Abstract
Quinone molecules are intracellular electron-transport carriers, as well as critical intra- and extracellular signals. However, transcriptional regulation of quinone signaling and its molecular basis are poorly understood. Here, we identify a thiol-stress-sensing regulator YodB family transcriptional regulator as a central component of quinone stress response of Staphylococcus aureus, which we have termed the quinone-sensing and response repressor (QsrR). We also identify and confirm an unprecedented quinone-sensing mechanism based on the S-quinonization of the essential residue Cys-5. Structural characterizations of the QsrR–DNA and QsrR–menadione complexes further reveal that the covalent association of menadione directly leads to the release of QsrR from operator DNA following a 10° rigid-body rotation as well as a 9-Å elongation between the dimeric subunits. The molecular level characterization of this quinone-sensing transcriptional regulator provides critical insights into quinone-mediated gene regulation in human pathogens.
Keywords: thiol alkylation, macrophage
Small molecule-mediated gene regulation is an essential component in cellular processes and cell-to-cell communication (1). For instance, quorum signals, such as homoserine lactones in Gram-negative bacteria and autoinducing peptides in Gram-positive bacteria, are adopted by quorum-sensing and regulatory systems to modulate gene expression (2–4).
As the critical intra- as well as extracellular signals, quinone molecules play important roles in cellular functions (5). Ubiquinone and menaquinone are considered essential for lipid electron transport (6, 7). More recently, vitamin K2, a derivative of menadione, was discovered to function as an important electron carrier in Drosophila mitochondria to help maintain normal ATP production (8). A disordered quinone pool would severely affect the respiratory systems of Drosophila, leading to subsequent changes in cellular metabolism (9–11). Meanwhile, quinone molecules are also considered a source of exogenous stress: a variety of toxins and antimicrobial compounds contain quinone, such as cercosporin and anthracycline (12–14). The toxicity of quinone molecules has been ascribed to their potential to serve as electrophiles and/or oxidants (14–16).
Although the biological functions of quinone molecules have been well explored, the role of quinone molecules in cellular signaling is less understood. In Escherichia coli, the anoxic redox control ArcAB is considered a quinone-sensing two-component system (17, 18). In Bacillus subtilis, the thiol-stress sensing regulator YodB is a prototypical transcriptional regulator responsible for quinone sensing and detoxification (19–21).
Redox signaling in bacteria has attracted extensive attention as a result of the importance in bacterial physiology and in defense against environmental and host stresses. The determination of structures of the reduced and oxidized bacterial peroxide-sensing regulators OxyR and OhrR has made possible proposals of intramolecular and intermolecular disulfide-mediated gene-regulation mechanisms (22–24). More recently, the crystal structures of NADH/NAD+ redox sensor Rex bound to NAD+ and DNA operator have revealed the NADH/NAD+-sensing mechanism of Rex family members (25). However, little is known about the exact molecular mechanisms responsible for quinone-mediated signaling pathways in pathogenic bacteria.
Staphylococcus aureus causes a variety of diseases from soft-tissue infections to life-threatening diseases including toxic shock syndrome, endocarditis, and necrotizing pneumonia (26, 27). The success of the bacterium in pathogenesis is mainly attributed to its sophisticated signaling and response systems, including global transcriptional regulators, and various two-component systems, which the bacterium uses to sense a variety of signals and environmental stimuli (28–34). Despite the recognized importance of quinone in cellular processes, its role in bacterial gene regulation and signal transductions in human pathogens remains elusive.
In this work, we present our discovery that the S. aureus transcriptional regulator quinone-sensing and response repressor (QsrR) mediates stress sensing and response to quinone through the use of its nucleophilic Cys-5 as a quinone sensor. We demonstrate that QsrR specifically recognizes a palindromic sequence, locating its target promoter regions in the absence of quinone stress. The presence of quinone molecules covalently modifies the N-terminal Cys-5 residue to abolish the protein–DNA interaction, thus leading to derepression of a set of quinone-detoxification genes. High-resolution crystal structures of both QsrR–DNA and QsrR–menadione complexes further allow us to picture the molecular basis for the signaling and regulatory mechanisms of this family of transcriptional regulators.
Results
QsrR Is Involved in Quinone Sensing and Response.
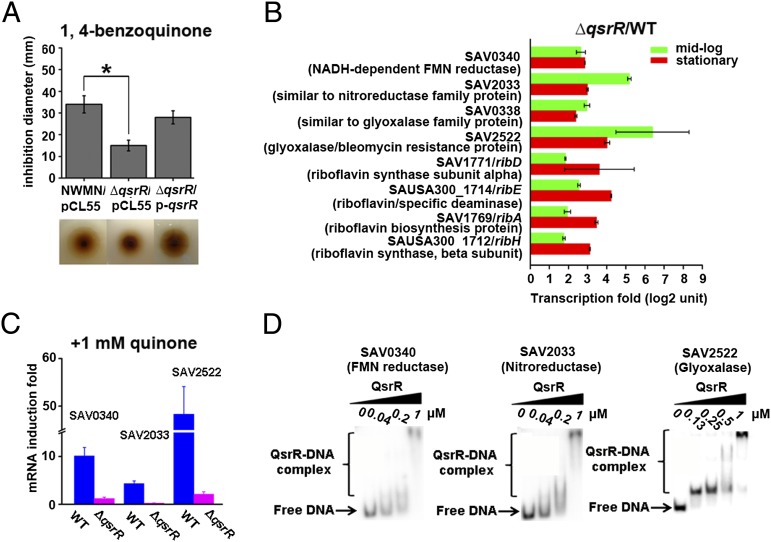
Quinone molecules represent a category of reactive oxygen species, which are often used by the human immune system to counter invading pathogens. Human pathogens such as S. aureus, on the other hand, must develop a certain counteractive defense mechanism to tackle quinone molecules. To study quinone signaling, we focused on S. aureus QsrR (SAV2123), a homolog of B. subtilis quinone stress-sensing regulator YodB (38% identities, 61% positives) (19, 21). We first performed a disk-diffusion assay to examine the role of QsrR in response to quinone stress. The qsrR in-frame deletion mutant strain (∆qsrR) exhibited high resistance to 2 M 1,4-benzoquinone (the major moiety of most cellular quinone molecules) with a growth inhibition area of 16 mm (Fig. 1A), whereas the Newman wild-type strain was more sensitive to 1,4-benzoquinone, with a growth inhibition area of 33 mm under the same conditions (Fig. 1A), indicating the role of QsrR in quinone detoxification. Similar results were obtained by using another quinone compound, methyl-p-benzoquinone (Fig. S1A). In both cases, introducing a single-copy plasmid p-qsrR containing an intact qsrR gene could restore the decreased quinone susceptibility of the ∆qsrR mutant. The decreased quinone susceptibility of the ∆qsrR mutant is also observed in USA300 and MW2 strains (Fig. S1B).
Fig. 1.
QsrR is involved in quinone signaling and response. (A) Disk-diffusion assays showing that QsrR impacts bacterial resistance toward 1,4-benzoquinone. The wild-type strain (NWMN/pCL55), ∆qsrR mutant (∆qsrR/pCL55), and complementation strain (∆qsrR/p-qsrR) were assayed with 2 M 1,4-benzoquinone and incubated overnight before data collection. The experiments were performed in triplicate with consistent results. *P < 0.01 (Student t test). (B) Selected genes differentially expressed in the ∆qsrR mutant compared with the wild-type strain from transcriptomic profiling. Transcriptomic profiling was performed at both of the midlog and stationary growth stages. The experiments were performed in duplicate with consistent results. (C) Quinone stress impacts the expression of QsrR target genes in a QsrR-dependent manner. The mRNA levels of SAV0340 (FMN reductase), SAV2033 (nitroreductase), and SAV2522 (glyoxalase) were monitored before and after treatment of 1 mM 1,4-benzoquinone at midlog growth stage by qRT-PCR. The experiments were performed in triplicate with consistent results. (D) QsrR directly binds to the promoter regions of SAV0340, SAV2033, and SAV2522. EMSAs were performed at a serial concentration of QsrR ranging from 0 to 1 μM with the 32P-labeled DNA.
To test the role of QsrR in gene regulation, we performed transcriptomic analysis on both the wild-type strain and ∆qsrR mutant. Fig. 1B lists genes that are up-regulated in the ∆qsrR mutant. The list consists mainly of putative antioxidant/quinone detoxification genes including: SAV0340, which encodes a NADH-dependent FMN reductase; SAV2033, which encodes a nitroreductase family protein; SAV0338, which encodes a glyoxalase family protein; SAV2522, which encodes a glyoxalase/bleomycin resistance protein, as well as riboflavin biosynthesis genes (SAV1771, SAUSA300_1714, SAV1769, and SAUSA300_1712). The microarray result was confirmed by quantitative real-time PCR (qRT-PCR) analysis of the selected genes (Table S1). In addition, the expression levels of putative quinone detoxification genes (SAV0340, SAV2033 and SAV2522) were highly induced in the wild-type strain in the presence of 1,4-benzoquinone, whereas the expression levels of these genes were unchanged (SAV0340 and SAV2522) or even slightly decreased (SAV2033) in ∆qsrR under the same conditions (Fig. 1C), further supporting the role of QsrR in quinone stress sensing and response.
Electrophoretic mobility-shift assays (EMSAs) showed that the QsrR protein is able to bind to the promoter regions of its target genes including SAV0340, SAV2033, and SAV2522. This result is highly indicative of a direct regulatory role of QsrR in controlling the expression of these genes (Fig. 1D). Footprinting assays further revealed a putative palindromic (inverted-repeat) binding box of “GTATAN{5}TATAC” located in the promoter region of SAV2033 (Fig. S1 C and D). This palindromic sequence is essential for the binding of QsrR because EMSAs showed that deletion of the binding box fully abolished the QsrR–DNA interaction (Fig. S1E). A genome-wide search of the invert repeats in S. aureus intergenic regions revealed a list of 13 genes that contain the putative QsrR binding box, including its own promoter region (qsrR, SAV2123) (Table S2 and Fig. S2A). EMSAs combined with β-galactosidase assays on the promoter of qsrR further confirmed the autoregulation of this gene (Fig. S2 B and C). As expected, genes identified from the whole-genome search based on the consensus binding sequence of QsrR are in accordance with the microarray result, thus supporting the notion that QsrR regulates the expression of these genes through a direct-binding mechanism.
Cys-5 Plays an Essential Role in Quinone Signaling.
Quinone molecules act as electrophiles as well as oxidants. As electrophiles, quinone molecules can react with cellular thiols to form Michael-addition products. As an oxidant, a quinone molecule can produce reactive oxygen species (ROS) by reducing itself. Because the one-electron reduction potential of 1,4-benzoquinone is far beyond the physiological relevant range, 1,4-benzoquinone is commonly regarded as an electrophile (35), which may form Michael-adducts with cysteine residues in QsrR as a way to tune the function of QsrR.
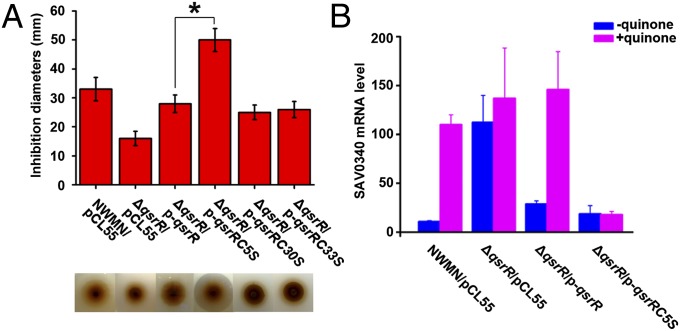
QsrR contains three cysteine residues (Cys-5, Cys-30, and Cys-33). To determine which Cys residue is essential for quinone sensing inside S. aureus, we mutated the residues to serine and introduced the mutated genes back to ΔqsrR, separately. We then performed a disk diffusion assay with 2 M 1,4-benzoquinone on the following six strains: the wild-type Newman (NWMN/pCL55), ΔqsrR mutant (ΔqsrR/pCL55), ΔqsrR mutant complemented with wild-type qsrR (ΔqsrR/p-qsrR), ΔqsrR mutant complemented with qsrRC5S (ΔqsrR/p-qsrRC5S), ΔqsrR mutant complemented with qsrRC30S (ΔqsrR/p-qsrRC30S), and ΔqsrR mutant complemented with qsrRC33S (ΔqsrR/p-qsrRC33S). As a result, the complementation strain carrying qsrRC5S showed an extremely high sensitivity with 50-mm inhibition area, whereas the other two complementation strains, ΔqsrR/p-qsrRC30S and ΔqsrR/p-qsrRC33S, did not exhibit any observable difference compared with ΔqsrR/p-qsrR (Fig. 2A). This difference was likely attributed to the loss of the quinone-signaling function of the variant protein QsrRC5S, resulting in the overrepression of QsrR-target quinone-detoxification genes, thus demonstrating the critical role of Cys-5 in quinone sensing and response. The various QsrR variants were also immunoblotted by using flag-tag expression vector pCL55-flag. No significant change of QsrR expression levels was observed, suggesting that various QsrR variants share similar stability and activity (Fig S3A).
Fig. 2.
Cys-5 is essential for quinone sensing and response of QsrR. (A) Disk-diffusion assays showing that Cys-5, but not Cys-30 or Cys-33, is critical to bacterial response and susceptibility toward 1,4-benzoquinone. Strains including wild-type Newman (NWMN/pCL55), ∆qsrR mutant (∆qsrR/pCL55), ∆qsrR mutant complemented with pCL55-qsrR (∆qsrR/p-qsrR), ∆qsrR mutant complemented with pCL55-qsrRC5S (∆qsrR/p-qsrRC5S), ∆qsrR mutant complemented with pCL55-qsrRC30S (∆qsrR/p-qsrRC30S), and ∆qsrR mutant complemented with pCL55-qsrRC33S (∆qsrR/p-qsrRC33S) were assayed under the same condition with 2 M 1,4-benzoquinone and incubated overnight before data collection. The experiments were performed in triplicate with consistent results. *P < 0.05 (Student t test). (B) Transcriptional analysis showing that Cys-5 in QsrR is essential for its response to quinone. Cells were grown to midlog phase before treatment of 1 mM 1,4-benzoquinone. The mRNA levels of SAV0340 were analyzed by qRT-PCR and 16S rRNA was used for normalization. The experiments were performed in triplicate with consistent results.
The importance of Cys-5 is further supported by transcriptional analysis of QsrR target genes (SAV0340 and SAV2522) in the four aforementioned strains NWMN/pCL55, ΔqsrR/pCL55, ΔqsrR/p-qsrR, and ΔqsrR/p-qsrRC5S. Bacteria were grown to midlog phase (OD600 = 0.6) and treated with or without 1 mM 1,4-benzoquinone for 10 min before RNA isolation. In the absence of quinone, the transcript levels of both genes were low in NWMN/pCL55, ΔqsrR/p-qsrR, and ΔqsrR/p-qsrRC5S compared with levels in ΔqsrR/pCL55 (Fig. 2B and Fig. S3B). Upon quinone treatment, a 10-fold increase of SAV0340 and a 50-fold increase of SAV2522 in gene expression were observed in the wild-type strain (Fig. 2B and Fig. S3B). In the ∆qsrR mutant, however, the addition of quinone either had no effect (SAV0340) or even slightly down-regulated the gene expression (SAV2522) (Fig. 2B and Fig. S3B). More importantly, complementation of ΔqsrR with qsrRC5S rendered the bacterium unresponsive to quinone stress and resulted in a constant repression of the expression of these two genes (Fig. 2B and Fig. S3B). This result further supports the critical role of Cys-5 in quinone signaling in QsrR.
EMSAs further substantiated the key role of Cys-5 in quinone signaling. As shown in Fig. S3C, 20 µM 1,4-benzoquinone fully abolished the DNA-binding ability of QsrR but hardly affected the DNA binding of QsrRC5S. Moreover, mass spectrum analysis on the QsbRC30S/C33S double-mutant protein (both Cys-30 and Cys-33 were mutated to Ser because they were not involved in quinone response based on the disk diffusion assay; Fig. 2A), and the QsbRC5S/C30S/C33S triple-mutant protein demonstrated the critical role of Cys-5 in quinone alkylation. A clear mass shift was observed around 112 Da (Fig. S3D) for the double-mutant protein QsbRC30S/C33S after incubation with 1 mM 1,4-benzoquinone (molecular weight 108), whereas the mass value of the triple mutant protein QsbRC5S/C30S/C33S remained unchanged (Fig. S3E) with the same treatment.
Structural Characterization of the QsrR–DNA Complex.
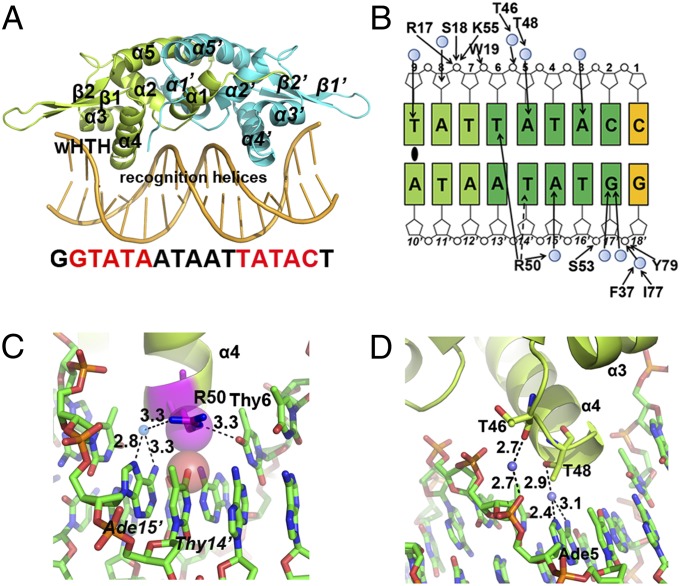
The high degree of sequence conservation (>30% sequence identities) among YodB/QsrR family members suggests a shared mode of protein–DNA interaction as well as quinone-mediated gene regulation (Fig. S4). However, the lack of structural information for YodB/QsrR family proteins prevents a deeper understanding of the detailed molecular mechanism. Therefore, we first determined the crystal structure of the QsrR–DNA complex (Fig. 3A), which was solved by maximum likelihood molecular replacement methods and refined to 2.30-Å resolution (Table S3). We used a 17-bp duplex DNA containing a pair of inverted repeats (bold) (5′-AGTATAATTATTATACC-3′) to perform cocrystallization. A dimer of QsrR bound to a double-stranded DNA is found in the asymmetric unit. The duplex DNA shares a canonical B-DNA shape as a C2′ endo sugar pucker, a 36° rotation per base pair, and a 3.3-Å rise per base pair along the axis are observed. Each subunit of the DNA-bound QsrR provides the classic winged helix–turn–helix (wHTH) domain (α3, α4, β1, and β2 in Fig. 3A) for the DNA binding, with the N-terminal two helices (α1 and α2 in Fig. 3A) and the C-terminal one helix (α5 in Fig. 3A) at the vertex for dimerization.
Fig. 3.
Structure of the QsrR–DNA complex depicting interactions between QsrR and the palindromic sequence. (A) Crystal structure of the QsrR–DNA complex. One subunit of the QsrR dimer is colored lemon and the other half is colored cyan. The secondary structure elements and DNA recognition helices are indicated. (B) Schematic diagram of contacts between one QsrR subunit and the half palindromic DNA. The operator DNA is a palindromic sequence except the side C:G pair (yellow), which is replaced by an A:T pair at the other half. The palindromic center is marked by an oval. Filled arrowheads represent hydrogen bonds and the dash arrowhead represents a van der Waals interaction. Filled blue circles represent water-mediated contacts. The inverted repeats are colored dark green and the central pairs are colored light green. (C and D) Close view of the interactions between the HTH motif of QsrR dimer and the operator DNA. Water molecules are blue spheres. van der Waals interactions are viewed as semitransparent spheres. Distances of putative hydrogen bonds are labeled in angstrom units.
The QsrR dimer presents a classic mode of DNA recognition with the wHTH domains; the recognition α helix contributes mainly to DNA binding. In total, 22 residues of the QsrR dimer make 26 DNA contacts, in which each subunit contributes a nearly identical interaction with the palindromic operator DNA (Fig. 3B).
The recognition α-helices (α4/α4′) of the HTH motif (α3, the turn, and α4) interact with two consecutive major grooves of the operator DNA where the highly conserved R50/R50′ residues (except that in B. subtilis, in which R is replaced by K; Fig. S4) at the N termini of the helices to make base-specific contacts: one hydrogen bond between Nε of R50 and O4 of thymine6, one water-mediated hydrogen bond between terminal nitrogen of R50 and N6 and N7 of adenine15′, as well as one van der Waals interaction between Cδ of R50 and thymine14′ (Fig. 3C). The turn region of the HTH motif provides two additional water-mediated hydrogen bonds via Oβ of T48 with N6 and N7 of adenine, as well as the carbonyl oxygen of T46 with the phosphate of thymine6 (Fig. 3D). Moreover, F37 at the N-terminal residue of α3 helix is involved in another water-mediated hydrogen bond through its backbone nitrogen with the phosphate of guanine18′ (Fig. S5A).
The second DNA-binding element, the wing, is composed of two β-sheets (β2 and β3), as well as their connecting loop, and contributes two additional contacts with the backbone phosphate of the operator DNA. The N-terminal residue of β2-sheet, I77, interacts with the backbone phosphate of guanine18′ through a water-mediated hydrogen bond with its carbonyl oxygen (Fig. S5A). Additionally, the highly conserved residue in YodB/QsrR family members, Y79 (Fig. S4), provides a direct hydrogen bond with the backbone phosphate of guanine18′ via its side-chain oxygen (Fig. S5A).
In addition to the wHTH motif, the loop region of the helix–helix (HH) motif (helices α1, α2, and the connecting residues) contributes a third element for DNA binding. The backbone nitrogen of S18 and N5 of R17 directly interacts with the phosphate of adenine8 through hydrogen bonding (Fig. S5B). Similar interactions are observed in OhrR-type regulators, in which the HH motif crosses the pseudodyad axis to interact with the DNA (23).
Structural Characterization of the Menadione-Bound QsrR.
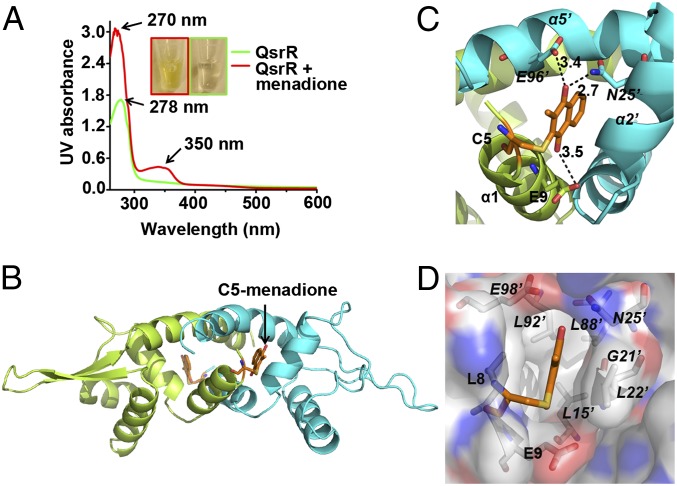
To crystallize quinone-bound QsrR, we tried three different kinds of quinone molecules: 1,4-benzoquinone, methyl-p-benzoquinone, and menadione. The menadione-bound QsrR complex yielded diffraction data of sufficient quality to determine the structure. The gel-filtration purified QsrR–menadione sample was analyzed by UV spectrum to confirm the formation of the QsrR–menadione complex, which revealed two characteristic peaks at 270 and 350 nm, which correspond to the absorbance of menadione (36). A burying peak at 280 nm corresponds to the absorbance of the protein itself. No peaks at 270 and 350 nm were observed in the QsrR protein sample without quinone treatment (Fig. 4A). The resulting QsrR–menadione complex sample was directly used for crystallization, which yielded stick-like crystals at pH 8.5. Maximum-likelihood molecular replacement methods were used to solve the structure of the menadione-bound QsrR and the structure was refined to 2.55-Å resolution (Table S3). The asymmetric unit of the crystal contains a QsrR dimer (Fig. 4B) covalently attached with a well-defined menadione molecule at the Cys-5 site of each monomeric subunit. The omit electron density for menadione molecule is consistent with the planar aromatic ring of naphthalene (Fig. S6A).
Fig. 4.
Structure of the QsrR–menadione complex. (A) QsrR covalently attached with menadione displays a UV absorbance distinct from apo-QsrR. Purified QsrR (80 μM) was incubated with 200 μM menadione, followed by gel filtration to remove unbounded menadione. (B) Crystal structure of the QsrR–menadione complex. One subunit of the QsrR dimer is colored lemon and the other subunit is colored cyan. The C5-menadione is colored orange. (C) Hydrogen bonds between menadione and QsrR. Distances of putative hydrogen bonds are labeled in angstrom units. (D) The menadione-binding pocket of QsrR. The pocket is viewed as a semitransparent surface. The hydrophobic surface is colored white. The positively charged surface is colored dark blue. The negatively charged surface is colored red.
The high reactivity of this Cys-5 residue toward menadione is likely attributed to the positive dipole of the first α-helix where Cys-5 is located. This positioning could effectively lower the pKa of the sulfhydryl group (-SH), which resides at the N-terminal of the helix with increased nucleophilicity. This type of positive macrodipole-mediated deprotonation of cysteine residues is also observed in the OhrR family proteins, in which the cysteine residues function as sensors for oxidative stresses via cysteine oxidation or the formation of disulfide bonds with low molecular weight thiols (23, 29).
Besides the covalent interaction, five residues of QsrR dimer directly contact the naphthalene ring, oxygen, and the methyl group of menadione through six noncovalent interactions (Fig. S6B; calculated by LigPlot+) (37). These interactions include a strong hydrogen bond between the side-chain nitrogen of N25′ (α2′ of subunit B′) and the O1 of menadione plus two weak hydrogen bonds including the interaction of the side-chain oxygen of E96′ (α5′ of subunit B′) with the O1 of menadione and the side-chain oxygen of E9 (α1 of subunit A) with the O2 of menadione (Fig. 4C). In addition, the two residues L22′ and G21′ at α2′ of subunit B′ provide three van der Waals interactions with the carbons of the naphthalene ring (Fig. S6C). Taken together, these interactions are likely to fix the orientation of menadione in the pocket surrounded by three helices that include α1 of subunit A, α2′ of subunit B′, and α5′ of subunit B′ at the dimeric interface. Close inspection of the pocket reveals a quite hydrophobic environment, suitable for the association of hydrophobic quinone molecules (Fig. 4D and Fig. S6 D and E).
Menadione Disrupts the QsrR–DNA Interaction by Perturbing QsrR Dimerization.
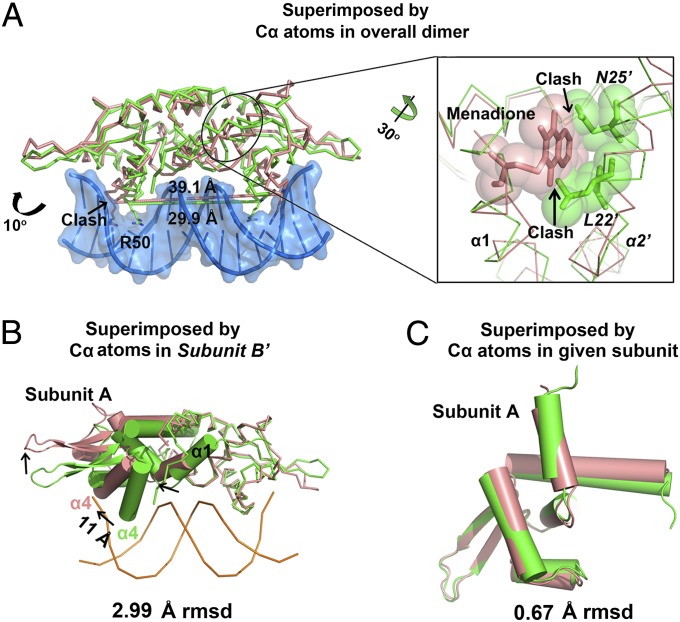
Menadione association causes significant conformation change of the DNA-bound QsrR. The recognition helices (α4 and α4′) in the DNA-bound QsrR are separated by 29.9 Å (between R50/R50′-Cα atoms; green in Fig. 5A) with a 106° rotation that allows the helices to recognize the consecutive major grooves of the duplex DNA. However, a 39.1-Å distance and a 117° rotation (salmon in Fig. 5A) are observed for α4 and α4′ in the menadione-bound QsrR structure, which prevent its binding to the operator DNA. Superimposition of the DNA-bound QsrR and menadione-bound QsrR structures by Cα atoms in subunit B′ reveals a rotation in the other subunit and a significant translation at the recognition helix (α4, 11 Å) as well as the wing parts (β1 and β2) and the menadione-binding helix (α1) (Fig. 5B and Fig. S7A). However, little difference is observed within the monomeric subunits with 0.67-Å rmsd of subunits A (Fig. 5C) and 1.32-Å rmsd of subunits B′ (Fig. S7B) compared with 2.99-Å rmsd between overall dimers. This movement is quite similar to most known repressors that take the DNA-binding domains of a dimeric pair spatially apart (38, 39). The translation of the first helix (α1) at the dimerization interface likely initiates the structural change. This change provides suitable space for menadione association and minimizes the clash between helices α1 and α2′ generated by menadione binding, thus leading to the rearrangement of the helices α1 as well as α2′ (Fig. 5A). The structural change is further transferred to the DNA-binding domain, leading to the dissociation of QsrR dimer from operator DNA.
Fig. 5.
Addition of menadione to QsrR causes a dramatic conformational change. (A) Structural comparison of the QsrR–DNA complex and QsrR–menadione by superimposing Cα atoms in overall dimer. The protein and DNA in QsrR–DNA are colored green and sky blue, respectively. The QsrR–menadione complex is colored salmon. Distances between the two recognition arginines (R50 Cα) are indicated. The clashes between menadione of QsrR–menadione complex and Asn25′ as well as Leu22′ of QsrR–DNA complex are viewed as semitransparent spheres. (B) Structural comparison of QsrR–DNA and QsrR–menadione by superimposing Cα atoms in subunit B′. The arrowheads point in the direction of the structural change after menadione association. The distance is measured between the Cα of R50 at the end of helix α4. The subunit A is viewed as cartoon and the subunit B′ is viewed as ribbon. (C) Overlay of Cα atoms in subunit A reveals the structural similarity of the QsrR–DNA (green) and QsrR–menadione (salmon) subunits.
The qsrR Mutant Favors S. aureus Survival within Macrophages.
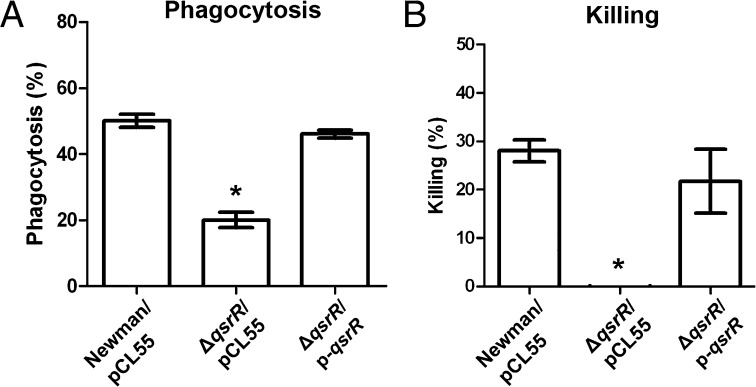
Because a set of quinone/ROS detoxification enzymes are highly expressed in the ΔqsrR mutant, we tested the role of qsrR in the S. aureus survival inside macrophages. The phagocytosis process of the Newman wild-type, qsrR mutant, and the complementation strains inside bone marrow-derived macrophages (BMDMs) was investigated, which showed more than twofold resistance of the qsrR mutant than its parental strain to phagocytosis (Fig. 6A). We also examined the survival of the three aforementioned strains inside macrophages. As shown in Fig. 6B, the qsrR mutant is much more resistant to macrophage killing than its parental strain, indicating the critical role of qsrR for S. aureus in macrophage resistance.
Fig. 6.
The qsrR mutant favors S. aureus survival in BMDMs. (A) The phagocytosis assay of the Newman wild-type, ΔqsrR mutant, and complementation strains within macrophages. Macrophages were infected as described in Experimental Procedures. The experiments were performed in triplicate. *P < 0.01. (B) Killing assay of the Newman wild-type, ΔqsrR mutant, and complementation strains within macrophages. The bacteria were infected for 1 h before being analyzed. The experiments were performed in triplicate. *P < 0.01.
Discussion
Although quinone molecules play important roles in various cellular processes (5), quinone-mediated gene regulation is poorly understood. We present here a quinone-sensing and response system in S. aureus and demonstrate the detailed mechanism of quinone signaling. The increased quinone resistance of the ∆qsrR mutant compared with the wild-type strain can be attributed to the enhanced expression of quinone-detoxification genes. SAV2522 and SAV0338, which encode the glyoxalase family proteins that may mediate the thiol-dependent ring cleavage of quinone-S-adducts (40–42). The homolog of SAV2522 in B. subtilis yfiE indeed possesses metabolic activity of catechol degradation (43). In addition, SAV0340, which encodes a NADH-dependent FMN reductase, could act on the detoxification of quinones (44, 45); its homolog yhdA in B. subtilis functions as a reductase for a variety of quinone molecules (46). Meanwhile, a nitroreductase, encoded by SAV2033, may contribute to a two-electron reduction of nitro groups in nitroaromatic compounds as well as quinones (47, 48).
In addition, the elevated expression of riboflavin biosynthesis genes in the ∆qsrR mutant (Fig. 1B) could also contribute to the detoxification of quinone molecules. Riboflavin is the central component of the cofactors FAD and FMN and is, therefore, required by all flavoenzymes (49). Flavoenzymes function in a wide variety of cellular processes, including the reduction and detoxification of quinones (49–51). We analyzed the total riboflavin content and found a threefold increase in the ∆qsrR mutant compared with the wild-type strain (Fig. S8A). The elevated riboflavin in the ∆qsrR mutant strain would contribute to the biosynthesis of cofactors FAD and FMN, consequently increasing the activity of flavoenzymes for potential detoxification functions.
To facilitate the crystallization of menadione-modified QsrR complex, we mutated both of the non–quinone-signaling cysteine residues, Cys30 and Cys33, to serine, thereby ensuring the homogeneity of S-quinonization. The crystal structure of the wild-type QsrR–DNA complex showed that these two cysteines are located in the loop region between helices α2 and α3. Therefore, the mutation of the cysteines will be less likely to interfere the overall structure of QsrR, although it may influence the local loop conformation. Our structural study proves that menadione, the precursor of menaquinone, is one of the signals for the quinone sensor QsrR. Because S. aureus relies on menaquinone, not ubiquinone, for electron transport, we suspect whether the precursor of ubiquinone, 2,3-dimethoxy-5-methyl-p-benzoquinone, is also a signal for QsrR. The gel-shift assay (Fig. S8B) confirms that it can disrupt QsrR–DNA interaction, indicating that this quinone molecule may also trigger response of QsrR. It is possible that QsrR can sense a variety of quinone molecules, including endogenous as well as exogenous quinone signals.
We show that quinones covalently modify QsrR through S-quinonization of the essential residue Cys-5. Cys-alkylation plays important roles in biology (52, 53); however, Cys-alkylation by quinones in signaling and transcription has not been revealed in the past. Comparison of the structures of QsrR–DNA and QsrR–menadione indicates that menadione association causes the two QsrR monomers to rotate, leading to the dissociation of QsrR from operator DNA. To support this mechanism, we found that the monomer conformation does not change between the QsrR–menadione complex and the QsrR–DNA complex. Therefore, we propose the following quinone-sensing mechanism (Fig. S8C) (1): quinones covalently modify Cys-5 of QsrR as a unique sensing mechanism (2); the two monomers of QsrR have to rotate and translate coordinately to accommodate or respond to quinone association (3); this conformational change is then transferred to the DNA-binding domains, thus causing these domains to rotate and translate apart, which induces dissociation of the protein from operator DNA (4); this dissociation derepresses the quinone-detoxification systems, thereby rebalancing the altered quinone pool. This S-quinonization–based quinone-sensing mechanism could be widely used in other organisms.
Experimental Procedures
Bacterial Strains, Primers, Plasmids, and Growth Conditions.
Bacterial strains and plasmids used in this study are listed in Table S4. Primers used in this study are listed in Table S5. E. coli strains were grown in Luria–Bertani broth (LB). S. aureus strains were grown in tryptic soy broth, except during transduction procedures, for which heart infusion broth supplemented with 5 mM CaCl2 was used. The transposon insertion ΔqsrR mutant was obtained from O. Schneewind and D. Missiakas (The University of Chicago, Chicago, IL) and further identified by phage transduction and PCR test. When necessary, antibiotics were added at the following concentrations: ampicillin, 100 μg/mL; kanamycin, 50 μg/mL; and chloramphenicol, 10 μg/mL. Vector pMCSG19 was used for expressing His-tagged proteins. E. coli–S. aureus shuttle vector pCL55 was used for complementation. E. coli–S. aureus shuttle vector pCL55-lacZ was used for lacZ assay. Cells were grown in 15-mL tubes with the tube-to-medium ratio of 10, 250 rpm of aeration, and 37 °C. OD600 of 0.6 represents midlog phase, whereas overnight culture represents stationary phase with OD600 ≈ 7.
Other Procedures.
Detailed procedures are available in SI Experimental Procedures.
Supplementary Material
Acknowledgments
We thank all beamline staff for data collection support. We thank Drs. O. Schneewind and D. Missiakas at The University of Chicago for providing transposon mutants. We thank Dr. Roman Laskowski at the European Bioinformatics Institute for LigPlot+ software assistance. We thank S. F. Reichard, MA, for editing the manuscript. This work was financially supported by National Institutes of Health (NIH) National Institute of Allergy and Infectious Diseases Grant AI074658 (to C.H.). C.H. is a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease Award. Q.J. is also supported by NIH Grant P50GM081892.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystal structures of QsrR–DNA and QsrR–menadione have been deposited in the Protein Data Bank (PDB), www.pdb.org (PDB ID codes 4HQE and 4HQM, respectively). The microarray data have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE35409).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219446110/-/DCSupplemental.
References
- 1.Camilli A, Bassler BL. Bacterial small-molecule signaling pathways. Science. 2006;311(5764):1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuqua C, Parsek MR, Greenberg EP. Regulation of gene expression by cell-to-cell communication: Acyl-homoserine lactone quorum sensing. Annu Rev Genet. 2001;35:439–468. doi: 10.1146/annurev.genet.35.102401.090913. [DOI] [PubMed] [Google Scholar]
- 3.Kleerebezem M, Quadri LE, Kuipers OP, de Vos WM. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol Microbiol. 1997;24(5):895–904. doi: 10.1046/j.1365-2958.1997.4251782.x. [DOI] [PubMed] [Google Scholar]
- 4.Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 5.Nohl H, Jordan W, Youngman RJ. Quinones in biology - functions in electron-transfer and oxygen activation. Adv Free Radical Biol Med. 1986;2(1):211–279. [Google Scholar]
- 6.Ernster L, Dallner G. Biochemical, physiological and medical aspects of ubiquinone function. Biochim Biophys Acta. 1995;1271(1):195–204. doi: 10.1016/0925-4439(95)00028-3. [DOI] [PubMed] [Google Scholar]
- 7.Suttie JW. Vitamin K-dependent carboxylase. Annu Rev Biochem. 1985;54:459–477. doi: 10.1146/annurev.bi.54.070185.002331. [DOI] [PubMed] [Google Scholar]
- 8.Vos M, et al. Vitamin K2 is a mitochondrial electron carrier that rescues pink1 deficiency. Science. 2012;336(6086):1306–1310. doi: 10.1126/science.1218632. [DOI] [PubMed] [Google Scholar]
- 9.Lider VA, Bogdanova SN. Role of P-quinones in regulation of glucose-6-phosphate metabolism. Bull Exp Biol Med. 1986;102(9):1193–1195. [PubMed] [Google Scholar]
- 10.Lohmann A, et al. Deficiency in phylloquinone (vitamin K1) methylation affects prenyl quinone distribution, photosystem I abundance, and anthocyanin accumulation in the Arabidopsis AtmenG mutant. J Biol Chem. 2006;281(52):40461–40472. doi: 10.1074/jbc.M609412200. [DOI] [PubMed] [Google Scholar]
- 11.Stites T, et al. Pyrroloquinoline quinone modulates mitochondrial quantity and function in mice. J Nutr. 2006;136(2):390–396. doi: 10.1093/jn/136.2.390. [DOI] [PubMed] [Google Scholar]
- 12.Doroshow JH. Effect of anthracycline antibiotics on oxygen radical formation in rat heart. Cancer Res. 1983;43(2):460–472. [PubMed] [Google Scholar]
- 13.Upchurch RG, Walker DC, Rollins JA, Ehrenshaft M, Daub ME. Mutants of Cercospora kikuchii Altered in Cercosporin Synthesis and Pathogenicity. Appl Environ Microbiol. 1991;57(10):2940–2945. doi: 10.1128/aem.57.10.2940-2945.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Brien PJ. Molecular mechanisms of quinone cytotoxicity. Chem Biol Interact. 1991;80(1):1–41. doi: 10.1016/0009-2797(91)90029-7. [DOI] [PubMed] [Google Scholar]
- 15.Monks TJ, Hanzlik RP, Cohen GM, Ross D, Graham DG. Quinone chemistry and toxicity. Toxicol Appl Pharmacol. 1992;112(1):2–16. doi: 10.1016/0041-008x(92)90273-u. [DOI] [PubMed] [Google Scholar]
- 16.Kumagai Y, et al. Oxidation of proximal protein sulfhydryls by phenanthraquinone, a component of diesel exhaust particles. Chem Res Toxicol. 2002;15(4):483–489. doi: 10.1021/tx0100993. [DOI] [PubMed] [Google Scholar]
- 17.Georgellis D, Kwon O, Lin EC. Quinones as the redox signal for the arc two-component system of bacteria. Science. 2001;292(5525):2314–2316. doi: 10.1126/science.1059361. [DOI] [PubMed] [Google Scholar]
- 18.Malpica R, Franco B, Rodriguez C, Kwon O, Georgellis D. Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. Proc Natl Acad Sci USA. 2004;101(36):13318–13323. doi: 10.1073/pnas.0403064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chi BK, et al. The redox-sensing regulator YodB senses quinones and diamide via a thiol-disulfide switch in Bacillus subtilis. Proteomics. 2010;10(17):3155–3164. doi: 10.1002/pmic.201000230. [DOI] [PubMed] [Google Scholar]
- 20.Chi BK, Kobayashi K, Albrecht D, Hecker M, Antelmann H. The paralogous MarR/DUF24-family repressors YodB and CatR control expression of the catechol dioxygenase CatE in Bacillus subtilis. J Bacteriol. 2010;192(18):4571–4581. doi: 10.1128/JB.00409-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leelakriangsak M, et al. Regulation of quinone detoxification by the thiol stress sensing DUF24/MarR-like repressor, YodB in Bacillus subtilis. Mol Microbiol. 2008;67(5):1108–1124. doi: 10.1111/j.1365-2958.2008.06110.x. [DOI] [PubMed] [Google Scholar]
- 22.Choi H, et al. Structural basis of the redox switch in the OxyR transcription factor. Cell. 2001;105(1):103–113. doi: 10.1016/s0092-8674(01)00300-2. [DOI] [PubMed] [Google Scholar]
- 23.Hong M, Fuangthong M, Helmann JD, Brennan RG. Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol Cell. 2005;20(1):131–141. doi: 10.1016/j.molcel.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 24.Newberry KJ, Fuangthong M, Panmanee W, Mongkolsuk S, Brennan RG. Structural mechanism of organic hydroperoxide induction of the transcription regulator OhrR. Mol Cell. 2007;28(4):652–664. doi: 10.1016/j.molcel.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 25.McLaughlin KJ, et al. Structural basis for NADH/NAD+ redox sensing by a Rex family repressor. Mol Cell. 2010;38(4):563–575. doi: 10.1016/j.molcel.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 26.Archer GL. Staphylococcus aureus: A well-armed pathogen. Clin Infect Dis. 1998;26(5):1179–1181. doi: 10.1086/520289. [DOI] [PubMed] [Google Scholar]
- 27.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339(8):520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 28.Fujimoto DF, et al. Staphylococcus aureus SarA is a regulatory protein responsive to redox and pH that can support bacteriophage lambda integrase-mediated excision/recombination. Mol Microbiol. 2009;74(6):1445–1458. doi: 10.1111/j.1365-2958.2009.06942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen PR, et al. An oxidation-sensing mechanism is used by the global regulator MgrA in Staphylococcus aureus. Nat Chem Biol. 2006;2(11):591–595. doi: 10.1038/nchembio820. [DOI] [PubMed] [Google Scholar]
- 30.Sun F, et al. Quorum-sensing agr mediates bacterial oxidation response via an intramolecular disulfide redox switch in the response regulator AgrA. Proc Natl Acad Sci USA. 2012;109(23):9095–9100. doi: 10.1073/pnas.1200603109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ji Q, et al. Staphylococcus aureus CymR is a new thiol-based oxidation-sensing regulator of stress resistance and oxidative response. J Biol Chem. 2012;287(25):21102–21109. doi: 10.1074/jbc.M112.359737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen PR, et al. A new oxidative sensing and regulation pathway mediated by the MgrA homologue SarZ in Staphylococcus aureus. Mol Microbiol. 2009;71(1):198–211. doi: 10.1111/j.1365-2958.2008.06518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun F, et al. AirSR, a [2Fe-2S] cluster-containing two-component system, mediates global oxygen sensing and redox signaling in Staphylococcus aureus. J Am Chem Soc. 2012;134(1):305–314. doi: 10.1021/ja2071835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol. 2003;48(6):1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez CE, Fukuto JM, Taguchi K, Froines J, Cho AK. The interactions of 9,10-phenanthrenequinone with glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a potential site for toxic actions. Chem Biol Interact. 2005;155(1-2):97–110. doi: 10.1016/j.cbi.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 36.Wilson I, Wardman P, Lin TS, Sartorelli AC. Reactivity of thiols towards derivatives of 2- and 6-methyl-1,4-naphthoquinone bioreductive alkylating agents. Chem Biol Interact. 1987;61(3):229–240. doi: 10.1016/0009-2797(87)90003-2. [DOI] [PubMed] [Google Scholar]
- 37.Laskowski RA, Swindells MB. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51(10):2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 38.Huffman JL, Brennan RG. Prokaryotic transcription regulators: More than just the helix-turn-helix motif. Curr Opin Struct Biol. 2002;12(1):98–106. doi: 10.1016/s0959-440x(02)00295-6. [DOI] [PubMed] [Google Scholar]
- 39.Lewis M. The lac repressor. C R Biol. 2005;328(6):521–548. doi: 10.1016/j.crvi.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 40.Ferguson GP, Tötemeyer S, MacLean MJ, Booth IR. Methylglyoxal production in bacteria: Suicide or survival? Arch Microbiol. 1998;170(4):209–218. doi: 10.1007/s002030050635. [DOI] [PubMed] [Google Scholar]
- 41.Ferguson GP, VanPatten S, Bucala R, Al-Abed Y. Detoxification of methylglyoxal by the nucleophilic bidentate, phenylacylthiazolium bromide. Chem Res Toxicol. 1999;12(7):617–622. doi: 10.1021/tx990007y. [DOI] [PubMed] [Google Scholar]
- 42.Booth IR, et al. Bacterial production of methylglyoxal: A survival strategy or death by misadventure? Biochem Soc Trans. 2003;31(Pt 6):1406–1408. doi: 10.1042/bst0311406. [DOI] [PubMed] [Google Scholar]
- 43.Tam T, et al. Differential gene expression in response to phenol and catechol reveals different metabolic activities for the degradation of aromatic compounds in Bacillus subtilis. Environ Microbiol. 2006;8(8):1408–1427. doi: 10.1111/j.1462-2920.2006.01034.x. [DOI] [PubMed] [Google Scholar]
- 44.Iyanagi T. On the mechanism of one-electron reduction of quinones by microsomal flavin enzymes: The kinetic analysis between cytochrome B5 and menadione. Free Radic Res Commun. 1990;8(4-6):259–268. doi: 10.3109/10715769009053359. [DOI] [PubMed] [Google Scholar]
- 45.Sollner S, Deller S, Macheroux P, Palfey BA. Mechanism of flavin reduction and oxidation in the redox-sensing quinone reductase Lot6p from Saccharomyces cerevisiae. Biochemistry. 2009;48(36):8636–8643. doi: 10.1021/bi900734a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Binter A, et al. A single intersubunit salt bridge affects oligomerization and catalytic activity in a bacterial quinone reductase. FEBS J. 2009;276(18):5263–5274. doi: 10.1111/j.1742-4658.2009.07222.x. [DOI] [PubMed] [Google Scholar]
- 47.Bryant C, DeLuca M. Purification and characterization of an oxygen-insensitive NAD(P)H nitroreductase from Enterobacter cloacae. J Biol Chem. 1991;266(7):4119–4125. [PubMed] [Google Scholar]
- 48.Nivinskas H, et al. Two-electron reduction of quinones by Enterobacter cloacae NAD(P)H:nitroreductase: Quantitative structure-activity relationships. Arch Biochem Biophys. 2002;403(2):249–258. doi: 10.1016/s0003-9861(02)00228-x. [DOI] [PubMed] [Google Scholar]
- 49.Massey V. The chemical and biological versatility of riboflavin. Biochem Soc Trans. 2000;28(4):283–296. [PubMed] [Google Scholar]
- 50.Li R, Bianchet MA, Talalay P, Amzel LM. The three-dimensional structure of NAD(P)H:quinone reductase, a flavoprotein involved in cancer chemoprotection and chemotherapy: Mechanism of the two-electron reduction. Proc Natl Acad Sci USA. 1995;92(19):8846–8850. doi: 10.1073/pnas.92.19.8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cavelier G, Amzel LM. Mechanism of NAD(P)H:quinone reductase: Ab initio studies of reduced flavin. Proteins. 2001;43(4):420–432. [PubMed] [Google Scholar]
- 52.Blum G, von Degenfeld G, Merchant MJ, Blau HM, Bogyo M. Noninvasive optical imaging of cysteine protease activity using fluorescently quenched activity-based probes. Nat Chem Biol. 2007;3(10):668–677. doi: 10.1038/nchembio.2007.26. [DOI] [PubMed] [Google Scholar]
- 53.Jessani N, Cravatt BF. The development and application of methods for activity-based protein profiling. Curr Opin Chem Biol. 2004;8(1):54–59. doi: 10.1016/j.cbpa.2003.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.