted to the present time in both humans and chimpanzees, i.e., is shared identical by descent between the two species. This outcome is not expected to occur by genetic drift alone, as it requires that neither human nor chimpanzee lineages coalesce before the human-chimpanzee ancestor, which is unlikely even in a large genome (16).
To date, two cases of human polymorphisms shared with other apes have been shown to be identical by descent (see (16) and Fig. S1 for additional background): variants in the MHC, a complex encoding cell surface glycoproteins that present peptides to T cells (17), and polymorphisms at ABO, a glycosyltransferase, that underlie the A and B blood groups (18). Ancient balancing selection leaves a narrow footprint in genetic variation (15, 18), however, which may be particularly difficult to detect without dense variation data (19). Thus, the recent availability of genome sequences for multiple humans and chimpanzees provides an opportunity to search comprehensively and with greater power for ancient balancing selection.
Identification of shared SNPs and haplotypes
We examined complete genome sequences from 59 humans from sub-Saharan Africa (Yoruba) (20) and 10 Western chimpanzees (Pan troglodytes verus) (21) in order to identify shared polymorphisms, namely high quality orthologous SNPs with identical alleles in the two species ((16), Table S1). In total, 33,906 autosomal and 492 X-linked single nucleotide polymorphisms (SNPs) passed our filters (Table S2). The lower proportion of shared SNPs found on the X (in humans, 0.36% of autosomal SNPs versus 0.19% of X-linked SNPs) is expected under neutrality, because of the lower mutation rate and the smaller effective population size of the X (22).
The set of shared SNPs has similar properties to those of non-shared SNPs in terms of mapping quality, depth of coverage and proportion in repeats (Fig. S2, Table S2), consistent with it containing few artifacts. The shared SNPs include a much higher proportion of CpGs, however: 71.5% of autosomal shared SNPs occur at CpG dinucleotides, whereas only 26.4% of all human SNPs have this property (Table S2). Since CpGs are known to have a higher mutation rate than other sites (23), this observation, along with the similarity in allele frequency distributions of shared and non-shared SNPs (Fig. S2), suggest that most instances of shared SNPs are due to the independent occurrence of the same mutation in both species – in other words, that most SNPs are identical by state rather than descent (16).
Nonetheless, SNPs are shared between humans and chimpanzees 1.3-fold more often than is expected by chance, after controlling for the composition of the adjacent base pairs (the sequence context thought to have the strongest effect on mutation rate variation (23)) (Fig. S3). This excess may be explained by residual effects on the mutation rate of the sequence context beyond the adjacent base pairs (Fig. S4) or by variation in selective constraint across sites, but could also reflect instances of balancing selection.
Within the set of shared SNPs, we sought to enrich for targets of balancing selection by two approaches (Fig. 1A): First, we considered shared coding SNPs (16), a set that a priori should contain more functional changes subject to purifying selection, so is less likely to include polymorphisms shared by chance alone. Second, to home in on cases with unequivocal evidence for balancing selection, we searched for polymorphisms shared due to identity by descent. Where balancing selection acted on a single site and maintained a polymorphism stably since the human-chimpanzee split, a short ancestral segment should persist until the present around the selected site, of expected length less than four kilobases (kb) (depending on the recombination rate (16)). This segment is likely to contain one or more neutral, shared polymorphisms that arose in the ancestral population of humans and chimpanzees and are in strong or complete linkage disequilibrium (LD) with the selected site (15, 18) (Fig. 1B). Thus, this scenario should produce specific patterns of haplotype sharing between species. Guided by these considerations, we focused on cases with two or more shared SNPs within four kb and in significant LD in humans and in chimpanzees, with the same coupling of alleles in the two species (henceforth “shared haplotypes”; (16)). These LD criteria should almost always be met when a neutral polymorphism has persisted due to close linkage with an ancient balanced polymorphism, and yet are expected to filter out the vast majority (>96%) of cases of neutral, recurrent mutations (Table S3, (16)). These LD criteria should also be met if balancing selection acted on two or more sites and there is epistasis between them (as is the case at ABO), in which case the shared haplotypes may be longer (Fig. 1B).
Figure 1. Analysis pipeline.
A) Diagram of the pipeline to identify shared coding SNPs and shared haplotypes. See (16) for details of the filtering and validation.
B) Two possible scenarios of ancient balancing selection that may be detected by our approach. In (i), only one site is under balancing selection and a second mutation is neutral, but persisted as a polymorphism until the present in both species because of tight linkage to the selected site. In (ii), two or more epistatically-interacting polymorphic sites are maintained by balancing selection from the ancestral population of human and chimpanzee to the present time. In this case, the ancestral segment could be substantially longer because there is selection against recombinant haplotypes.
Importantly, we imposed stringent quality control filters on the shared haplotypes and coding SNPs (Fig. 1A) in order to exclude regions with highly similar paralogs present in the reference genomes of humans or chimpanzees as well as artifacts arising from duplicates that either fixed or are polymorphic in the two species but for which one copy is absent from both references genomes (these filters should also weed out regions that experience paralogous gene conversion; see (16) for details). After filtering, we considered pairs of shared SNPs to belong to the same shared haplotypes if they had a SNP in common (Tables S4, S5).
Protein variants
Across the genome, the MHC stood out (Fig. S5), with 11 shared non-synonymous and seven shared synonymous SNPs, including six non-synonymous and three synonymous that were not among the many cases of shared haplotypes in this region (Table S6, (16)).
Unexpectedly, given that the basis for A and B blood groups is shared between humans and gibbons but not chimpanzees (who lack the B type) (18), we found two SNPs shared between humans and chimpanzees in ABO, approximately four kb from the sites that distinguish A and B blood types in humans (Fig. S6). Neither shared SNP is non-synonymous (one is synonymous, the other intronic) and they do not meet our criteria for creating shared haplotypes, but there is a peak of diversity around them within both humans and chimpanzees, suggesting that they may be ancient variants (Fig. S6).
In addition, we found 199 synonymous SNPs, 135 non-synonymous SNPs and 1 premature stop shared between humans and chimpanzees, distributed among 324 genes (Fig. 2B, Table S5). Notable among these is a non-synonymous SNP in GP1BA, a gene encoding a glycoprotein present on the membrane of platelets, which is responsible for binding to the ABO antigens expressed on the Von Willebrand Factor (VWF) (24). The specific polymorphism in GP1BA shared between humans and chimpanzees, corresponding to the human platelet alloantigen 2 (HPA-2) polymorphism, affects the binding affinity to VWF and is associated with platelet count (25). More generally, the blood glycoprotein VWF is used as a bridge to anchor platelets to injured blood vessels for coagulation, and variants in ABO are strongly associated with protein levels of VWF (24). These findings suggest that two genes associated with the same complex may have been targets of long-lived balancing selection.
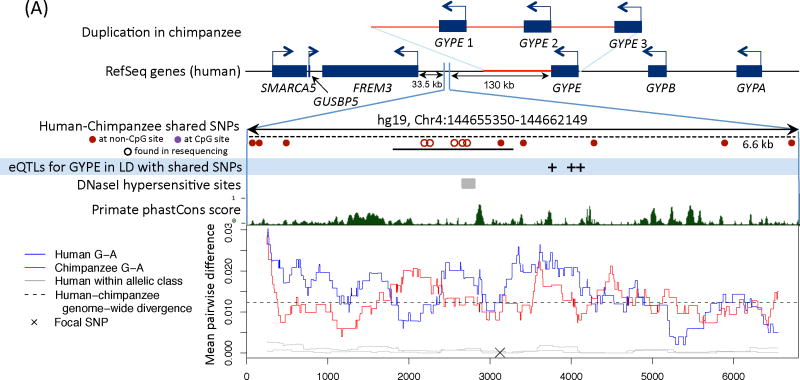
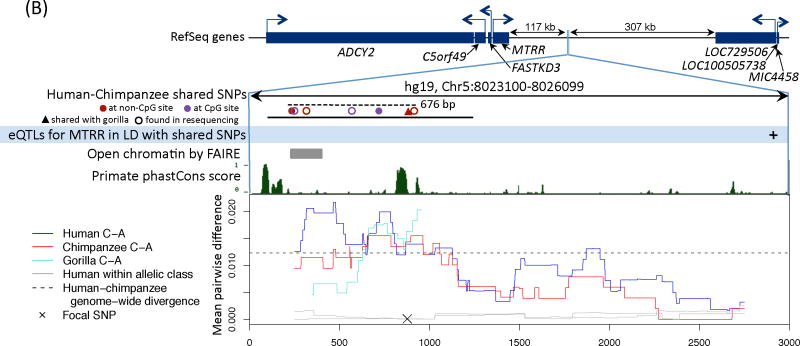
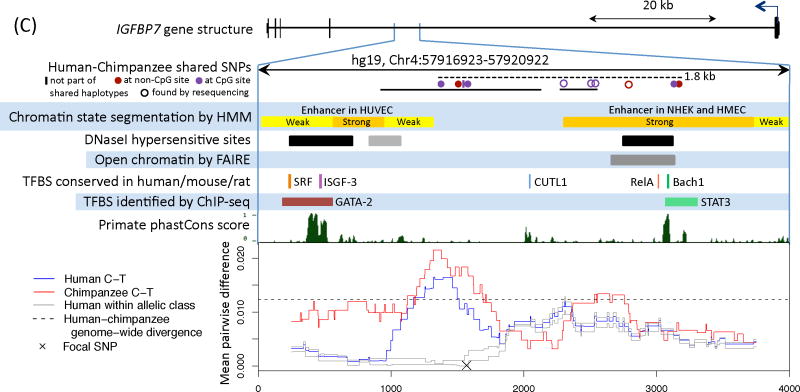
Figure 2. Functional information for three regions with a polymorphism shared identical by descent in humans and chimpanzees.
We show the nearby genes and direction of transcription, then a close up of the region with shared polymorphisms between humans and chimpanzees. The original shared SNPs used to identify shared haplotypes are shown as solid circles. The region resequenced in the validation experiment is indicated with a solid black bar and the length of the shared haplotypes with a dashed black bar (16). For sources of the functional annotation tracks shown, see (16). In the last panel, we focus on a shared SNP (hg19, chr4:144658471, chr5:8023976, and chr4:57918492, respectively) and show the average pairwise difference between allelic classes for humans (in blue) and chimpanzees (in red), for a 500 bp sliding window; the average pairwise difference within an allelic class in humans is in gray. We further indicate the average genome-wide divergence between human and chimpanzee (1.2%; (39)) with a dotted black line. For divergence between more distant ape species and a zoom out of diversity levels in each region, see Figs. S7 and S9.
A) FREM3. A duplication in chimpanzees that includes the GYPE gene is shown above the gene structure in humans (26). The shared SNPs and eQTLs for GYPE in monocytes (40) are in almost perfect LD, with a pairwise r2 ranging from 0.98 to 1.
B) MTRR. The shared SNP represented by a triangle is also seen in a sample of seven gorillas obtained by Sanger resequencing (see (16)); pairwise differences between allelic classes in gorillas is shown in turquoise for the resequenced region. The maximum pairwise r2 between a shared SNP and the eQTL for MTRR in monocytes is 0.47 (40) (16). The FAIRE signal is enriched in six cell lines.
C) IGFBP7. In the scan for shared haplotypes, five shared SNPs were found in the four kb region, occurring in two clusters with three and two SNPs, respectively, which are not in LD with each other in humans. Two of the shared SNPs found in the resequencing and a SNP outside the resequenced region constitute an additional instance of shared haplotypes. The FAIRE signal is enriched in four cell lines. Using a focal SNP in the second cluster yields similar results (see Fig. S7).
Regions with shared haplotypes
We identified 125 regions outside the MHC with shared haplotypes between humans and chimpanzees, whose total lengths span 4 bp to 6649 bp (Table S4). In five of the regions (nearest FREM3, MTRR, PROKR2 and in HUS1 and IGFBP7), there are more than two pairs of shared SNPs in significant LD, which simulations suggest should never occur in the genome by neutral recurrent mutations alone (16).
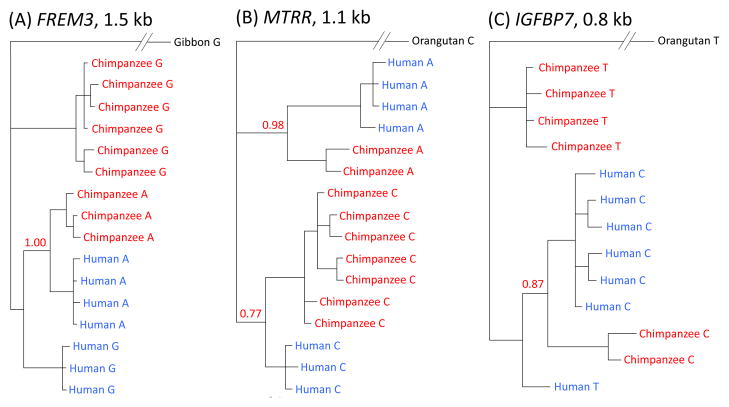
In the regions nearest FREM3, MTRR, and in IGFBP7, there is a peak of diversity in humans and chimpanzees around the shared SNPs that is comparable or in excess of the average divergence between the two species (and yet no evidence for elevated mutation rates in the region, as assessed by the levels of divergence between more distant outgroup species), consistent with the polymorphisms predating the human-chimpanzee split (Figs. 2, S7). Furthermore, when we built a phylogenetic tree based on these regions, haplotypes from different species that carry the same allele are more closely related to each other than they are to haplotypes from the same species with the other allele (with high posterior probability, and based on 800 bps or more; Fig. 3A–C, (16)). This clustering pattern establishes that these cases cannot be explained solely by recurrent mutation (16).
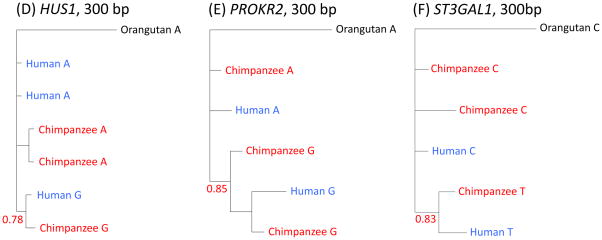
Figure 3.
Phylogenetic trees of haplotypes labeled with the same focal SNP considered in Fig. 2 or Fig. S8 for (A) FREM3, (B) MTRR, (C) IGFBP7, (D) HUS1, (E) PROKR2 and (F) ST3GAL1. Trees were generated from our resequencing data using MrBayes, with the median posterior probability of the clade over two runs reported in red (16). Results are for the entire resequenced regions for FREM3 and MTRR, and for the largest regions for which we found strong support in other cases. For FREM3, MTRR and IGFBP7, the regions on which the trees are based are long (>800 bps), providing strong support for a polymorphism shared identical by descent (16). For HUS1, the tree still clusters by allele when considering 1 kb (with posterior probability 0.58), but for ST3GAL1 and PROKR2, this is not the case (for more details, see (16)).
Interestingly, the shared SNPs nearest FREM3 are in almost perfect LD with several eQTLs for GYPE (~130 kb away) in monocytes (Fig. 2A). Along with GYPA and GYPB, GYPE originated from one copy in the common ancestor of African apes (26). GYPA is a known receptor for Plasmodium falciparum proposed to be under balancing selection in humans, which, together with GYPB, codes for the MNS blood group (26); much less is known about GYPE, but it may also specify the M blood group antigen (27). The shared SNPs ~117 kb from MTRR, a gene involved in the production of methionine and implicated in the regulation of folate metabolism, are also in significant LD with an eQTL in monocytes, for MTRR (Fig. 2B). In turn, the shared SNPs in an intron of IGFBP7 occur in a likely enhancer (Fig. 2C). IGFBP7 has been shown to regulate cell proliferation, cell adhesion and angiogenesis in cancer cell lines, and plays a role in innate immunity by interacting with chemokines implicated in the regulation of lymphocyte trafficking (28).
In the two other regions (in HUS1 and nearest PROKR2) as well as in a region with only one pair of shared SNPs in significant LD (nearest ST3GAL1), diversity levels are only unusually high in humans, but nonetheless a phylogenetic tree for a small subset of the region (300 bps) clusters by allele and not by species (Figs. 3D–F, S8). These patterns are consistent with the presence of an ancient balanced polymorphism on an ancestral segment that has been highly eroded by recombination (for a more in-depth discussion, see (16)). PROKR2 is a receptor that functions as a pro-inflammatory mediator and whose ligand is able to modulate immune response (29). In turn, ST3GAL1 is a sialyltransferase that modifies the cell surface glycan structure of dendritic cells (30) and for which knockout mice lack peripheral CD8+ T lymphocytes (31).
To check for possible sequencing or mapping errors, we resequenced the six regions with evidence for a polymorphism shared identical by descent (summarized in Table S7) in 11–12 humans, 10–12 chimpanzees and four to seven gorillas. In all cases, we confirmed the presence of the expected shared SNPs and the predicted LD patterns among them (16). Additionally, we found that, in the MTTR and ST3GAL1 regions, one of the SNPs in the shared haplotypes is also segregating in gorilla (Fig. 2B, S8, (16)).
Common properties of ancient balanced polymorphisms
The narrow signature of ancient balancing selection allows the possible causal sites to be delimited to a few kilobases. Of the six regions with evidence for a long-lived balanced polymorphism, those in HUS1 and IGBFBP7 and nearest ST3GAL1 likely have regulatory activity (Figs. 2, S8). More generally, only two of the 125 candidate regions include a shared SNP that is coding (in both cases, synonymous), but at least ten regions appear to have a regulatory role (Table S8, (16)). Our findings therefore suggest that balancing selection has targeted regulatory variation in the human genome. The possible mechanisms underlying the maintenance of such polymorphisms are unclear, but could involve allele-specific properties that lead to differences in levels of expression, in response to stimuli, or in patterns of expression across tissues (as is the case for B4galnt2 in mice (32)).
To further assess the commonalities among the set of 125 regions, we tested for an enrichment of gene categories for the nearest protein-coding gene (Tables 1, S9, (16)). We found significant enrichments of a number of overlapping categories, driven by the presence of 24 membrane glycoproteins in the test set of 54 genes (p < 10−3, corresponding to a 2.4 fold enrichment of glycoproteins over the background and a 1.2 fold enrichment of membrane glycoproteins over a background of only glycoproteins; Tables 1, S10–S12). Five of the 24 membrane glycoproteins have an immunoglobulin I-set domain (p=0.006; a 6.3 fold enrichment over a background of membrane glycoproteins). The same trends are seen when considering an almost completely independent set of 335 coding SNPs (only two occur in shared haplotypes, neither of which contributes to these trends): glycoprotein and cell adhesion are top categories among shared coding SNPs (p < 0.02; Tables S13–S14). Though the number of genes involved is small, there is also an enrichment of gene ontology categories related to galactosyltransferase activity among genes near shared haplotypes and for categories related to glycosylation among genes with a shared coding SNP (Tables S9, S14).
Table 1.
Enrichment analysis.
Gene category enrichment of the closest gene within 20 kb of shared human-chimpanzee haplotypes (16). We show only the top categories, for which p <10−3 (see Table S9 for a longer list), noting that, because the categories overlap, the Bonferroni correction is conservative. The heading “Count” refers to the number of genes from the gene set with the given property (Term); “List Total” to number of genes from the gene set that can be annotated in the category; “Pop Hits” to the number of genes in the background with the given property and “Pop Total” to the number of genes from the background that can be annotated in the category.
| Category | Term | Count | List Total | Pop Hits | Pop Total | Fold Enrichment | P-value | Bonferroni corrected P-value |
|---|---|---|---|---|---|---|---|---|
| SP_PIR_KEYWORDS | glycoprotein | 27 | 50 | 3733 | 16731 | 2.42 | 2.56×10−6 | 3.33×10−4 |
| GOTERM_CC_FAT | GO:0031224~intrinsic to membrane | 31 | 42 | 4719 | 11298 | 1.77 | 4.48×10−5 | 0.0051 |
| INTERPRO | IPR013098:Immunoglobulin I-set | 6 | 46 | 133 | 14738 | 14.45 | 5.08×10−5 | 0.0076 |
| INTERPRO | IPR007110:Immunoglobulin-like | 7 | 46 | 373 | 14738 | 6.01 | 8.93×10−4 | 0.1254 |
Given that viruses frequently utilize host glycans to gain entry into host cells and some bacteria imitate host glycans to evade the host immune system (e.g., (33–35)), these enrichments suggest that the targets of balancing selection that we identified likely evolved in response to pressures exerted by human and chimpanzee pathogens, mirroring what is known about other genes under balancing selection in humans (see (1, 17, 18, 36) and references therein). Moreover, the observation that variation at loci that lie at the interface of host-pathogen interactions was stably maintained for millions of years is consistent with the hypothesis that arms races between hosts and pathogens can result not only in transient polymorphisms but also, in the presence of a cost to resistance, to a stable limit cycle in allele frequencies in the host (4, 9, 37).
In summary, we found several instances of ancient balancing selection in humans in addition to the two previously known cases. Our analysis suggests that this mode of selection has not only involved protein changes but also the regulation of genes involved in the interactions of humans and chimpanzees with pathogens, and point to membrane glycoproteins as frequent targets. Since we deliberately focused on the subset of cases of balancing selection that are least equivocal – requiring variation at two or more sites to be stably maintained in the two species from their split to the present – we likely missed balanced polymorphisms with a high mutation rate to new selected alleles (i.e., with high allelic turnover (38)), in which the ancestral segment has been too heavily eroded by recombination, as well as any instance where balancing selection pressures are more recent than the human-chimpanzee split. Thus, it seems likely that many more cases of balancing selection in the human genome remain to be found.
Supplementary Material
Acknowledgments
Thanks to D. Conrad, Y. Lee, M. Nobrega, J. Pickrell, H. Shim as well as A. Kermany, A. Venkat and other members of the PPS labs for helpful discussions; to I. Aneas, M. Çalışkan, M. Nobrega, and C. Ober for their assistance with experiments; and to G. Coop for discussions and comments on an earlier version of this manuscript. E. M. L. was supported in part by NIH training grant T32 GM007197. This work was supported by NIH HG005226 to J.D.W.; Israel Science Foundation grant 1492/10 to G.S.; a Wolfson Royal Society Merit Award, a Wellcome Trust Senior Investigator Award (095552/Z/11/Z), Wellcome Trust Grants 090532/Z/09/Z and 075491/Z/04/B to P.D.; Wellcome Trust grant 086084/Z/08/Z to G.M. and NIH GM72861 to M.P. M. P. is a Howard Hughes Medical Institute Early Career Scientist.
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
The data set of shared SNPs is available from http://przeworski.uchicago.edu/wordpress/?page_id=20. Data from the validation experiment are available from GenBank under accession numbers KC541701-KC542146. The biological material obtained from the San Diego Zoo and used in this study is subject to an MTA.
References
- 1.Hedrick PW. Heredity (Edinb) 2011 Oct;107:283. doi: 10.1038/hdy.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reid DG. Biological Journal of the Linnean Society. 1987;30:1. [Google Scholar]
- 3.Gigord LD, Macnair MR, Smithson A. Proc Natl Acad Sci U S A. 2001 May 22;98:6253. doi: 10.1073/pnas.111162598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stahl EA, Dwyer G, Mauricio R, Kreitman M, Bergelson J. Nature. 1999 Aug 12;400:667. doi: 10.1038/23260. [DOI] [PubMed] [Google Scholar]
- 5.Wright S. Genetics. 1939 Jun;24:538. doi: 10.1093/genetics/24.4.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hiwatashi T, et al. Mol Biol Evol. 2010 Feb;27:453. doi: 10.1093/molbev/msp262. [DOI] [PubMed] [Google Scholar]
- 7.Heliconious Genome Consortium. Nature. 2012;487:94. [Google Scholar]
- 8.Ghosh R, Andersen EC, Shapiro JA, Gerke JP, Kruglyak L. Science. 2012 Feb 3;335:574. doi: 10.1126/science.1214318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charlesworth B, Charlesworth D. Elements of Evolutionary Genetics. Roberts and Company; 2010. [Google Scholar]
- 10.Dobzhansky T. Genetics of the evolutionary process. Columbia University Press; 1970. [Google Scholar]
- 11.Lewontin RC. The genetic basis of evolutionary change. Columbia University Press; New York: 1974. [Google Scholar]
- 12.Gillespie JH. The causes of molecular evolution. Oxford University Press; 1991. [Google Scholar]
- 13.Hudson RR, Kaplan NL. Genetics. 1988 Nov;120:831. doi: 10.1093/genetics/120.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charlesworth D. PLoS Genet. 2006 Apr;2:e64. doi: 10.1371/journal.pgen.0020064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiuf C, Zhao K, Innan H, Nordborg M. Genetics. 2004 Dec;168:2363. doi: 10.1534/genetics.104.029488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.See Supplementary Online Materials
- 17.Klein J, Satta Y, O’HUigin C, Takahata N. Annu Rev Immunol. 1993;11:269. doi: 10.1146/annurev.iy.11.040193.001413. [DOI] [PubMed] [Google Scholar]
- 18.Segurel L, et al. Proc Natl Acad Sci U S A. 2012 Nov 6;109:18493. doi: 10.1073/pnas.1210603109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bubb KL, et al. Genetics. 2006 Aug;173:2165. doi: 10.1534/genetics.106.055715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The 1000 Genomes Project Consortium. Nature. 2010 Oct 28;467:1061. [Google Scholar]
- 21.Auton A, et al. Science. 2012 Apr 13;336:193. [Google Scholar]
- 22.Patterson N, Richter DJ, Gnerre S, Lander ES, Reich D. Nature. 2006 Jun 29;441:1103. doi: 10.1038/nature04789. [DOI] [PubMed] [Google Scholar]
- 23.Hodgkinson A, Eyre-Walker A. Nat Rev Genet. 2011 Nov;12:756. doi: 10.1038/nrg3098. [DOI] [PubMed] [Google Scholar]
- 24.Franchini M, Capra F, Targher G, Montagnana M, Lippi G. Thromb J. 2007;5:14. doi: 10.1186/1477-9560-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gieger C, et al. Nature. 2011 Dec 8;480:201. [Google Scholar]
- 26.Ko WY, et al. Am J Hum Genet. 2011 Jun 10;88:741. doi: 10.1016/j.ajhg.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kudo S, Fukuda M. J Biol Chem. 1990 Jan 15;265:1102. [PubMed] [Google Scholar]
- 28.Nagakubo D, et al. J Immunol. 2003 Jul 15;171:553. doi: 10.4049/jimmunol.171.2.553. [DOI] [PubMed] [Google Scholar]
- 29.Monnier J, Samson M. FEBS J. 2008 Aug;275:4014. doi: 10.1111/j.1742-4658.2008.06559.x. [DOI] [PubMed] [Google Scholar]
- 30.Videira PA, et al. Glycoconj J. 2008 Apr;25:259. doi: 10.1007/s10719-007-9092-6. [DOI] [PubMed] [Google Scholar]
- 31.Priatel JJ, et al. Immunity. 2000 Mar;12:273. doi: 10.1016/s1074-7613(00)80180-6. [DOI] [PubMed] [Google Scholar]
- 32.Johnsen JM, et al. Mol Biol Evol. 2009 Mar;26:567. doi: 10.1093/molbev/msn284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gagneux P, Varki A. Glycobiology. 1999 Aug;9:747. doi: 10.1093/glycob/9.8.747. [DOI] [PubMed] [Google Scholar]
- 34.Olofsson S, Bergstrom T. Ann Med. 2005;37:154. doi: 10.1080/07853890510007340. [DOI] [PubMed] [Google Scholar]
- 35.Day CJ, Semchenko EA, Korolik V. Front Cell Infect Microbiol. 2012;2:9. doi: 10.3389/fcimb.2012.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruwende C, et al. Nature. 1995 Jul 20;376:246. doi: 10.1038/376246a0. [DOI] [PubMed] [Google Scholar]
- 37.Tellier A, Brown JK. Proc Biol Sci. 2007 Mar 22;274:809. doi: 10.1098/rspb.2006.0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahata N. Proc Natl Acad Sci U S A. 1990 Apr;87:2419. doi: 10.1073/pnas.87.7.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chimpanzee Sequencing and Analysis Consortium. Nature. 2005 Sep 1;437:69. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 40.Zeller T, et al. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.