Significance
Membrane proteins (MPs) perform a variety of essential cellular functions, account for about one-third of encoded proteins in genomes, and comprise more than one-half of human drug targets. High-resolution structures are essential to understand the underlying molecular mechanisms of MPs and facilitate structure-based drug design efforts. Detergents are indispensible in the solubilization of MPs, but they tend to destabilize MPs and often impede the growth of well-ordered protein crystals. We describe a class of structurally unique detergents, designated as facial amphiphiles, which improved MP stability and success in the crystallization of different families of MPs.
Abstract
Amphiphile selection is a critical step for structural studies of membrane proteins (MPs). We have developed a family of steroid-based facial amphiphiles (FAs) that are structurally distinct from conventional detergents and previously developed FAs. The unique FAs stabilize MPs and form relatively small protein–detergent complexes (PDCs), a property considered favorable for MP crystallization. We attempted to crystallize several MPs belonging to different protein families, including the human gap junction channel protein connexin 26, the ATP binding cassette transporter MsbA, the seven-transmembrane G protein-coupled receptor-like bacteriorhodopsin, and cytochrome P450s (peripheral MPs). Using FAs alone or mixed with other detergents or lipids, we obtained 3D crystals of the above proteins suitable for X-ray crystallographic analysis. The fact that FAs enhance MP crystallizability compared with traditional detergents can be attributed to several properties, including increased protein stability, formation of small PDCs, decreased PDC surface flexibility, and potential to mediate crystal lattice contacts.
Membrane proteins (MPs) perform essential roles in cellular physiology, account for about one-third of encoded proteins in genomes, and comprise more than one-half of human drug targets. High-resolution molecular structures provide insight into the underlying molecular mechanisms of MPs and a context for organizing the results of functional studies, and they facilitate structure-based drug design. Because of their hydrophobic domains, MPs require the use of detergents and/or lipids for solubilization from membranes. Detergents commonly used to solubilize and stabilize MPs contain long alkyl chains [e.g., dodecyl-β-D-maltoside (DDM)] and typically form large protein–detergent complexes (PDCs) comprised of tens to hundreds of detergent molecules. MP crystals grown from large PDCs typically diffract less well than crystals grown from smaller PDCs (1) and/or suffer lattice defects (i.e., anisotropy and twinning), likely because of poor packing in the lattice and high solvent content (2, 3). In addition, detergent molecules in PDCs are largely disordered and usually do not contribute to rigid crystal contacts. The reduced polar surface area in MPs compared with soluble proteins also impedes crystallization.
In recent years, there has been a substantial increase in the success of crystallization of MPs using novel amphiphiles, such as cubic phase-forming lipids or bilayered micelles (bicelles) (4), and fusions with soluble proteins to facilitate crystal contacts (5, 6). Nevertheless, the success rate for growing well-diffracting 3D crystals has been much lower for MPs than soluble proteins, and MPs account for less than 2% of all current entries in the Protein Data Bank (PDB). Consequently, there continues to be a need for identification of amphiphiles that confer stability and crystallizability of MPs. For instance, a variety of novel amphiphiles, including protein-based nanodiscs (7), amphiphilic polymers (8), peptide-based surfactants (9, 10), and maltose-neopentyl glycol (11) or Di-Mal detergents (12), has been widely used for the solubilization and stabilization of MPs for functional or biophysical studies. However, there has been limited success using some of these novel amphiphiles to crystallize MPs (11, 13–15).
Another class of widely studied amphiphiles exhibits facial amphiphilicity that is distinct from the end polarity present in conventional head-to-tail detergents. Representative facial amphiphiles (FAs) include bile acids and their derivatives, such as CHAPS and CHAPSO, which have been widely used to solubilize and stabilize integral MPs but have yielded little success in crystallization, despite many years of effort. Here, we present the synthesis and evaluation of newly designed steroid-based FAs, which are structurally different from conventional detergents and previously developed FAs. We show that these unique FAs stabilized MPs from different protein families and yielded diffraction-quality 3D crystals.
Results
Structures and Properties of FAs.
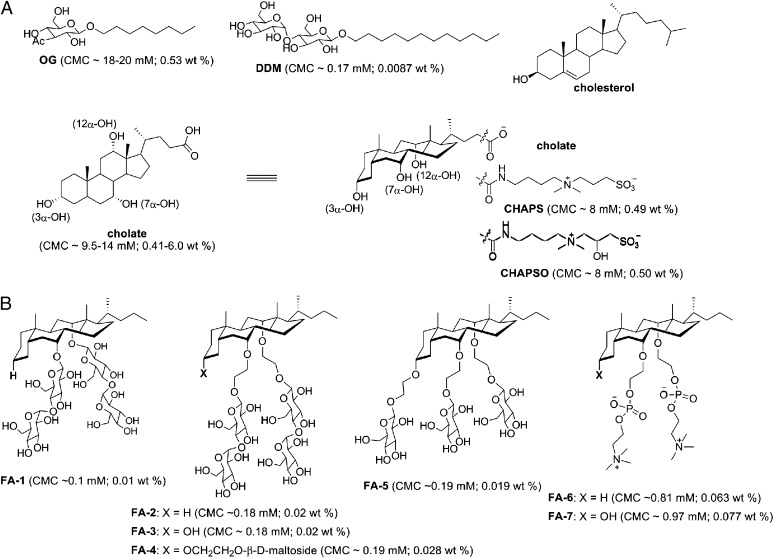
We previously reported the synthesis of an FA (hereafter referred to as FA-1) with β-D-maltoside attached to the 7α- and 12α-positions in cholate (16). This unique FA design removed the flexible terminal polar head groups in cholate, CHAPS, and CHAPSO and attached polar side groups under the steroid surface. We have now synthesized and evaluated an expanded series of FAs of this design (Fig. 1). We found that the identity, number, and position of the polar groups significantly affected the detergent properties and their capacity to stabilize MPs. This report focuses specifically on FAs bearing nonionic sugar (glucoside or maltoside) or zwitterionic (phosphocholine) polar groups, which occur commonly in conventional detergents and lipids.
Fig. 1.
Chemical structures of (A) conventional head-to-tail detergents (OG and DDM), the natural steroid lipid cholesterol, and previously described steroid-based FAs (cholate and its derivatives CHAPS and CHAPSO) and (B) newly designed FAs. Detergent CMC values are listed under the respective chemical structures.
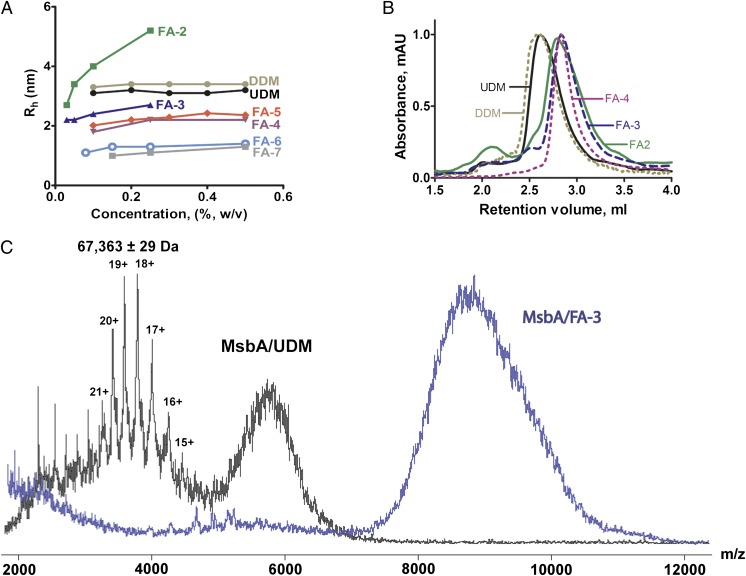
The 7α- and 12α-OH groups in cholate were of low reactivity (e.g., for glycosylation) because of steric hindrance, and therefore, we extended each position with an ethylene oxide linker to improve synthetic efficiency (Fig. 1). The newly synthesized FAs were highly soluble in water (>10% wt/vol), except FA-2 (<2% wt/vol), a structure in which, like FA-1, the 3α-OH group was eliminated. FA-3 and FA-4 are structurally analogous to FA-2 but retain 3α-polar groups (hydroxyl or maltoside). Whereas FA-2 formed large micelles in a concentration-dependent manner, FA-3 and FA-4 formed small micelles over a broad range of concentrations (Fig. 2A), indicating that the presence of a 3α-polar group limited the growth of micelle aggregates. FA-4 and FA-5 have three sugar (maltoside or glucoside) attachments on the steroid backbone and formed micelles of similar size. Interestingly, FA-6 and FA-7, containing phosphocholine groups, formed even smaller micelles than the FAs modified with nonionic sugar groups (Fig. 2A). This property may result from electrostatic repulsion by phosphocholine. With the exception of FA-2, the hydrodynamic radii (Rh) of micelles formed by this FA series were between 1.3 and 2.5 nm, smaller than micelles of DDM (∼3.2 nm) or octyl-β-D-glucoside (∼2.7 nm), two of the most widely used detergents in MP structural studies (17). The aggregation number (AN) of FA micelles was conveniently estimated based on Rh values using a calibration curve of molecular weight (MW) -Rh (Materials and Methods). The estimated AN values were >30, 20–30, 10–14, 16–25, 6–10, and 5–10 for the micelles formed by FA-2, FA-3, FA-4, FA-5, FA-6, and FA-7, respectively. This range of AN values was indicative of the concentration-dependent changes in micellar size (Rh) shown in Fig. 2A. In addition, the critical micellar concentration (CMC) values of the FAs (Fig. 1) ranged from 0.01% (0.1 mM) to 0.077% (1.03 mM). These CMCs were markedly lower than traditional FAs, such as cholic acid, CHAPS, and CHAPSO, with CMCs ranging from 8 to 14.5 mM.
Fig. 2.
Properties of FAs in micelles and PDCs. (A) Hydrodynamic radii (Rh) of FA micelles in water vs. detergent concentration as determined by DLS at RT. The DLS profiles of DDM and UDM micelles are shown for comparison. (B) Analytical SEC profiles of dimeric MsbA solubilized in FAs indicated the formation of smaller PDCs compared with UDM and DDM. (C) ESI–mass spectra of MsbA (monomer molecular mass of 67,228 Da) solubilized in UDM or FA-3; both spectra were acquired at the same collision energy (200 V). Peaks indicative of the MsbA monomer were observed in MsbA/UDM but not MsbA/FA-3.
PDC Properties of FAs.
Compared with conventional long-chain detergents, the steroid backbone of FAs presents a distinct hydrophobic arrangement and a much larger hydrophobic surface area that may allow tighter and more effective packing around the hydrophobic surfaces of MPs. For the ATP binding cassette (ABC) transporter MsbA, we showed previously that the FA-1/MsbA molar binding ratio in PDCs was approximately fivefold smaller than the undecyl-β-D-maltoside (UDM) /MsbA binding ratio (16). To further explore the properties of FAs in PDCs, we compared size exclusion chromatography (SEC) profiles of MsbA complexed with FAs (FA-2, FA-3, and FA-4) that differ in micelle size (Figs. 1 and 2). The Stokes radii of MsbA measured in these three FAs were consistently smaller than for UDM or DDM (Fig. 2B), confirming that FAs enabled the formation of small PDCs. The smaller micelle, reduced PDC size, and lower detergent:protein ratio obtained with FAs may result in tighter crystal packing of MPs than would occur in larger PDCs, and thus, they would be expected to improve the diffraction quality and reduce crystal mosaicity.
To characterize the binding strength between FAs and MPs, we used electrospray ionization MS (ESI-MS). In this application of ESI-MS, loosely bound detergents can be stripped from MP surfaces in the gas phase by increasing the energy in the collision cell of a mass spectrometer (18). At the maximal collision energy (200 V) applied to an MsbA/UDM sample, the spectrum displayed a broad distribution of charged protein peaks that were deconvoluted to a molecular mass of 67 kDa for the MsbA monomer (Fig. 2C). An additional unresolved broad peak at m/z ∼5,500 was attributed to protein embedded (fully or partially) in detergent micelles. However, at the same collision energy, we did not detect release of the 67 kDa MsbA monomer in the MsbA/FA-3 spectrum. Instead, only a large, unresolved protein–detergent micelle peak was detected (Fig. 2C). The MS results clearly indicate that, compared with UDM, FA-3 bound more tightly to MsbA, thus protecting the protein from dissociation to nonfunctional monomers. Such tight binding would likely help stabilize MPs solubilized in FAs.
Extraction, Purification, and Stabilization of MPs Using FAs.
To explore the general use of the unique FAs, we evaluated their ability to solubilize and stabilize selected MP targets, including bacteriorhodopsin (BR) from Halobacterium salinarum, the ABC transporter MsbA from Escherichia coli, and the human gap junction channel composed of connexin 26 (Cx26) expressed in Sf9 insect cells. FAs effectively extracted MsbA and Cx26 from their respective E. coli and Sf9 cell membranes and allowed significant purification (Figs. S1 and S2). However, FAs failed to extract BR from purple membranes, possibly because of the unusual lipid composition in the archaebacteria. Therefore, BR was first solubilized and purified in octyl-β-D-glucoside (OG), a detergent commonly used in its purification. The purified protein was then exchanged to the appropriate FA before stability measurements using the absorption spectrum of its endogenous ligand retinal (16). By this criterion, ligand absorption was nearly abolished in about 30 d when BR was maintained at room temperature (RT) in OG (Fig. 3B). In contrast, all sugar-based FAs effectively stabilized BR at RT for more than 4 mo without significant changes in the absorption spectrum (Fig. 3 A and B). Interestingly, the phosphocholine-derived FA-7 stabilized BR as well as sugar-based FAs, faring significantly better than FA-6 that lacks the 3α-OH group.
Fig. 3.
Stability of MPs in FAs. (A) BR in FA-3 maintained its characteristic absorption spectrum for over 4 mo at RT. (B) Absorbance at 550 nm of BR in OG or FAs over a 4-mo period at RT. (C) The ATPase activity of MsbA incubated in FAs at RT was maintained for more than 1 mo compared with the ATPase activity of MsbA in UDM or DDM. (D) Isothermal denaturation of Cx26 at 37 °C as measured by accessibility of Cys202 in transmembrane helix 4 to the prefluorescent dye cpm. Detergents were at 2× CMC.
To determine the stability of MsbA in various amphiphiles, ATPase activity was periodically assayed as described (16, 19). MsbA retained activity for more than 1 mo at RT in sugar-based FAs. In contrast, there was a dramatic loss of activity within 2 d in maltoside detergents (UDM or DDM), which are commonly used for purification of MsbA and other transporters (16, 20) (Fig. 3C). Moreover, the initial activity of MsbA in FA-3, FA-4, or FA-5 was two to three times greater than when purified in UDM or DDM (∼2.6 μmol/mg per minute). Although the initial activity in FA-2 was comparable with UDM and DDM, the activity remained stable during the time course compared with the dramatic loss of activity in UDM and DDM. Consistent with the ATPase activity measurements, MsbA visibly precipitated within 2 d in UDM or DDM but remained completely soluble in the FAs throughout these experiments. Mixing UDM-purified MsbA with FAs also resulted in higher ATPase activity and increased the stability of MsbA, suggesting a direct stabilizing effect of FAs on MsbA in the PDC.
We tested the isothermal stability of Cx26 (37 °C) in the presence of FAs by measuring cysteine accessibility to the fluorescent dye 7-diethylamino-3-(4′-maleimidylphenyl)-4-methylcoumarin (cpm) (14). Compared with DDM, all sugar-based FAs endowed Cx26 with similar or marginally improved stability (Fig. 3D). We also used cpm to measure the Cx26 transition temperatures (Tm) (21) by heating samples in FA-3 or DDM from 10 °C to 85 °C; both yielded a comparable Tm value (∼52 °C). Finally, the oligomeric stability of Cx26 purified in FA-3 was assessed by negative stain EM. We observed a field of relatively uniform particles with the characteristic doughnut appearance, similar to Cx26 in DDM (Fig. S2C), showing that Cx26 in FAs existed in the native oligomeric structure, most likely a hexameric hemichannel.
3D Crystallization of Cx26 in FAs.
On the basis of the detergent properties and the solubility and stability screens, we focused on the sugar-based facial amphiphiles FA-3, FA-4, and FA-5 for the MP crystallization experiments. The successful crystallization of each of the chosen targets reinforces the conclusion that protein stability was enhanced in selected FAs.
Crystals of Cx26 in DDM diffracted weakly to 4.0 Å resolution and suffered from severe anisotropy and radiation damage (Table S1). In an earlier study using branch-chained maltosides (14), we improved the diffraction slightly but could not solve the anisotropy problem. We then pursued crystallization of Cx26 using the FAs. FA-3 and FA-5 were selected for more extensive studies based on a preliminary screen (FA-4 also gave crystals).
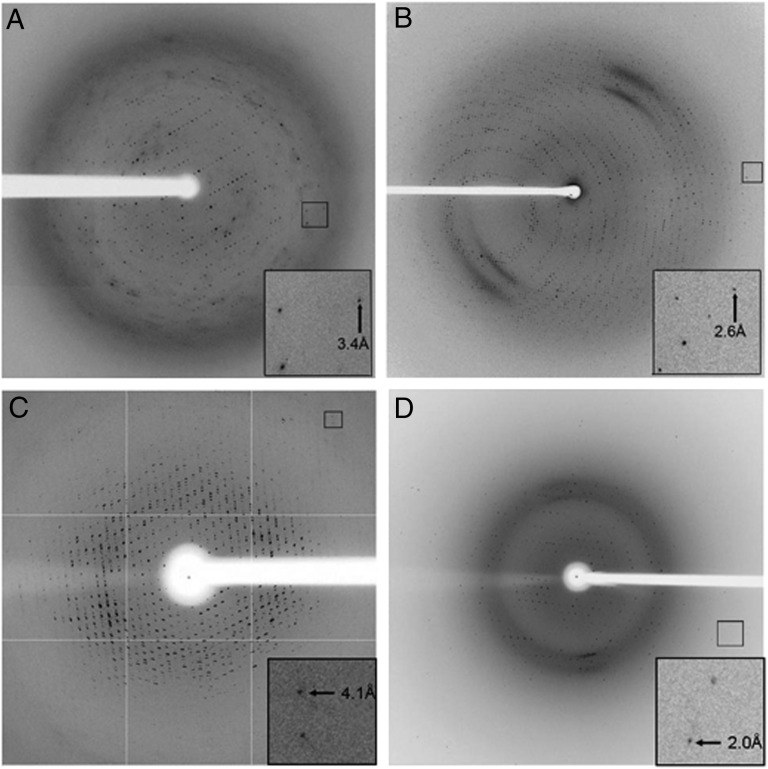
Cx26 in FA-3 readily crystallized in a number of sparse matrix screen conditions by both hanging- and sitting-drop vapor diffusions. Crystals could be reproducibly grown at RT, typically appeared within 2 wk of setup, and attained an average size of 50 microns in at least two dimensions. Crystallization occurred faster (within 5–7 d) if the reservoir buffer included Ca2+, which is known to induce closure of gap junction channels by an unclear mechanism. Initial indexing of the crystals suggested a rhombohedral Bravais lattice and a maximal diffraction resolution of 3.0 Å. A complete dataset was collected to an isotropic diffraction limit of 3.2 Å resolution (Fig. 4A). Ultimately, the datasets were scaled in space group R32 to a resolution cutoff of 3.3 Å, and the crystallographic statistics are given in Table S1. The structure was solved by molecular replacement, with two molecules in the asymmetric unit. In addition, we also crystallized Cx26 in the absence of Ca2+, which yielded the same crystal form, albeit with a resolution limit of 3.6 Å. Refinement and rebuilding of the Ca2+-bound and -free structures at 3.3 Å and 3.8 Å resolutions, respectively, have been completed, and the description and analysis of these structures will be published elsewhere. The Fo-Fc maps showed several large difference electron density peaks near hydrophobic regions within the interior of the channel pore and in pockets on the channel exterior that would face membrane lipids. We have preliminarily modeled the FA-3 steroid backbone and parts of the side chains into some of these peaks, resulting in small drops in Rfree in subsequent rounds of refinement. Although the fit of FA-3 into some of them is quite satisfactory, additional analysis is needed to verify the identity of these peaks.
Fig. 4.
MP crystals grown in FAs displayed isotropic diffraction at moderate to high resolution. X-ray diffraction patterns from crystals of (A) Cx26/FA-3, (B) Cx26/FA-5, (C) MsbA/FA-5, and (D) BR in FA-7/DMPC bicelles. Insets are enlargements of the boxed regions in the diffraction patterns, showing the highest resolution reflections recorded. The brightness and contrast of Insets were adjusted to allow easier observation of these reflections.
Improved diffraction was observed from Cx26/FA-5 crystals, although these crystals typically required 2–3 mo to appear and 4–6 mo to reach their maximum size. Diffraction could be recorded to 2.4 Å resolution, and a complete dataset was collected to 2.6 Å resolution, with a limiting resolution for data processing set at 3.1 Å. However, data processing and structure determination have been difficult, although the crystal lattice is most likely cubic (Fig. 4B).
Crystallization of MsbA Using FAs.
A wide open conformation of E. coli MsbA has been reported to relatively low resolution (5.3–6.2 Å) from crystals grown in dodecyl-α-D-maltoside in the absence of nucleotide (20). The conformations of MsbA and other ABC efflux transporters are highly dynamic and flexible, which exacerbates the challenge of growing ordered crystals from this family of proteins. Interestingly, MsbA crystals were readily grown by the vapor diffusion method in the presence of all FAs examined, including FA-3, FA-4, and FA-5, at RT or 4 °C. These crystals reached their maximum size in 2 mo, ∼100–400 μm in one dimension. Thus far, the best diffracting crystals (∼4.1 Å resolution) were of MsbA in a complex with FA-3 or FA-5 (Fig. 4C). The protein crystallized in the P212121 space group and was different from the crystal form obtained in dodecyl-α-D-maltoside (20). The self-rotation function of the processed data suggested 32-point group, noncrystallographic symmetry with a trimer of MsbA dimers (i.e., three dimers with their twofold axes normal to a central threefold axis). Thus, we expect that the structure of the asymmetric unit is a hexamer of MsbA with 32 symmetry. This hexameric packing was confirmed by examining electron density maps calculated from low-resolution datasets collected on MsbA crystals in the presence of FA-3 or FA-5 (Table S2). In this work, we have shown the use of FAs to crystallize MsbA. Meanwhile, additional efforts, such as the suppression of the conformational dynamics of MsbA, are necessary to achieve structures at higher resolution.
FA-Dimyristoylphosphatidylcholine Bicelle Crystallization of BR.
FAs could also be used to generate bicelles, which are useful for MP structure determination by NMR and X-ray crystallography (22). We used WT BR as a model protein to test FA–bicelle systems for the crystallization and X-ray analysis of integral MPs. Of note, both lipidic cubic phase (LCP) and bicelle methodologies were developed from the crystallization of BR (23–25) before applications to other MP systems.
Unlike bicelle matrices consisting of dimyristoylphosphatidylcholine (DMPC) :CHAPSO or dihexanoyl phosphatidylcholine (DMPC:DHPC), the bicelle milieu of DMPC:FAs (using FA-4 or FA-7) could accommodate a higher lipid content (q > 10, where q corresponds to the lipid:detergent molar ratio). Both FA-4 and FA-7 allowed us to synthesize bicelles with up to 40% (wt/vol) DMPC. Similar to DMPC:CHAPSO and DMPC:DHPC bicelles, DMPC:FA bicelles underwent a transition from the liquid to the gel phase above RT. Interestingly, it was straightforward to grow the previously reported triclinic, diamond-shaped type I crystals of BR using DMPC-FA bicelles from RT to ∼30 °C (26). The BR crystals that grow in DMPC:FA-4 and DMPC:FA-7 bicelles [q = 10 and CL = 9% (wt/vol), where CL is total lipid content] diffracted X-rays in a comparable fashion with crystals grown in a DMPC:CHAPSO [q = 2.8 and CL = 8% (wt/vol)] bicelle (25) (Fig. 4D and Table S3). The space group, unit cell dimensions, and molecular packing were very similar to the template BR crystal structure (PDB ID code 1KME) (25), with a 0.12–0.24 Å rmsd between Cα atoms in the structures. However, the BR structure solved in DMPC:FA-4 bicelles (PDB ID code 4HYX) showed that several unstructured residues near the C-terminal region (from E234 to S239; G231–A233 not modeled) were clearly resolved in one asymmetric unit of two per unit cell. Interestingly, these residues were not apparent in dozens of other BR structures deposited in the PDB (solved mostly in LCP). Overall, the fact that the structure is not altered indicates that MP structures may be largely unaffected by the composition of the bicelle matrix. Bicelle-based crystallization with unique FAs will help glean molecular details of MPs, because these FAs have been shown to satisfy one of the challenging prerequisites for their structural study, namely stabilizing MPs after their removal from the native lipid bilayer environment.
Crystallization of Cytochrome P450 Enzymes and Peripheral MPs Using FA Commercial Detergent Mixtures.
More superfamilies of monotopic/peripheral MPs have been described than polytopic MPs, which is documented in the well-maintained MP database (http://opm.phar.umich.edu/). Solubilization and crystallization of the former group of MPs also require the use of detergents to sequester hydrophobic protein patches and prevent protein aggregation. By mixing them with commercial detergents, FAs have been successfully used to crystallize several mammalian P450 enzymes [CYP24A1 (27), CYP2B4 (28–32), and CYP2B6 (32–34)]. Here, we provide a summary of these results (Table S4). Altogether, four FAs, including FA-3, FA-4, FA-6, and FA-7, have been tested. Statistically, FA-4 and FA-7 have yielded more success. Overall, we found that the addition of FAs at 1× CMC dramatically improved the crystallization propensity and crystal quality of these P450s. In some cases, FA addition was crucial to crystal growth (Table S4).
The case of CYP24A1 crystallization has provided interesting results for comparison between CHAPS and FAs (27). CYP24A1 crystals grown in CHAPS alone diffracted to a resolution of only ∼7 Å, whereas mixing CHAPS and FA (1:1 at 1× CMC) yielded a monoclinic crystal that diffracted to 2.0 Å resolution (FA-3) (27) and an orthorhombic form at 3.4 Å resolution (with FA-6).
In the recent CYP2B4 structure complexed with the antiplatelet drug ticlopidine (PDB ID code 3KW4) (28), two resolved FA-4 molecules mediated crystal contacts between hydrophobic regions of two symmetry-related protein molecules (Fig. 5). A molecule of FA-4 also protected a hydrophobic region of CYP2B4 that inserts into endoplasmic reticulum membranes. Electron density for the terminal alkyl chain and the terminal sugar rings at the 7α- and 12α-positions were missing because of disorder, suggesting that restricting the flexibility of amphiphiles may further improve ordered packing. Although P450 enzymes are peripheral MPs, high-resolution X-ray structures of P450s in FAs suggest a tentative mechanism, whereby FAs potentially aid stabilization and crystallization of other MPs.
Fig. 5.
FAs protect hydrophobic protein surfaces and mediate crystal lattice contacts in the CYP2B4:ticlopidine complex structure. (A) A view of the twofold symmetry-related crystal-packing interface shows that two FA-4 molecules (blue and green sticks) mediate crystal contacts between hydrophobic regions of the two protein molecules (gray and dark blue). (B) An enlargement of the boxed region in A. (C) FA-4, having three maltoside polar groups, is shown glycosylating a hydrophobic region of 2B4 that inserts into cellular endoplasmic reticulum membranes. (D) An enlargement of the boxed region in C shows that the steroid segment of FA-4 is in direct contact with several hydrophobic 2B4 side chains. 2B4 amino acids found within a 5 Å radius of FA-4 are labeled. A 2Fo-Fc omit map for FA-4 is shown as a gray mesh contoured at 1σ.
Discussion
Cholic acid and other bile salts present unique and versatile platforms for the engineering of broadly useful amphiphiles, because their rigid, hydrophobic steroid moiety is thought to be less denaturing than flexible alkyl chains present in conventional detergents. However, previously developed cholate-based detergents, such as CHAPS and CHAPSO, show some resemblance to conventional head-to-tail detergents, with a large polar head group at one end and the three hydroxyl groups providing only weak facial amphiphilicity. By removing the terminal carboxylate group of cholate and retaining the short alkyl chain attached to the steroid nucleus, we created an FA design that mimics the structural profile of cholesterol. We also found that retaining the 3α-OH group in FAs (present in the β-configuration in cholesterol) produced detergents with better chemical behavior. In addition, placing polar groups below the rigid steroid surface increased facial amphiphilicity and also imposed a degree of constraint on their flexibility. We propose that this structural improvement over the cholate/CHAPS/CHAPSO family is key to the improved success of FAs in our current tests of MP crystallization. Of note, a related deoxycholate-based tandem FA design has been recently described for stabilization of MPs (35) and may also be useful for crystallization.
Steroid-based FAs form small PDCs, associate tightly with MPs (as shown by ESI-MS), and likely exhibit decreased PDC surface flexibility. Coupled with their shown enhancement of protein stability, FAs possess several favorable attributes for successful MP crystallization. It is particularly interesting that two FAs mediate crystal lattice contacts in one of the P450 structures. Although rare, there are MP structures in which amphiphilic molecules could be modeled and were revealed to mediate crystal contacts, and some prominent examples are the use of steroid-based amphiphiles in bilayer-based MP crystallizations. For instance, CHAPSO molecules (structure shown in Fig. 1) mediate surface contacts of BR, which concurs with the report that BR more readily crystallizes in CHAPSO-DMPC bicelles than DHPC-DMPC bicelles (26). Novel steroid–steroid packing has also been observed in the structures of the β2-adrenergic receptor crystallized in LCP, where the addition of cholesterol was crucial for growing crystals (36). Cholesterol also mediates 2D crystal contacts of metarhodopsin I, for which addition of cholesterol likewise increased protein crystallizability and stability (37). These examples highlight the possibility of developing novel FAs not only for stabilizing MPs but also, for increasing the crystallization propensity by mediating MP surface interactions.
Here, we have shown the use of newly designed FAs in the stabilization and crystallization of several different classes of MPs. These results argue that steroid-based FAs should be considered alongside traditional detergents in the solubilization, purification, and crystallization of MPs. Given the significant success of using FAs as additives to conventional detergents for crystallization of cytochrome P450s, this approach could represent a convenient and efficient way to screen and use FAs for crystallizing other types of MPs. Such a strategy has produced a notable success in the crystallization of NorM, a multidrug and toxin extrusion transporter, where the addition of FA-3 reduced protein aggregation and improved crystal diffraction (38). Along with a larger toolbox of detergents, success in the next phase of MP structure determination will be facilitated by using a combination of recently developed technologies: unconventional crystallization methods such as LCP and bicelle crystallization, efficient use of highly intense synchrotron X-ray microfocused beams, and use of structure stabilizing agents such as soluble domain fusions, engineered disulfide bonds, and antibody fragments.
Materials and Methods
Synthetic Procedures and Characterization Data for FAs.
FAs were synthesized stepwise starting from cholic acid. Details of the synthesis and characterization are in SI Materials and Methods. The CMC of each FA was determined by a standard fluorescent dye binding protocol using 8-anilino-1-naphthalenesulfonic acid as the probe (39). Dynamic light scattering (DLS) was measured at ambient temperature in glass-bottomed optical 384-well plates (Greiner) on a Dynapro Plate Reader (Wyatt Technology) (14). The MW of FA micelles was estimated based on an MW-Rh calibration curve constructed from globular protein standards, a feature incorporated in the Dynapro instrument software. AN was then calculated by dividing the estimated MW of FA micelles by the molar MW of composed single FA.
Analytical SEC.
MsbA purified in FAs or maltoside detergents (UDM or DDM) was injected onto a Sepax SRT SEC 300 column using an AKTA Purifier FPLC System (GE Healthcare). The column was preequilibrated with a mobile phase containing 20 mM Tris (pH 7.5), 20 mM NaCl, and 3× CMC of the same detergent as the protein sample. Samples have also been analyzed using a Superdex 200 10/300 GL Column (GE Healthcare). On both columns, MsbA in FAs eluted at a longer retention time than in maltoside detergents.
MS.
MsbA purified in UDM or FA-3 was introduced into the nano-ESI source through gold-coated capillaries prepared in house (40). The Synapt-HDMS G1 Mass Spectrometer (Waters) was modified for transfer of high m/z complexes as described previously (41). Backing pressure was between 6 and 9 mbar to optimize micelle transfer in the ion-focusing region. Other parameters were as follows: the collision cell was set at 200 V, argon flow was 5.6 mL/min, and capillary, sampling, and extracting cone voltages were set at 1.7 kV, 80 V, and 0.8 V, respectively. The mobility cell was operated at 50 V with a helium flow of 50 mL/min. All spectra were smoothed using MassLynx software and calibrated with 100 mg/mL cesium iodide.
BR Preparation, Stabilization, and Crystallization.
BR was prepared using a standard protocol (42). Briefly, purple membranes from H. salinarium were solubilized with 50 mM sodium phosphate buffer (pH 6.9) containing 1.2% (wt/vol) OG and purified by SEC using a Superdex 200 16/60 Column (GE Healthcare) in solubilization buffer adjusted to pH 5.6. BR-containing fractions were concentrated to 10 mg/mL using a 50 kDa MWCO Amicon Centrifugal Device (Millipore). BR concentrations were determined by absorbance at 550 nm on a NanoDrop spectrophotometer using the extinction coefficient ε550 = 5.8 × 104 M−1 cm−1. To investigate the stability of BR in the presence of FAs, BR was exchanged from OG using a dilution/concentration method (10) by the addition of 50–100 volumes of buffer containing FA at 5× CMC. The absorption spectrum of BR was monitored using an HP 8453 Diode Array UV-Visible Spectrophotometer (Agilent).
Bicelles were prepared as described (25, 26), with a minor modification to the ratio of DMPC to FA-4 or FA-7 (q = 10). Crystals of BR in bicelles were grown using vapor diffusion hanging-drop methods by combining the protein/bicelle mixture with an equal volume of well solution containing 3.6 M sodium phosphate buffer (pH 3.4), 3.5% (vol/vol) triethyleneglycol, and 180 mM hexanediol. X-ray diffraction data were collected at the Stanford Synchrotron Radiation Lightsource beamline 11–1 (43). Data were integrated with iMOSFLM (44) and scaled with SCALA (45). The structures were solved by molecular replacement using PHASER (46) using the BR structure (25) as a search model and REFMAC (47) for refinement. The structures were compared with a previously reported structure (PDB ID code 1KME) using the structure superposition function of the MIFit program (48).
MsbA Preparation, Stabilization, and Crystallization.
MsbA was prepared as described previously with slight modification (20). Briefly, bacterial cell pellets containing MsbA were directly solubilized in 20 mM Tris (pH 8.0), 20 mM NaCl, 1% (wt/vol) detergent, 10% (vol/vol) glycerol, 0.1 mg/mL DNase I, and a proteinase inhibitor mixture (Roche). After centrifugation at 38,000 × g for 45 min, MsbA was purified from the supernatant using an immobilized metal ion affinity chromatography (IMAC) column followed by SEC using a Superdex 200 16/60 Column. Detergent at 3× CMC was used in the wash and elution steps. MsbA-containing eluate fractions were concentrated to 5–10 mg/mL for ATPase activity assays and crystallization trials. The purity of MsbA was analyzed using 4–20% SDS/PAGE gels stained with Coomassie.
The ATPase activity of MsbA was measured at 37 °C using the ATP-regenerating system described by Vogel and Steinhart (49) as modified by Urbatsch et al. (50). Briefly, 0.5–1 μg MsbA were added to 100 μL 50 mM Tris⋅HCl (pH 7.5) buffer containing 10 mM ATP, 12 mM MgCl2, 6 mM phosphoenolpyruvate, 1 mM NADH, 10 units lactate dehydrogenase, and 10 units pyruvate kinase. ATP hydrolysis was determined by the decrease in NADH absorbance at 340 nm using a Beckman Coulter DXT880 Multiplate Spectrophotometer. ATPase activity was calculated using the following equation: ΔOD/(ε × [protein] × time), where ΔOD is the change in absorbance and ε is the extinction coefficient of NADH. The concentration of purified MsbA was estimated by comparing the SDS/PAGE intensity of Coomassie-stained protein bands with known amounts of BSA.
MsbA crystals were grown using sitting-drop vapor diffusion methods by mixing equal volumes of protein with a well solution containing 0.1 M Tris/0.2 M trisodium citrate buffer (pH 5.7), 120 mM LiSO4, and 27.5% (vol/vol) PEG300. X-ray diffraction data were collected at the Stanford Synchrotron Radiation Lightsource beamline 11–1 (43). Data were integrated with iMOSFLM (44) and scaled with SCALA (45).
Cx26 Preparation, Stabilization, and Crystallization.
Cx26 was prepared as described previously with modifications (14). Briefly, Sf9 insect cell membranes containing Cx26 were solubilized by the addition of 1% (wt/vol) detergent (DDM, FA-3, FA-4, or FA-5) in a buffer containing 20 mM Tris (pH 8.0) and 1 M NaCl. After centrifugation at 100,000 × g for 1 h, the supernatant was applied to an IMAC Column. Detergents were used at 3–10× CMC in the wash and elution steps. Isothermal stability of Cx26 was measured as described previously (14). Cx26 in each FA was diluted in buffer to 5× CMC in the presence of 5 μM cpm. This solution was maintained at 37 °C while the fluorescence (405 nm/485 nm) was monitored over time using a Beckman Coulter DXT880 Multiplate Spectrofluorimeter. EM of Cx26 was performed after negative staining with uranyl acetate as described (14).
For crystallization experiments, Cx26 was first solubilized in decyl-β-D-maltoside. IMAC was used to exchange the detergent into each FA as monitored by thin layer chromatography (TLC). Crystals of Cx26 in FA-3 were grown at RT by hanging-drop vapor diffusion by mixing 1 µL protein in 0.02% FA-3 with 1 µL reservoir solution composed of 100 mM Hepes (pH 7), 15 mM sodium formate, 20 mM CaCl2, and 12% (wt/vol) PEG 3350. Cryoprotectant (either glycerol or ethylene glycol) in Harvesting Buffer [100 mM Hepes (pH 7), 20 mM sodium formate, 25 mM CaCl2, 14% (wt/vol) PEG3350] was added directly to the crystallization drop at 5-min intervals to increase the concentration of glycerol or ethylene glycol in the drop by 5% increments. Crystals were then harvested directly from the drop into loops, flash-frozen by plunging into liquid N2, and loaded into ALS-style pucks for synchrotron X-ray data collection. Diffraction data were collected at the Southeastern Regional Collaborative Access Team beam line 22-ID at the Advanced Photon Source at Argonne National Laboratory. Initial indexing of the crystals by HKL2000 (51) suggested a rhombohedral Bravais lattice (R3), a maximal diffraction resolution of 3.0 Å, and a mosaicity of 0.5°. Using a 50-μm beam and 1° oscillations, several wedges of data were collected from one crystal. Ultimately, three datasets were integrated separately and merged after scaling (in the R32 space group), with an overall Rmerge of 7.1% and a resolution cutoff at 3.3 Å. Crystallographic statistics are listed in Table S1.
Supplementary Material
Acknowledgments
We thank A. J. Annalora, S. Gay, and M. B. Shah for crystallization of the cytochrome P450s, G. Chang for providing the MsbA construct and crystallizing NorM, S. Wang for assistance in ESI-MS experiments, and V. Cherezov and M. G. Finn for helpful discussions. Crystal diffraction data were collected at the Stanford Synchrotron Radiation Lightsource (SSRL). The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research; the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program; and the National Institute of General Medical Sciences. The Cx26 X-ray diffraction datasets were collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID (or 22-BM) beamline at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. W-31-109-Eng-38. This work was supported by National Institutes of Health Grants R01 ES003619 (to J.R.H.), P50 GM073197 (to R.C.S. and Q.Z.), R01 HL048908 (to M.J.Y.), and R01 GM098538 (to Q.Z.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4HYX and 4HWL).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1221442110/-/DCSupplemental.
References
- 1.Sonoda Y, et al. Benchmarking membrane protein detergent stability for improving throughput of high-resolution X-ray structures. Structure. 2011;19(1):17–25. doi: 10.1016/j.str.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tate CG. Practical considerations of membrane protein instability during purification and crystallisation. Methods Mol Biol. 2010;601:187–203. doi: 10.1007/978-1-60761-344-2_12. [DOI] [PubMed] [Google Scholar]
- 3.Bill RM, et al. Overcoming barriers to membrane protein structure determination. Nat Biotechnol. 2011;29(4):335–340. doi: 10.1038/nbt.1833. [DOI] [PubMed] [Google Scholar]
- 4.Johansson LC, Wöhri AB, Katona G, Engström S, Neutze R. Membrane protein crystallization from lipidic phases. Curr Opin Struct Biol. 2009;19(4):372–378. doi: 10.1016/j.sbi.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 5.Rosenbaum DM, et al. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318(5854):1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 6.Chun E, et al. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure. 2012;20(6):967–976. doi: 10.1016/j.str.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bayburt TH, Grinkova YV, Sligar SG. Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Lett. 2002;2(8):853–856. [Google Scholar]
- 8.Tribet C, Audebert R, Popot JL. Amphipols: Polymers that keep membrane proteins soluble in aqueous solutions. Proc Natl Acad Sci USA. 1996;93(26):15047–15050. doi: 10.1073/pnas.93.26.15047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiley P, et al. Self-assembling peptide detergents stabilize isolated photosystem I on a dry surface for an extended time. PLoS Biol. 2005;3(7):e230. doi: 10.1371/journal.pbio.0030230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGregor CL, et al. Lipopeptide detergents designed for the structural study of membrane proteins. Nat Biotechnol. 2003;21(2):171–176. doi: 10.1038/nbt776. [DOI] [PubMed] [Google Scholar]
- 11.Chae PS, et al. Maltose-neopentyl glycol (MNG) amphiphiles for solubilization, stabilization and crystallization of membrane proteins. Nat Methods. 2010;7(12):1003–1008. doi: 10.1038/nmeth.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Q, Tao H, Hong WX. New amphiphiles for membrane protein structural biology. Methods. 2011;55(4):318–323. doi: 10.1016/j.ymeth.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Theisen MJ, Potocky TB, McQuade DT, Gellman SH, Chiu ML. Crystallization of bacteriorhodopsin solubilized by a tripod amphiphile. Biochim Biophys Acta. 2005;1751(2):213–216. doi: 10.1016/j.bbapap.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 14.Hong WX, et al. Design, synthesis, and properties of branch-chained maltoside detergents for stabilization and crystallization of integral membrane proteins: Human connexin 26. Langmuir. 2010;26(11):8690–8696. doi: 10.1021/la904893d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matar-Merheb R, et al. Structuring detergents for extracting and stabilizing functional membrane proteins. PLoS One. 2011;6(3):e18036. doi: 10.1371/journal.pone.0018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Q, et al. Designing facial amphiphiles for the stabilization of integral membrane proteins. Angew Chem Int Ed Engl. 2007;46(37):7023–7025. doi: 10.1002/anie.200701556. [DOI] [PubMed] [Google Scholar]
- 17.Newstead S, Ferrandon S, Iwata S. Rationalizing alpha-helical membrane protein crystallization. Protein Sci. 2008;17(3):466–472. doi: 10.1110/ps.073263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrera NP, Di Bartolo N, Booth PJ, Robinson CV. Micelles protect membrane complexes from solution to vacuum. Science. 2008;321(5886):243–246. doi: 10.1126/science.1159292. [DOI] [PubMed] [Google Scholar]
- 19.Davidson AL, Chen J. ATP-binding cassette transporters in bacteria. Annu Rev Biochem. 2004;73:241–268. doi: 10.1146/annurev.biochem.73.011303.073626. [DOI] [PubMed] [Google Scholar]
- 20.Ward A, Reyes CL, Yu J, Roth CB, Chang G. Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc Natl Acad Sci USA. 2007;104(48):19005–19010. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexandrov AI, Mileni M, Chien EY, Hanson MA, Stevens RC. Microscale fluorescent thermal stability assay for membrane proteins. Structure. 2008;16(3):351–359. doi: 10.1016/j.str.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 22.Faham S, Ujwal R, Abramson J, Bowie JU. Practical Aspects of Membrane Proteins Crystallization in Bicelles. 2009. Chapter 5, ed Larry D, Current Topics in Membranes (Academic, London), Vol 63, pp 109–125. [Google Scholar]
- 23.Landau EM, Rosenbusch JP. Lipidic cubic phases: A novel concept for the crystallization of membrane proteins. Proc Natl Acad Sci USA. 1996;93(25):14532–14535. doi: 10.1073/pnas.93.25.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pebay-Peyroula E, Rummel G, Rosenbusch JP, Landau EM. X-ray structure of bacteriorhodopsin at 2.5 angstroms from microcrystals grown in lipidic cubic phases. Science. 1997;277(5332):1676–1681. doi: 10.1126/science.277.5332.1676. [DOI] [PubMed] [Google Scholar]
- 25.Faham S, Bowie JU. Bicelle crystallization: A new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure. J Mol Biol. 2002;316(1):1–6. doi: 10.1006/jmbi.2001.5295. [DOI] [PubMed] [Google Scholar]
- 26.Faham S, et al. Crystallization of bacteriorhodopsin from bicelle formulations at room temperature. Protein Sci. 2005;14(3):836–840. doi: 10.1110/ps.041167605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Annalora AJ, et al. Crystal structure of CYP24A1, a mitochondrial cytochrome P450 involved in vitamin D metabolism. J Mol Biol. 2010;396(2):441–451. doi: 10.1016/j.jmb.2009.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gay SC, et al. Structures of cytochrome P450 2B4 complexed with the antiplatelet drugs ticlopidine and clopidogrel. Biochemistry. 2010;49(40):8709–8720. doi: 10.1021/bi100914z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilderman PR, et al. Plasticity of cytochrome P450 2B4 as investigated by hydrogen-deuterium exchange mass spectrometry and X-ray crystallography. J Biol Chem. 2010;285(49):38602–38611. doi: 10.1074/jbc.M110.180646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gay SC, et al. Structural analysis of mammalian cytochrome P450 2B4 covalently bound to the mechanism-based inactivator tert-butylphenylacetylene: Insight into partial enzymatic activity. Biochemistry. 2011;50(22):4903–4911. doi: 10.1021/bi200482g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilderman PR, et al. Investigation by site-directed mutagenesis of the role of cytochrome P450 2B4 non-active-site residues in protein-ligand interactions based on crystal structures of the ligand-bound enzyme. FEBS J. 2012;279(9):1607–1620. doi: 10.1111/j.1742-4658.2011.08411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shah MB, et al. Conformational adaptation of human cytochrome P450 2B6 and rabbit cytochrome P450 2B4 revealed upon binding multiple amlodipine molecules. Biochemistry. 2012;51(37):7225–7238. doi: 10.1021/bi300894z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gay SC, et al. Crystal structure of a cytochrome P450 2B6 genetic variant in complex with the inhibitor 4-(4-chlorophenyl)imidazole at 2.0-A resolution. Mol Pharmacol. 2010;77(4):529–538. doi: 10.1124/mol.109.062570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shah MB, Pascual J, Zhang Q, Stout CD, Halpert JR. Structures of cytochrome P450 2B6 bound to 4-benzylpyridine and 4-(4-nitrobenzyl)pyridine: Insight into inhibitor binding and rearrangement of active site side chains. Mol Pharmacol. 2011;80(6):1047–1055. doi: 10.1124/mol.111.074427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chae PS, et al. Tandem facial amphiphiles for membrane protein stabilization. J Am Chem Soc. 2010;132(47):16750–16752. doi: 10.1021/ja1072959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cherezov V, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318(5854):1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruprecht JJ, Mielke T, Vogel R, Villa C, Schertler GF. Electron crystallography reveals the structure of metarhodopsin I. EMBO J. 2004;23(18):3609–3620. doi: 10.1038/sj.emboj.7600374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He X, et al. Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature. 2010;467(7318):991–994. doi: 10.1038/nature09408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Vendittis E, Palumbo G, Parlato G, Bocchini V. A fluorimetric method for the estimation of the critical micelle concentration of surfactants. Anal Biochem. 1981;115(2):278–286. doi: 10.1016/0003-2697(81)90006-3. [DOI] [PubMed] [Google Scholar]
- 40.Hernández H, Robinson CV. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat Protoc. 2007;2(3):715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 41.Bush MF, et al. Collision cross sections of proteins and their complexes: A calibration framework and database for gas-phase structural biology. Anal Chem. 2010;82(22):9557–9565. doi: 10.1021/ac1022953. [DOI] [PubMed] [Google Scholar]
- 42.Dencher NA, Heyn MP. Preparation and properties of monomeric bacteriorhodopsin. Methods Enzymol. 1982;88:5–10. [Google Scholar]
- 43.Soltis SM, et al. New paradigm for macromolecular crystallography experiments at SSRL: Automated crystal screening and remote data collection. Acta Crystallogr D Biol Crystallogr. 2008;64(Pt 12):1210–1221. doi: 10.1107/S0907444908030564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leslie AG. Integration of macromolecular diffraction data. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 10):1696–1702. doi: 10.1107/s090744499900846x. [DOI] [PubMed] [Google Scholar]
- 45.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63(Pt 1):32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McRee DE. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125(2–3):156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 49.Vogel G, Steinhart R. ATPase of Escherichia coli: Purification, dissociation, and reconstitution of the active complex from the isolated subunits. Biochemistry. 1976;15(1):208–216. doi: 10.1021/bi00646a032. [DOI] [PubMed] [Google Scholar]
- 50.Urbatsch IL, Sankaran B, Weber J, Senior AE. P-glycoprotein is stably inhibited by vanadate-induced trapping of nucleotide at a single catalytic site. J Biol Chem. 1995;270(33):19383–19390. doi: 10.1074/jbc.270.33.19383. [DOI] [PubMed] [Google Scholar]
- 51.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.