Abstract
Pheochromocytomas (PHEOs) and paragangliomas (PGLs) related to mutations in the mitochondrial succinate dehydrogenase (SDH) subunits A, B, C, and D, SDH complex assembly factor 2, and the von Hippel-Lindau (VHL) genes share a pseudohypoxic expression profile. However, genotype-specific differences in expression have been emerging. Development of effective new therapies for distinctive manifestations, e.g., a high rate of malignancy in SDHB- or predisposition to multifocal PGLs in SDHD patients, mandates improved stratification. To identify mutation/location-related characteristics among pseudohypoxic PHEOs/PGLs, we used comprehensive microarray profiling (SDHB: n = 18, SDHD-abdominal/thoracic (AT): n = 6, SDHD-head/neck (HN): n = 8, VHL: n = 13). To avoid location-specific bias, typical adrenal medulla genes were derived from matched normal medullas and cortices (n = 8) for data normalization. Unsupervised analysis identified two dominant clusters, separating SDHB and SDHD-AT PHEOs/PGLs (cluster A) from VHL PHEOs and SDHD-HN PGLs (cluster B). Supervised analysis yielded 6937 highly predictive genes (misclassification error rate of 0.175). Enrichment analysis revealed that energy metabolism and inflammation/fibrosis-related genes were most pronouncedly changed in clusters A and B, respectively. A minimum subset of 40 classifiers was validated by quantitative real-time polymerase chain reaction (quantitative real-time polymerase chain reaction vs. microarray: r = 0.87). Expression of several individual classifiers was identified as characteristic for VHL and SDHD-HN PHEOs and PGLs. In the present study, we show for the first time that SDHD-HN PGLs share more features with VHL PHEOs than with SDHD-AT PGLs. The presented data suggest novel subclassification of pseudohypoxic PHEOs/PGLs and implies cluster-specific pathogenic mechanisms and treatment strategies.
Introduction
Predispositions to certain tumors have been linked to an ever-increasing number of mutations. To date, mutations in 11 different genes have been associated with development of paragangliomas (PGLs), which are catecholamine-producing, chromaffin cell tumors, including adrenal pheochromocytomas (PHEOs). Initially, discovery of mutations was guided by syndromic presentation and family history; however, more recently discovered mutations can present in seemingly sporadic fashion. Known PHEO/PGL susceptibility genes are von Hippel-Lindau (VHL) and neurofibromatosis 1 (NF1) in the homonymous syndromes (VHL and NF1, respectively), RET proto-oncogene in multiple endocrine neoplasia type 2, succinate dehydrogenase D (SDHD) in familial PGL type 1 (PGL1) and Carney-Stratakis Syndrome (CSS), SDHC in PGL3 and CSS, SDHB in PGL4 and CSS, SDH complex assembly factor 2 (SDHAF2) in PGL2, prolyl hydroxylase 2 (PHD2/EGLN1), transmembrane protein 127 (TMEM127), kinesin family member 1B, SDHA, and MYC-associated factor X (reviewed in [1]). Most recently, activating mutations in hypoxia-inducible factor 2α (HIF2α) have been associated with PGL and polycythemia [2].
Notwithstanding the multitude of susceptibility genes, mutation-derived PHEOs/PGLs separate into merely two main clusters, one containing SDHA, SDHB, SDHC, SDHD, SDHAF2, and VHL and the other consisting of NF1, RET proto-oncogene, TMEM127, kinesin family member 1B, and MYC-associated factor X mutation-derived PHEOs/PGLs (reviewed in [1,3]). The PHEOs/PGLs of the first mentioned cluster are characterized by a pseudohypoxic phenotype, i.e., inappropriate stabilization of HIFα subunits under normoxia (reviewed in [1,3]).
Under normoxia, hydroxylation of HIFα by PHDs (PHD1/EGLN2, PHD2/EGLN1, and PHD3/EGLN3) designates them for VHL-dependent ubiquitylation and subsequent degradation [4]. Accordingly, VHL mutations can promote HIFα stabilization. Similarly, SDH dysfunction causes HIF stabilization by succinate or reactive oxygen species accumulation-mediated PHD inhibition [5,6].
Despite increasing evidence for differences within the pseudohypoxic cluster [7–9], the molecular basis for distinct clinical behaviors including the preferential site of tumor development, biochemical phenotype, or metastatic potential remains largely unknown. Mutations in SDHB have been associated with extra-adrenal PGLs and high risk of malignancy [1,3]. SDHD mutations, however, predispose to multifocal PHEOs/PGLs, primarily from the head and neck (HN) region, with low metastatic risk [3]. However, HN PGL can be inoperable, or surgery can lead to severe side effects due to close vicinity to major blood vessels and nerves. VHL mutation-derived PHEOs/PGLs are almost always adrenal, non-metastatic, but frequently bilateral and/or recurrent, thus adrenal sparing treatment options are of high importance [3]. SDHA, SDHC, and SDHAF2 mutation-derived PHEOs/PGLs are rare and have not yet been characterized in detail. Large cohort studies including PHEO/PGL patients revealed that SDHA, SDHC, and SDHAF2 are extremely rare (0.1–0.5%, 0.4–2.2%, and non-detectable, respectively) [9–13]. In a Dutch population of HN PGL patients, SDHAF mutations were found in 4% and SDHC mutations in 0.4% of cases (SDHA not tested) [14].
To date, genetic testing presents an important diagnostic tool for risk assessment of PHEO/PGL patients and their families. However, genetic testing is cost intensive and targeted therapeutic options for the distinct subtypes of PHEOs/PGLs are not yet available. Detection of novel therapeutic targets for the particular PHEO/PGL subtypes mandates the identification of subtype-specific characteristics, e.g., by gene expression profiling.
In the present study, gene expression profiles of pseudohypoxic PHEOs/PGLs were compared among each other and to normal adrenal medulla (NAM) to identify new signatures for disease stratification. The study was designed to decrease variability as much as possible by using homogeneous sample groups of SDHB, SDHD, and VHL mutation-derived PHEOs/PGLs as has been previously recommended [15]. Due to limited access to an appropriate number of samples, no SDHA, SDHC, and SDHAF2 mutation-derived PHEOs/PGLs were included. Association of specific pathways or genes with the distinct phenotypes of pseudohypoxic PHEOs/PGLs may lead to identification of new diagnostic markers and potential therapeutic targets.
Materials and Methods
Tissue
PHEO/PGL tissue was collected with informed consent at Dutch and US centers: the National Institutes of Health (Bethesda, MD), Tufts Medical Center (Boston, MA), Baylor College of Medicine (Houston, TX), Radboud University Nijmegen Medical Centre (Nijmegen, The Netherlands), and Erasmus MC (Rotterdam, The Netherlands). NAMs were collected from anonymous tissue donors at Tufts Medical Center and Comenius University (Bratislava, Slovakia) after confirmed brain death or during radical nephrectomy. Tissues were collected under a protocol that was approved by the Institutional Review Board of the respective institutions. Genetic testing was performed with informed consent, following established guidelines for testing [1].
Normal medulla and cortex from the same adrenals (n = 8) were separated as previously described [16]. In total, snap-frozen tissue of 45 histopathologically confirmed PHEOs/PGLs was used. The samples were grouped according to genetic/syndromic background and tumor location: SDHB (n = 18), SDHD-abdominal/thoracic (AT; n = 6), SDHD-HN (n = 8), and VHL PGLs (n = 13). Patient information is summarized in Table 1.
Table 1.
Patient Information.
| Sample ID | Chip ID | Genetic Backgr. | Sex | Age at Surgery | Age at First Presentation | Age at Dgn. | Loc. | Status | MA | qRT-PCR |
| 01M/01C | PKh_03* | dna | m | 61.0 | dna | dna | dna | dna | x | 01M1 |
| 02M/02C | PKh_04* | dna | nk | nk | dna | dna | dna | dna | x | 02M* |
| 03M/03C | PKh_05* | dna | f | 53.0 | dna | dna | dna | dna | x | 03M*,† |
| 04-1M/04-1C | PKh_06* | dna | f | 72.0 | dna | dna | dna | dna | x | 04-1M* |
| 04-2M/04-2C | PKh_07* | dna | f | 72.0 | dna | dna | dna | dna | x | 04-2M* |
| 05M/05C | PKh_42* | dna | m | 65.0 | dna | dna | dna | dna | x | |
| 06M/06C | PKh_43* | dna | m | 56.0 | dna | dna | dna | dna | x | |
| 07M/07C | PKh_44* | dna | m | 62.0 | dna | dna | dna | dna | x | |
| 08B | PKh_40 | SDHB | f | 30.0 | 27 | 30 | A | Pr-NM | x | |
| 09B | PKh_10 | SDHB | m | 28.5 | 12 | 29 | E | Met | x | |
| 10B | PKh_11 | SDHB | m | 24.0 | 20 | 23 | E | Pr-M | x | |
| 11B | PKh_13 | SDHB | f | 46.0 | 28 | 46 | A | Pr-NM | x | |
| 12B | PKh_18 | SDHB | f | 9.0 | 8 | 9 | E | Mlt-NM | x | x |
| 13B | PKh_19 | SDHBP | m | 22.8 | 15–16 | 22 | E | Pr-M | x | |
| 14B | PKh_20 | SDHB | m | 26.6 | 8 | 9 | E | Met | x | x |
| 15B | PKh_21 | SDHB | m | 38.2 | 37 | 38 | E | Pr-M | x | |
| 16B | PKh_22 | SDHB | f | 45.6 | 41 | 42 | E | Mlt-NM | x | |
| 17-1B | PKh_27 | SDHB | f | 36.0 | 32 | 33 | E | Met | x | x |
| 17-2B | PKh_28 | SDHB | f | 36.2 | 32 | 33 | E | Met | x | |
| 18B | PKh_31 | SDHB | m | 52.0 | 40 | 41 | E | Met | x | |
| 19B | PKh_32 | SDHB | m | 12.0 | 13 | 13 | E | Pr-NM | x | |
| 20B | PKh_33 | SDHB | m | 31.0 | 30 | 30 | E | Pr-NM | x | x |
| 21B | PKh_34 | SDHB | m | 55.0 | 50 | 53 | E | Mlt-NM | x | x |
| 22B | PKh_36 | SDHB | m | 35.0 | no symptoms | 34 | E | Mlt-NM | x | |
| 23B | PKh_38 | SDHB | f | 34.8 | 10 | 18 | E | Met | x | |
| 24B | PKh_39 | SDHB | m | 17.1 | 15 | 15 | A | Mlt-NM | x | |
| 25D(HN) | PKh_37 | SDHD | m | 24.3 | 23 | 23 | HN | Pr-NM | x | |
| 26D(HN) | PKh_60 | SDHD | f | 34.0 | 34 | 34 | HN | Bi-M | x | |
| 27D(HN) | PKh_01 | SDHD | f | 49.3 | 28 | 28 | HN | Pr-NM | x | x |
| 28D(HN) | PKh_02 | SDHD | f | 61.1 | 60 | 60 | HN | Pr-NM | x | |
| 29D(AT) | PKh_08 | SDHD | m | 16.0 | 16 | 16 | A | Pr-NM | x | x |
| 30D(AT) | PKh_09 | SDHD | f | 31.0 | 31 | 31 | A | Pr-NM | x | x |
| 31-1D(AT) | PKh_24 | SDHD | f | 26.6 | 23 | 24 | A | Mlt-NM | x | |
| 31-2D(HN) | PKh_25 | SDHD | f | 26.6 | 23 | 24 | HN | Mlt-NM | x | x |
| 31-3D(HN) | PKh_29 | SDHD | f | 27.1 | 23 | 24 | HN | Mlt-NM | x | |
| 32D(HN) | PKh_58 | SDHD | m | 48.0 | 48 | 48 | HN | Bi-NM | x | x |
| 33D(AT) | PKh_26 | SDHD | m | 61.4 | 58 | 58 | A | Pr-NM | x | x |
| 34D(HN) | PKh_59 | SDHD | f | 29.0 | 29 | 29 | HN | Mlt-NM | x | x |
| 35-1D(AT) | PKh_35 | SDHD | m | 31.8 | 31 | 25 | A | Mlt-NM | x | x |
| 35-2D(AT) | PKh_41 | SDHD | m | 32.6 | 31 | 25 | E | Mlt-NM | x | |
| 36V | PKh_57 | VHL | f | 24.8 | no symptoms | 17 | A | Bi-NM | x | |
| 37-1V | PKh_55 | VHL | m | 22.7 | 18 | 22 | A | Bi/Mlt-NM | x | |
| 37-2V | PKh_56 | VHL | m | 23.1 | 18 | 22 | A | Bi/Mlt-NM | x | |
| 38V | PKh_54 | VHL | m | 16.1 | 16 | 16 | A | Bi/Rec-NM | x | |
| 39V | PKh_53 | VHL | m | 28.6 | no symptoms | <28 | A | Pr-NM | x | |
| 40V | PKh_52 | VHL | m | 13.1 | no symptoms | 12 | A | Bi-NM | x | |
| 41V | PKh_45 | VHL | f | 42.8 | nk | <41 | A | Pr-NM | x | |
| 42V | PKh_46 | VHL | m | 29.4 | 29 | 29 | A | Bi/Mlt-NM | x | |
| 43V | PKh_47 | VHL | f | 42.6 | nk | <15 | A | Bi-NM | x | x |
| 44V | PKh_48 | VHL | m | 39.2 | nk | 39 | A | Pr-NM | x | x |
| 45V | PKh_49 | VHL | m | 30.6 | <30 | 30 | A | Bi-NM | x | |
| 46V | PKh_50 | VHL | m | 33.1 | 4 | 5 | A | Bi/Mlt/Rec-NM | x | x |
| 47V | PKh_51 | VHL | m | 18.9 | no symptoms | 16 | A | Bi/Mlt-NM | x | |
| 48V | dna | VHL | m | 33.5 | nk | nk | A | Bi-NM | x |
A indicates adrenal; f, female; backgr., background; Bi, bilateral; dgn., diagnosis; dna, does not apply; E, extra-adrenal; ID, identifier; loc., location; m, male; M, metastatic; MA, microarray; met, metastases; Mlt, multifocal primary; nk, not known; NM, non-metastatic; Pr, primary; Rec, recurrent; SDHDP, SDHD-polymorphism; x indicates sample used in experiment specified by column heading; patients 25, 31, and 40 were first-degree relatives to 35, 33, and 47, respectively.
Applies to normal medulla samples.
Included for ACTR10, EGLN3, F8, LGR5, MPP6, NEFM, SLC18A1, TMEM130, and VSNL1.
Microarray Processing
RNA extraction was performed as previously described [16]. RNA quality and quantity was ensured using Bioanalyzer (Agilent Technologies, Inc, Santa Clara, CA) and NanoDrop (Thermo Scientific, Wilmington, DE), respectively. Samples were processed following the recommended Affymetrix protocol. Fragmented and labeled cDNA was hybridized onto GeneChip Human Gene 1.0 ST Arrays (Affymetrix, Santa Clara, CA). Staining of biotinylated cDNA and scanning of arrays were performed according to the manufacturer's recommendations. All data are available at the GEO database (GSE 39716).
Bioinformatic Analysis
Raw CEL files were imported into Expression Console 1.0 (Affymetrix). Only “core” probe sets were used to perform “gene-level” probe set summarization, background subtraction, and quantile normalization using the RMA option in the software. Data analysis was performed using R packages from the Bioconductor project (http://www.bioconductor.org) [17].
To avoid bias due to potential cortical contamination, genes with at least two-fold greater expression in cortex than medulla were determined cortex-specific and removed from the data sets, resulting in 31,202 transcripts. For each sample, a ratio of the expression profile to the mean expression profile of normal medulla was computed. Gene-wise Z-score normalization across all samples was applied to adjust for technical variation [18]. Differential expression analysis was performed using significance analysis of microarray (SAM) [19] with two- or multiclass option. False discovery rates were estimated with 1000-fold sample permutations and significant gene changes were arbitrarily selected at ≤ .02. Hierarchical clustering, principal component analysis, between-group analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation, and overrepresentation analysis (P value cutoff, .05) were performed using the R packages “stats,” “made4”- and “GSA.” Gene network analysis and functional categorization was performed with Ingenuity Pathway Analysis (IPA; www.ingenuity.com). Class prediction analysis using prediction analysis for microarray (PAMR) [20] was done to identify 10-fold cross-validated gene expression predictors for the four subtypes of PHEOs/PGLs.
Quantitative Real-Time Polymerase Chain Reaction
RNA from a subset of tissues as indicated (Table 1) was used to generate cDNA (SuperScript III First-Strand Synthesis SuperMix; Life Technologies, Grand Island, NY) according to the manufacturer's recommendations. The following TaqMan primer probe sets (Life Technologies) were used: 18S: 4319413E, ABCC9: hs00245832_m1, ALDH1A1: hs00946916_m1, ARP11: hs00218475_m1, C7: hs00175109_m1, CARTPT: hs00182861_m1, CFH: hs00962373_m1, CGNL1: hs00262671_m1, DNAH14: hs00376554_m1, EGLN3: hs00222966_m1, elongation of very long chain fatty acid protein 7 (ELOVL7): hs00405150_m1, F13A1: hs00173388_m1, F8: hs00252034_m1, FCGR1A: hs00174081_m1, FCGR1B: hs00417598_m1, FGF11: hs00182803_m1, GABRA1: hs00168058_m1, GPC3: hs00170471_m1, GYG2: hs00418496_m1, HOXC6: hs00171690_m1, KCNN2: hs00222059_m1, LGR5: hs00173664_m1, MPP6: hs00212785_m1, NEFM: hs00606024_m1, OLFML2B: hs00295836_m1, PLTP: hs01067287_m1, RAB3B: hs01001137_m1, RARRES2: hs00161209_g1, SCIN: hs00263961_m1, SLC18A1: hs00161839_m1, SLC9A7: hs00261601_m1, SUCNR1: hs00908230_m1, TFAP2C: hs00231476_m1, TFPI: hs00196731_m1, TMEM130: hs00905715_m1, VSNL1: hs02386966_m1. Cycling conditions were as previously described [16]. Relative expression to 18S and normal medulla was estimated by the ΔΔCt method. Concordance between quantitative real-time polymerase chain reaction (qRT-PCR) validation data and microarray data was tested using the correlation of correlation index [21]. Statistical evaluation of expression differences for each of the classifiers between the groups were analyzed by analysis of variance (ANOVA) with Student-Neuman-Keuls post-hoc analysis adjusted for multiple comparisons.
Results
Purity of Adrenal Medulla Tissue
Separating human adrenal medulla from cortex is challenging due to a convoluted border and cortical cell islands within the medulla. To determine and exclude genes of cortical origin from the medulla samples, we used a computational approach. Of the total transcripts 4.73% (1551/32,740) were associated with adrenal cortex (P < .0001; Figure 1A). Gene set enrichment analyses for cortex and medulla samples with KEGG pre-compiled gene modules were performed [22]. Significant enrichment (at P ≤ .05) of steroid hormone, androgen, and estrogen metabolism was evident for cortex, while tyrosine metabolism and neuronal functions were associated with medulla (Figure 1B).
Figure 1.
Proportion of cortical and medullary genes and their associated functions in normal control samples. (A) Venn diagram of the two-fold differentially expressed probes identified in normal cortex with reference to normal medulla. For each subset of genes, a Pearson correlation coefficient was calculated between the two sets of controls and shown within the parenthesis. (B) Heatmap of pathways associated with normal cortex and normal medulla.
Gene Expression Profiles Classify Mutation/Location Subtypes
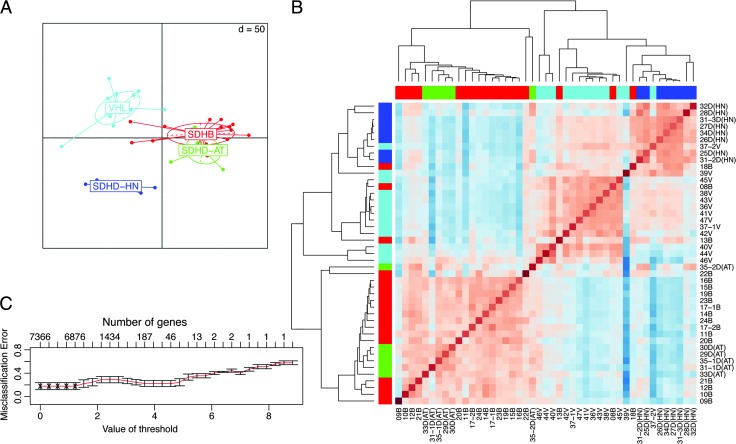
Initial screening by SAM yielded a total of 7366 differentially expressed transcripts. Principal component analysis of samples grouped by mutation/location showed clear separation of VHL and SDHD-HN from each other as well as from the partly overlapping SDHB and SDHD-AT PGLs (Figure 2A). In agreement, unsupervised average linkage hierarchical clustering of significant genes using pairwise Pearson correlation revealed separation of the tumors into two main clusters (Figure 2B), cluster A consisting of SDHB and SDHD-AT PGLs and cluster B containing VHL and SDHD-HN PHEOs/PGLs. Cluster B is further divided into separate VHL and SDHD-HN PHEO/PGL subclusters.
Figure 2.
Unsupervised analysis of tumors with SDHB, SDHD, and VHL mutations. (A) Group level principal component analysis showing association of the different tumors. Projection lines from each group correspond to individual sample locations on the resulting axes. (B) Heatmap of a pairwise correlation of genes from significance analysis for microarrays. Hierarchical cluster analysis was performed by average linkage and correlation distance metrics. (C) Class prediction results of the nearest shrunken centroid model. Cross-validation errors of the classifiers are shown as a function of the threshold parameter. The value giving minimum errors was chosen to give a selected list of genes.
Supervised PAMR was used to narrow down the genes of interest under maintenance of optimal classification capabilities. Ten-fold cross-validation yielded 6937 transcripts with an overall misclassification rate of 0.175 (Figure 2C and Table W1). SDHD-HN PGLs were classified with high confidence (zero misclassifications). Sample discrimination by cluster analysis based on the 6937 genes maintained the assignment into two major clusters (data not shown).
Enriched Functions in PAMR Analysis
To determine which functional themes corresponded to the gene expression profiles of the two main clusters, we performed a series of comparisons using IPA. Cluster A was associated with 425 and cluster B with 1270 upregulated genes. Canonical pathway analysis by IPA identified 17 and 121 significantly changed pathways (P < .05), respectively (Table W2). Fourteen pathways (82%) selected for cluster A were found to be involved in metabolism. The top cluster A pathways included oxidative phosphorylation (OXPHOS), mitochondrial dysfunction, ubiquinone biosynthesis, and citrate cycle (Table W2).
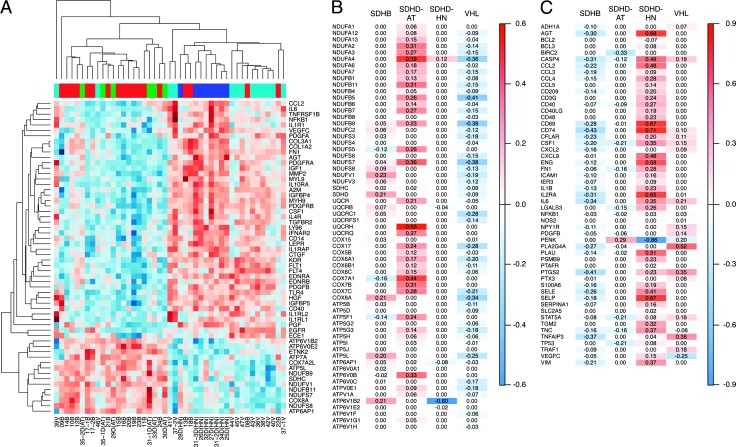
In cluster B, 91 (75%) of the 121 significantly changed pathways were found to be involved in hepatic fibrosis and immune function (Table W2). Hierarchical clustering using the differentially expressed genes from the top IPA functions, i.e., 14 OXPHOS (cluster A) and 44 hepatic fibrosis genes (cluster B), led to clear separation of PHEOs/PGLs according to mutation/location (Figure 3A).
Figure 3.
Canonical pathways associated with the tumor subgroups. (A) Heatmap of oxidative phosphorylation and hepatic fibrosis-related transcripts selected by IPA. (B) Mean expression score of oxidative phosphorylation complex subunits. (C) Mean expression of NFκB target genes in the tumor subgroups. Red indicates increased and blue decreased expression relative to normal medulla and the other PHEO/PGL subtypes.
Differential Expression of Selected OXPHOS Genes
To elaborate the differences in OXPHOS complex expression, we compared the shrunken centroid scores from the PAMR model between the PHEO/PGL subtypes (Figure 3B). Overall, expression of OXPHOS genes appeared decreased in VHL, close to normal in SDHD-HN, and increased in SDHD-AT and SDHB PHEOs/PGLs. Seventeen and seven of 25 mitochondrial complex I genes were above normal in SDHD-AT and SDHB PHEOs/PGLs, respectively, while decreased expression prevailed in VHL PHEOs (22/25 genes). Similarly, mitochondrial complex IV genes were below normal in VHL (8/10) and above normal in SDHD-AT (8/10) and SDHB (2/10). Patterns for the other complexes were analogous.
Differential Expression of Selected Hepatic Fibrosis Genes
Upregulated genes typically associated with tissue fibrosis from cluster B (Table W2) included fibrillar collagens COL1A2 and COL3A1, matrix metalloprotease 2 (MMP2), metalloprotease inhibitor A2M, the chemotactic protein CCL2, the contractile proteins MYL9, growth factor signaling-related transcripts (IGF1, IGFBP4, IGFBP5, CTGF/IGFBP8, VEGFC, FLT1/VEGFR1, FLT4/VEGFR3, KDR/VEGFR2, PGF, PDGFA, PDGFRA, PDGFB, PDGFRB, HGF, and EGFR), and the transcriptional regulator NFKB1, including several players of nuclear factor κB(NFκB) activation (IL1RI, IL1RAP, IL1RL2, EGFR, and TLR4). As mentioned above, 44 genes of the hepatic fibrosis pathway have been identified to be upregulated in cluster B by enrichment analysis, including several genes involved in NFκB signaling. For this reason, we matched the list of 6937 genes from PAMR to a published list of NFκB target genes (source: http://bioinfo.lifl.fr/NF-KB/). In total, 53 genes from our PAMR data were identified as NFκB targets, most of which were upregulated in cluster B tumors (Figure 3C).
Among others, activation of NFκB has been shown to induce epithelial-to-mesenchymal transition (EMT). EMT markers have been recognized as indicators of poor prognosis or aggressive behavior and recent reports have proposed important EMT genes such as TWIST1 and SNAI1 as markers of malignancy in PHEOs/PGLs [23,24]. Thus, we performed a cluster analysis of our samples based on a list of 92 EMT genes [24], 44 of which mapped to our SAM list. Several key players in EMT (e.g., TWIST1, TCF3, and MMP1) were not part of our SAM list, because a difference in expression between the groups was not present or not sufficient to withstand permutation analysis. On the basis of the heatmap of 44 EMT genes, we found that up-regulation of certain EMT genes may play a role in some metastatic SDHB tumors; however, the same EMT genes are elevated in a number of non-metastatic samples and thus do not qualify as stand-alone markers of malignancy in our sample set (Figures W2 and W3). Cluster analysis based on EMT genes essentially confirmed the pattern described above, although group separation was not quite as clear as when based on all SAM genes or OXPHOS and hepatic fibrosis (Figure W3).
Minimum Subset of Classifiers
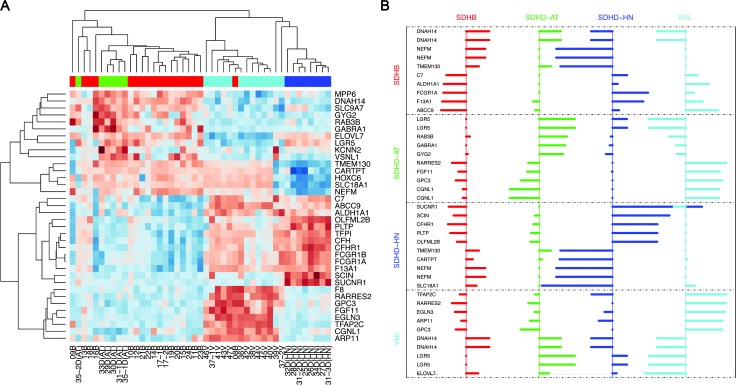
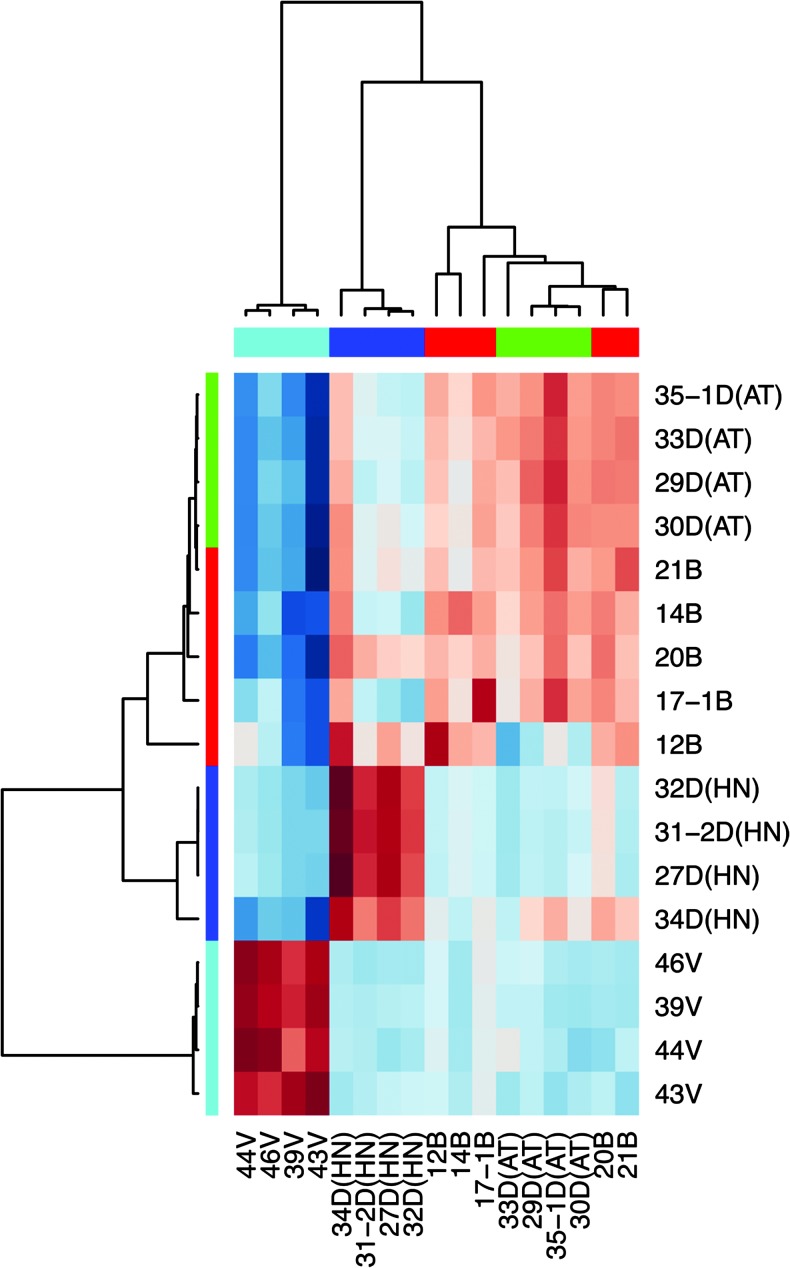
To identify the most characteristic genes for each of the PHEO/PGL subtypes, we selected a minimum set of top 10 genes with strongest group association for each of the mutation/location classes from the PAMR data (including upregulated and downregulated genes). The choice of this selection is heuristic and we found that less than 10 genes from any class resulted in higher misclassifications (data not shown). Surprisingly, the minimum subset of classifiers improved classification capabilities compared to the 6937 PAMR-selected genes(overallerror rate:0.066, SDHB: 0.05, SDHD-AT:0.16, SDHD-HNP: 0.00, VHL: 0.07). A heatmap (Figure 4A) and a profile plot (Figure 4B) show relative expression scores of the 40 classifiers (36 unique genes). To validate proper characterization and to potentially identify stand-alone markers for each group, qRT-PCR for 35 of the 36 genes of interest was performed. Comparison between micro-array and qRT-PCR data showed an overall correlation of correlation coefficient of 0.87 (SDHB: 0.87, SDHD-AT: 0.97, SDHD-HN: 0.92, VHL: 0.57). Thus, the qRT-PCR results essentially confirmed the microarray data (Figure 5A).
Figure 4.
Minimum subset of classifiers for separation of the different mutation/location types. (A) Unsupervised hierarchical clustering of the gene expression data of the minimum subset of 40 classifiers. The different tumor types (red, SDHB; blue, SDHD-HN; green, SDHD-AT; cyan, VHL) are distinguished by the pattern of up-regulation (red) and down-regulation (blue). Separation into two main clusters, a shared SDHB and SDHD-AT cluster and one that further subclusters into VHL and SDHD-HN PGLs, is evident. (B) Profile plot depicting expression levels (shrunken centroid scores) for the minimum subset of classifiers, i.e., 10 top scoring genes for each class. Bars facing right represent positive and those facing left represent negative shrunken centroid scores indicating increased and decreased expression relative to normal medulla and the other PHEO/PGL subtypes.
Figure 5.
Correlation heatmap of Pearson correlation results for 35 genes of the minimum subset of classifiers between micro-array and qRT-PCR data. The mutation types are shown as colored bars below the dendrograms (red, SDHB; blue, SDHD-HN, green, SDHD-AT; cyan, VHL).
Subgroup Classification Based on Individual Genes
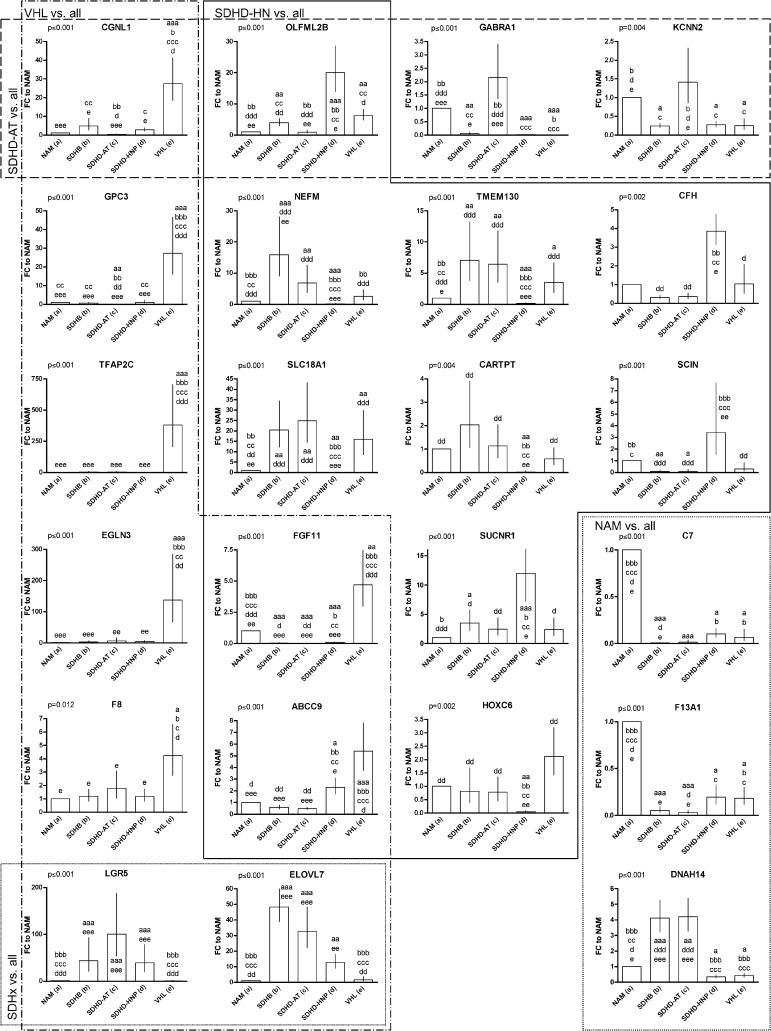
Classification of cluster B tumors based on individual genes was feasible for both subgroups. qRT-PCR confirmed discriminatory capacity of 8/10 candidates for VHL PHEOs. Six of them were more highly expressed in VHL (TFAP2C, EGLN3, GPC3, FGF11, CGNL1, and F8), and two were least expressed in VHL PHEOs (LGR5 and ELOVL7; Figure 6). Interestingly, the latter were significantly elevated in all SDHx mutation-derived PHEOs/PGLs. In addition, one gene predicted to characterize SDHB was most highly expressed in VHL tumors (ABCC9). Thus, we identified nine genes that showed distinct expression in VHL PHEOs from the other groups. Three of them also qualified as classifiers for another tumor group (CGNL1 for SDHD-AT; ABCC9 and FGF11 for SDHD-HN).
Figure 6.
Relative expression of selected genes of the minimum subset of 35 genes to NAM. The genes are arranged into clusters according to their capabilities as classifiers (VHL vs. all, SDHD-HN vs. all, SDHD-AT vs. all, NAM vs. all). Overlapping indicates that the respective genes show characteristic expression in two of the tumor groups. Overall significance has been evaluated for each gene by ANOVA (value given for each gene in the upper left corner). Significant differences between each group and (a) NAM, (b) SDHB, (c) SDHD-AT, (d) SDHD-HN, and (e) VHL are indicated above each applicable column. Replications of the letter specify the level of significance (xxx, P < .001; xx, P < .01; x, P < .05).
Significantly different expression in SDHD-HN PGLs compared to all other groups was evident for seven of nine candidates. Two genes showed increased (SUCNR1 and OLFML2B) and five genes showed decreased expression (SLC18A1, NEFM, CARTPT, TMEM130, and HOXC6) in SDHD-HN PGLs (Figure 6). As mentioned above, FGF11 and ABCC9 levels also distinguished SDHD-HN PGLs from all other groups. SCIN and CFH levels in SDHD-HN were significantly different from the other PHEO/PGL subtypes. Thus, in total, we identified 11 characteristic genes for SDHD-HN PGLs.
Identification of characteristic individual genes was less successful for cluster A PHEOs/PGLs. While CGNL1, OLFML2B, GABRA1, and KCNN2 expression of SDHD-AT PHEOs/PGLs was distinct from that of all groups except normal medulla (Figure 6), expression of none of the top ranked genes for SDHB was significantly different from SDHD-AT PHEOs/PGLs. However, DNAH14, C7, and NEFM were significantly differentially expressed in SDHB PHEOs/PGLs compared to NAMs, SDHD-HN PGLs, and VHL PHEOs (Figure 6).
Expression levels of three evaluated genes were characteristic for NAM (C7, F13A1, and DNAH14; Figure 6). In addition, DNAH14 clearly separated cluster A and B tumors. Graphs of additional tested genes are depicted in Figure W1.
Discussion
Previous studies revealed divergent expression patterns in pseudohypoxic PHEOs/PGLs, which allowed genotype discrimination [7–9]. Commonly, SDHx PHEOs/PGLs have been grouped together, preventing detection of differences. Mining for disease-relevant characteristics requires normalization to healthy tissue. Thus, we evaluated the gene expression profiles of pseudohypoxic PHEOs/PGLs associated with SDHB, SDHD, and VHL mutations in relation to NAM. Our major new findings are that SDHD-HN PGLs are different from SDHD-AT PGLs and more closely resemble VHL PHEOs. These observations indicate that expression profiles of pseudohypoxic PHEOs/PGLs depend on tumor location as well as underlying mutation.
Purity of Adrenal Medulla Tissue
Computational analysis determined the content of cortical genes within the medulla samples at 4.73%. This agrees with our previous study, where cortical contamination of the medulla samples was estimated to be less than 5% [16]. Although this represents a reasonable degree of purity, in “-omics” type studies ∼5% contamination may cause significant bias. Thus, we excluded cortical genes from the medulla data sets.
Mutation and Location Determine Expression Profiles
Unsupervised analysis revealed similarities in the expression patterns of SDHB-with SDHD-AT PHEOs/PGLs and SDHD-HN PGLs with VHL-derived PHEOs, respectively. This indicates that SDHD-derived PHEOs/PGLs differ depending on the tissue of origin. Remarkably, two SDHD-HN PGLs and one PHEO from the same patient clearly separated into the different clusters.
In agreement with our findings, HN PGLs clustered in close vicinity to VHL PHEOs upon unsupervised analysis in a previous study involving sporadic and hereditary PHEOs/PGLs [8]. Contrary to our results, no separation into distinct VHL PHEO and HN PGL sub-clusters has been observed. The good separability of our sample set may be due to the stringent sample selection criteria, i.e., well-defined, homogenous subgroups of pseudohypoxic PHEOs/PGLs. The study of Lopez-Jimenez et al. involved HN PGLs of different genetic backgrounds (SDHB, SDHC, and SDHD mutations and apparently sporadic), which clustered in close proximity to each other but apart from AT PGLs with underlying mutations in the same genes [8]. Consistently, a different study reported that gene expression of HN PGLs appeared uniform, independent of hereditary background (PGL1/SDHD, PGL2/SDHAF2, or sporadic) [25]. Thus, mutational background appears to have less influence on the expression pattern of PHEOs/PGLs than their location.
While distinct expression profiles in SDHD-HN and SDHD-AT PHEOs/PGLs are unexpected because of mutual SDHD malfunction, prominent differences in their biochemical phenotypes are well known. Only a subset of HN PGLs may secrete catecholamines (29%) [26], while almost all AT PHEOs/PGLs are biochemically active [27]. The distinct secretory characteristics probably reflect different cells of origin for sympathetic AT PHEOs/PGLs and parasympathetic HN PGLs. Thus, despite the same genetic background, differences in gene expression patterns seem plausible. However, parallels between sympathetic VHL PHEOs and parasympathetic HN PGLs currently appear limited.
To our surprise, SDHD-AT and SDHB PHEOs/PGLs did not separate into distinct subclusters. In addition, the different types of SDHB samples involved did not cluster according to their characteristics (metastases, primary metastatic, or primary non-metastatic). Seven of nine SDHB-metastatic samples fell into cluster A. However, the same cluster contained 14 non-metastatic SDHB and SDHD-AT PHEOs/PGLs. Thus, we do not see an indication that the composition of cluster A reflects aggressive or malignant potential of the included PHEOs/PGLs. Despite involving 50% metastatic SDHB samples, no gene signature of metastatic PHEOs/PGLs could be identified. Sub-grouping SDHB samples into metastatic (n = 9, i.e., n = 6 metastases and n = 3 primary metastatic) and primary non-metastatic (n = 9) gave inacceptable error rates of 44.4% for SDHB-non-metastatic and 55.6% for SDHB-metastatic (see Table W3). We thus concluded that the differences between SDHB-metastatic and SDHB-non-metastatic samples were less pronounced than those to the other groups, possibly because only a few genes discriminate SDHB-metastatic and SDHB-non-metastatic samples. However, we cannot exclude that the poor classification was based on the relatively small sample number for each of the subgroups (n = 9 each). Since a very large proportion of SDHB PHEO/PGL patients develop metastases, and classification of SDHB tumors as non-metastatic can only be based on the fact that metastases have not (yet) presented, we decided to abstain from subgrouping of the SDHB samples.
Enriched Biologic Functions in Cluster A
Cluster A was associated with metabolic pathways, such as OXPHOS, mitochondrial dysfunction, ubiquinone biosynthesis, and citric acid cycle. Disturbances in the mitochondrial energy metabolism are anticipated in tumors with mutations in the mitochondrial SDH complex. As enzyme of the citric acid cycle, SDH converts succinate into fumarate. Simultaneously, electrons are donated to ubiquinone, an essential electron carrier of the OXPHOS. Here, we report a potentially compensatory up-regulation of pathways that are directly connected to SDH function in AT PGLs with underlying SDHB and SDHD mutations.
Differential Expression of Selected OXPHOS Genes
Up-regulation of OXPHOS transcripts was prevalent in SDHB and SDHD-AT PGLs, supporting our previous finding of increased expression of several OXPHOS subunits in SDHB compared to VHL mutation-derived PHEOs/PGLs [28]. Which subunits were upregulated was rarely mutual between both subgroups. Thus, SDHB and SDHD mutations apparently both influence OXPHOS gene expression in AT PHEOs/PGLs but not SDHD-HN PGLs.
Favier et al. reported low expression of several OXPHOS genes in SDHx PHEOs/PGLs [7]. Discrepancy with our data may again be due to differences in sample selection. The SDHx group of Favier et al. contained HN PGLs, which in our study did not show increased OXPHOS subunit expression. Furthermore, SDHB, SDHC, and SDHD mutation-derived PGLs were grouped together. The limited agreement in OXPHOS subunit expression between SDHB, SDHD-AT, and SHD-HN PGLs mentioned above may have concealed their distinct expression profiles.
Decreased expression of OXPHOS subunits related to VHL mutations has been previously reported and indicates decoupling of glycolysis and OXPHOS [7,29].
Compensatory up-regulation to rescue an impaired energy metabolism in consequence of SDHx dysfunction seems probable. However, the regulatory mechanism remains to be elaborated. Possibly up-regulation occurs in a disconcerted manner, which may fail to enhance OXPHOS capacity or even increase reactive oxygen species generation. Special attention should be given to the finding that despite underlying SDHD mutations, no up-regulation of OXPHOS genes was evident in HN PGLs.
Enriched Biologic Functions in Cluster B
In cluster B, hepatic fibrosis and immune functions were among the top enriched pathways. Tissue fibrosis is excessive accumulation of extracellular matrix (ECM) components, which is the physiologic response to injury and is facilitated by inflammatory mediators. Tissue fibrosis and inflammation initiation have been shown to be direct or HIF stabilization-dependent consequences of VHL mutations. In hepatic cells, VHL mutations have reportedly caused fibrosis and promoted inflammation in an HIF-2α-dependent manner [30]. Transgenic HIF-2α stabilization has also been shown to promote renal fibrosis [31]. In addition, direct interaction of pVHL with the ECM component collagen IV has been described [32,33]. VHL mutations disrupt this interaction and cause improper collagen matrix assembly. Faulty ECM assembly due to VHL mutations has been accompanied by increased expression of MMP2 [34], which was also elevated in our cluster B tumors (Figure 4C). Kurban et al. reported that inappropriate ECM assembly in consequence of VHL mutations facilitates angiogenesis, tumor development, and invasion. Thus, disrupted interaction between VHL and ECM components appears to be a crucial HIF-independent tumorigenic trait. Tissue fibrosis and inflammation genes were overexpressed in VHL and, even more so, SDHD-HN PGLs. To our knowledge, these pathways are currently entirely unexplored in SDHD-HN PGLs. HIF-2α-mediated initiation of tissue fibrosis and inflammation may be common to all pseudohypoxic PHEOs/PGLs. Similar expression of HIF-2α in VHL- and SDHx-derived PHEOs/ PGLs [7] and predominance of HIF-1α in the latter have both been observed [35]. In any case, the reason for normal expression of tissue fibrosis and inflammation genes in SDHx-AT compared to SDHDHN and VHL PHEOs/PGLs remains to be determined.
Consideration of Selected Hepatic Fibrosis-Related Genes
The hepatic fibrosis pathway is interrelated with several other pathways, including NFκB, vascular endothelial growth factor (VEGF), platelet-derived growth factor (PDGF), and insulin-like growth factor 1 (IGF1) signaling, which have been previously shown to play a role in PHEOs/PGLs or other tumors.
For instance, VHL negatively regulates NFκB [36], and accordingly, VHL mutations strengthen NFκB activity, which causes apoptosis resistance, increased angiogenesis, and therapy resistance in renal cell carcinomas. Recently, inhibition of NFκB signaling in a murine PHEO model has been shown to cause cell death and significant decrease in metastatic spread [37]. Similarly, NFκB inhibition showed promising results in treatment of renal cell carcinomas [38]. Our data indicate that particularly in cluster B PHEOs/PGLs targeting the NFκB pathway provides a promising new therapeutic approach.
Pseudohypoxic PHEOs/PGLs often appear highly vascular, and anti-angiogenic treatment has been suggested [39]. Induction of angiogenic transcripts/VEGF signaling has been described in response to NFκB as well as HIF activation. Here, we report increased expression of VEGF signaling-related transcripts in cluster B tumors, i.e., VEGFC, FLT1/VEGFR1, KDR/VEGFR2, and FLT4/VEGFR3. Increased expression of VEGF, FLT1/VEGFR1, and KDR/VEGFR2 has been previously reported for a group of PHEOs/PGLs, including one VHL case [40]. While potentially all pseudohypoxic PHEOs/PGLs show elevated expression of angiogenesis genes, the particular genes mentioned here appear to be exclusively upregulated in VHL and SDHD-HN PGLs. Thus, targeting VEGFR1, 2, and 3 in cluster B PHEOs/PGLs appears to be a promising approach.
As an aspect of hepatic fibrosis, PDGF signaling showed several elevated players in cluster B. PDGFB is a well-described oncogene [41] and targeting PDGFRB and VEGF led to symptom relieve and tumor shrinkage in a VHL patient with multiple tumors, including a metastatic PGL [42]. Activating mutations in PDGFRA is often seen in gastrointestinal stromal tumors.
IGF signaling has been previously shown to play a role in the pathogenesis of PHEOs/PGLs. Here, we show elevated expression of IGF1 and several binding proteins (IGFBP4, IGFBP5, and CTGF/IGFBP8) in cluster B tumors. Fottner et al. have previously reported increased expression of IGF1 receptor (IGFR1/FCGR1A) in human PHEOs, including three VHL cases [43]. Inactivation of VHL has been shown to cause increased IGFR1/FCGR1A expression [44], which contributes to renal tumor development. Increased IGFR1/FCGR1A levels have been associated with chemoresistance and IGF1 signaling inhibition improved treatment outcome in renal cancer [45]. Differences in IGFR1/FCGR1A expression between the two clusters was not evident; however, IGFR1/FCGR1A was selected into the minimum subset of classifiers for characterization of SDHB PHEOs/PGLs. QRT-PCR revealed significantly elevated expression in SDHDHN compared to SDHD-AT PGLs (Figure W1). Expressions of IGF1 itself as well as several regulating IGF-binding proteins however were elevated in cluster B tumors. Up-regulation of IGFBP4 in nine PHEOs of unknown genetic background has been reported [46]. Fernandez et al. observed that IGF1 signaling is crucial in cell proliferation and tumor maintenance in a mouse model based on mouse PHEO cells [47]. Further studies are mandated to explore the role of IGF signaling in cluster B tumors and its potential value as a therapeutic target.
Minimum Subset of Genes to Characterize PHEO/PGL Subtypes
Selected genes of the minimum subset will be discussed here for their potential role in the distinct pseudohypoxic PHEOs/PGLs. In agreement with previous reports, we detected increased EGLN3/PHD2 transcript in VHL PHEOs compared to the other groups [8,48]. EGLN3 protein level however did not differ between VHL and SDHB PHEOs/PGLs [8]. Inhibition of EGLN3-mediated culling has been proposed as a common mechanism in PHEO/PGL development [49]. Thus, despite its elevated mRNA expression in VHL-derived PHEOs, EGLN3 function may be inhibited, either due to JUNB-mediated inhibition or inadequate protein expression.
In addition to a role in mediating apoptosis, EGLN3/PHD2 also acts as HIF PHD. As mentioned above, VHL mutations can in appropriately stabilize HIFα. As HIF target gene, particularly responding to HIF2α stabilization, EGLN3 induction is thus not surprising in VHL-derived tumors [50]. However, despite HIF stabilization EGLN3/PHD2 transcript levels appear normal in other pseudohypoxic PHEOs/PGLs [7,9]. Elevation of EGLN3/PHD2 mRNA levels exclusively in VHL PHEOs/PGLs further supports distinct manifestations of pseudohypoxia.
In further agreement with our data, differential expression between VHL and SDHB PHEOs/PGLs has been previously reported for RARRES2, CGNL1, and ELOVL7 [8].
Two genes that we found to be significantly decreased in SDHD-HN PGLs compared to all other groups were SLC18A1, the chromaffin granule amine transporter also known as VMAT1, and the intermediate neurofilament NEFM (also called NFM, NF-M, or NF3). Previously, varying levels of SLC18A1/VMAT1 expression have been reported for PHEOs/PGLs [51,52]. In agreement with our data, SLC18A1/VMAT1 has been previously shown to be more highly expressed in VHL PHEOs than normal adrenal [53]. NEFM may be mandatory in carotid body maturation under hypoxia [54] and has been proposed as marker for neuroendocrine tumors [55].
In combination, the minimum subset of classifiers could not clearly differentiate tumors within cluster A. Nevertheless, four individual genes were characteristic for SDHD-AT relative to all other PHEOs/PGLs but not normal medulla. Overall, our findings indicate that the differences in expression profiles of SDHB and SDHD-AT PHEOs/PGLs are subtle and identification of distinguishing features may require large analysis groups, perhaps homogenous to the level of particular mutation and/or location.
In disregard of their failure as characteristic genes for one particular subgroup, two candidate genes were more highly expressed in all SDHx cases relative to adrenal medulla and VHL-derived PHEOs. First, LGR5 (aliases: GPR49, HG38, FEX, and GPR67) has been recognized as stem cell marker in multiple adult tissues and cancer [56]. Its increased expression specifically in SDHB-derived PHEOs/PGLs has been previously reported [9]. Contrarily, our data indicate similarly elevated LGR5 levels in SDHB and SDHD PGLs (Figure 6). Due to its cell surface location and potential role in tumor-initiating stem cells, this receptor may provide an excellent therapeutic target in SDHx PHEOs/PGLs. Second, the lipogenic ELOVL7 has been reported to be involved in prostate cancer growth [57].
Summary
Pseudohypoxic PHEOs/PGLs can be stratified according to different hereditary backgrounds and/or tumor locations. Surprisingly, SDHD-AT and SDHD-HN PHEOs/PGLs do not cluster together but with SDHB (cluster A) and VHL PHEOs/PGLs (cluster B), respectively. Despite their distinct risks of malignancy, SDHB and SDHD-AT PHEOs/PGLs failed to separate into distinct subclusters. This indicates surprising agreement in their expression profiles and prevented the identification of distinct gene signatures.
The majority of upregulated pathways were related to energy metabolism for cluster A and to tissue fibrosis and immune function for cluster B. Overall, OXPHOS genes were more highly expressed in cluster A tumors; however, which particular subunits were overexpressed differed for SDHD-AT and SDHB PHEOs/PGLs. Cluster B tumors showed increased tissue fibrosis, including NFκB-, PDGF-, IGF-, IL1-, and VEGF-related gene expression.
Classification based on a minimum subset of 36 genes maintained separability of the groups into two main clusters, with cluster B further dividing into subclusters in agreement with the respective mutation types. Subgroup classification based on the minimum subset of 36 genes improved error rates. Expression levels of 35 genes from the minimum subset of classifiers were essentially confirmed by qRT-PCR. Evaluation of the potential of each of those genes to correctly classify the different subtypes of pseudohypoxic PHEOs/PGLs identified expression of nine transcripts as characteristic for VHL, 11 for SDHD-HN, and four for SDHD-AT PHEOs/PGLs; however, none successfully classified SDHB PHEOs/PGLs.
In conclusion, pseudohypoxic PHEOs/PGLs show distinct expression profiles, which should be taken into account for improvement of diagnosis as well as the development of customized therapeutic options. Special consideration should be given to the apparently distinct manifestations of SDHD mutations in sympathetic and parasympathetic tissues.
Supplementary Material
Acknowledgments
We thank Robert Wesley for in-depth discussion of the data.
Abbreviations
- AT
abdominal/thoracic
- CSS
Carney-Stratakis Syndrome
- HIF2α
hypoxia-inducible factor 2α
- HN
head/neck
- IPA
Ingenuity Pathway Analysis
- NF1/NF1
neurofibromatosis 1 syndrome/gene
- OXPHOS
oxidative phosphorylation
- PAMR
prediction analysis for microarray
- PGL
paraganglioma
- PGL1, 2, 3, 4
familial PGL types 1, 2, 3, 4
- PHD
prolyl hydroxylase
- PHD2/EGLN1
prolyl hydroxylase 2
- PHEO
pheochromocytoma
- qRT-PCR
quantitative real-time polymerase chain reaction
- SAM
significance analysis of microarray
- SDH
succinate dehydrogenase
- SDHA
SDH subunit A
- SDHAF2
SDH complex assembly factor 2
- SDHB
SDH subunit B
- SDHC
SDH subunit C
- SDHD
SDH subunit D
- TMEM127
transmembrane protein 127
- VHL/VHL
von Hippel-Lindau syndrome/gene
Footnotes
This study was funded by the Eunice Kennedy Shriver National Institute of Child Health and Human Development and the National Human Genome Research Institute, National Institutes of Health (Bethesda, MD). The authors have nothing to disclose.
This article refers to supplementary materials, which are designated by Tables W1 to W3 and Figures W1 to W3 and are available online at www.neoplasia.com
References
- 1.Welander J, Soderkvist P, Gimm O. Genetics and clinical characteristics of hereditary pheochromocytomas and paragangliomas. Endocr Relat Cancer. 2011;18:R253–R276. doi: 10.1530/ERC-11-0170. [DOI] [PubMed] [Google Scholar]
- 2.Zhuang Z, Yang C, Lorenzo F, Merino M, Fojo T, Kebebew E, Popovic V, Stratakis CA, Prchal JT, Pacak K. Somatic HIF2A gain-of-function mutations in paraganglioma with polycythemia. N Engl J Med. 2012;367:922–930. doi: 10.1056/NEJMoa1205119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fishbein L, Nathanson KL. Pheochromocytoma and paraganglioma: understanding the complexities of the genetic background. Cancer Genet. 2012;205:1–11. doi: 10.1016/j.cancergen.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaelin WG., Jr The von Hippel-Lindau tumor suppressor gene and kidney cancer. Clin Cancer Res. 2004;10:6290S–6295S. doi: 10.1158/1078-0432.CCR-sup-040025. [DOI] [PubMed] [Google Scholar]
- 5.Guzy RD, Sharma B, Bell E, Chandel NS, Schumacker PT. Loss of the SdhB, but not the SdhA, subunit of complex II triggers reactive oxygen species-dependent hypoxia-inducible factor activation and tumorigenesis. Mol Cell Biol. 2008;28:718–731. doi: 10.1128/MCB.01338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pollard PJ, Briere JJ, Alam NA, Barwell J, Barclay E, Wortham NC, Hunt T, Mitchell M, Olpin S, Moat SJ, et al. Accumulation of Krebs cycle intermediates and over-expression of HIF1alpha in tumours which result from germ-line FH and SDH mutations. Hum Mol Genet. 2005;14:2231–2239. doi: 10.1093/hmg/ddi227. [DOI] [PubMed] [Google Scholar]
- 7.Favier J, Briere JJ, Burnichon N, Riviere J, Vescovo L, Benit P, Giscos-Douriez I, De Reynies A, Bertherat J, Badoual C, et al. The Warburg effect is genetically determined in inherited pheochromocytomas. PLoS One. 2009;4:e7094. doi: 10.1371/journal.pone.0007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lopez-Jimenez E, Gomez-Lopez G, Leandro-Garcia LJ, Munoz I, Schiavi F, Montero-Conde C, de Cubas AA, Ramires R, Landa I, Leskela S, et al. Research resource: transcriptional profiling reveals different pseudohypoxic signatures in SDHB and VHL-related pheochromocytomas. Mol Endocrinol. 2010;24:2382–2391. doi: 10.1210/me.2010-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burnichon N, Vescovo L, Amar L, Libe R, de Reynies A, Venisse A, Jouanno E, Laurendeau I, Parfait B, Bertherat J, et al. Integrative genomic analysis reveals somatic mutations in pheochromocytoma and paraganglioma. Hum Mol Genet. 2011;20:3974–3985. doi: 10.1093/hmg/ddr324. [DOI] [PubMed] [Google Scholar]
- 10.Buffet A, Venisse A, Nau V, Roncellin I, Boccio V, Le Pottier N, Boussion M, Travers C, Simian C, Burnichon N, et al. A decade (2001-2010) of genetic testing for pheochromocytoma and paraganglioma. Horm Metab Res. 2012;44:359–366. doi: 10.1055/s-0032-1304594. [DOI] [PubMed] [Google Scholar]
- 11.Lefebvre S, Borson-Chazot F, Boutry-Kryza N, Wion N, Schillo F, Peix JL, Brunaud L, Finat A, Calender A, Giraud S. Screening of mutations in genes that predispose to hereditary paragangliomas and pheochromocytomas. Horm Metab Res. 2012;44:334–338. doi: 10.1055/s-0032-1306308. [DOI] [PubMed] [Google Scholar]
- 12.Mannelli M, Castellano M, Schiavi F, Filetti S, Giacche M, Mori L, Pignataro V, Bernini G, Giache V, Bacca A, et al. Clinically guided genetic screening in a large cohort of Italian patients with pheochromocytomas and/or functional or nonfunctional paragangliomas. J Clin Endocrinol Metab. 2009;94:1541–1547. doi: 10.1210/jc.2008-2419. [DOI] [PubMed] [Google Scholar]
- 13.Neumann HP, Erlic Z, Boedeker CC, Rybicki LA, Robledo M, Hermsen M, Schiavi F, Falcioni M, Kwok P, Bauters C, et al. Clinical predictors for germline mutations in head and neck paraganglioma patients: cost reduction strategy in genetic diagnostic process as fall-out. Cancer Res. 2009;69:3650–3656. doi: 10.1158/0008-5472.CAN-08-4057. [DOI] [PubMed] [Google Scholar]
- 14.Hensen EF, Siemers MD, Jansen JC, Corssmit EP, Romijn JA, Tops CM, van der Mey AG, Devilee P, Cornelisse CJ, Bayley JP, et al. Mutations in SDHD are the major determinants of the clinical characteristics of Dutch head and neck paraganglioma patients. Clin Endocrinol (Oxf) 2011;75:650–655. doi: 10.1111/j.1365-2265.2011.04097.x. [DOI] [PubMed] [Google Scholar]
- 15.Kim K, Zakharkin SO, Allison DB. Expectations, validity, and reality in gene expression profiling. J Clin Epidemiol. 2010;63:950–959. doi: 10.1016/j.jclinepi.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fliedner SM, Breza J, Kvetnansky R, Powers JF, Tischler AS, Wesley R, Merino M, Lehnert H, Pacak K. Tyrosine hydroxylase, chromogranin A, and steroidogenic acute regulator as markers for successful separation of human adrenal medulla. Cell Tissue Res. 2010;340:607–612. doi: 10.1007/s00441-010-0965-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.R Development Core Team, author. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2012. ISBN 3-900051-07-0, Available at: http://www.R-project.org. [Google Scholar]
- 18.Schuetz CS, Bonin M, Clare SE, Nieselt K, Sotlar K, Walter M, Fehm T, Solomayer E, Riess O, Wallwiener D, et al. Progression-specific genes identified by expression profiling of matched ductal carcinomas in situ and invasive breast tumors, combining laser capture microdissection and oligonucleotide microarray analysis. Cancer Res. 2006;66:5278–5286. doi: 10.1158/0008-5472.CAN-05-4610. [DOI] [PubMed] [Google Scholar]
- 19.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci USA. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shankavaram UT, Reinhold WC, Nishizuka S, Major S, Morita D, Chary KK, Reimers MA, Scherf U, Kahn A, Dolginow D, et al. Transcript and protein expression profiles of the NCI-60 cancer cell panel: an integromic micro-array study. Mol Cancer Ther. 2007;6:820–832. doi: 10.1158/1535-7163.MCT-06-0650. [DOI] [PubMed] [Google Scholar]
- 22.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waldmann J, Slater EP, Langer P, Buchholz M, Ramaswamy A, Walz MK, Schmid KW, Feldmann G, Bartsch DK, Fendrich V. Expression of the transcription factor snail and its target gene twist are associated with malignancy in pheochromocytomas. Ann Surg Oncol. 2009;16:1997–2005. doi: 10.1245/s10434-009-0480-y. [DOI] [PubMed] [Google Scholar]
- 24.Loriot C, Burnichon N, Gadessaud N, Vescovo L, Amar L, Libe R, Bertherat J, Plouin PF, Jeunemaitre X, Gimenez-Roqueplo AP, et al. Epithelial to mesenchymal transition is activated in metastatic pheochromocytomas and paragangliomas caused by SDHB gene mutations. J Clin Endocrinol Metab. 2012;97:E954–E962. doi: 10.1210/jc.2011-3437. [DOI] [PubMed] [Google Scholar]
- 25.Hensen EF, Goeman JJ, Oosting J, Van der Mey AG, Hogendoorn PC, Cremers CW, Devilee P, Cornelisse CJ. Similar gene expression profiles of sporadic, PGL2-, and SDHD-linked paragangliomas suggest a common pathway to tumorigenesis. BMC Medical Genomics. 2009;2:25–35. doi: 10.1186/1755-8794-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Duinen N, Steenvoorden D, Kema IP, Jansen JC, Vriends AH, Bayley JP, Smit JW, Romijn JA, Corssmit EP. Increased urinary excretion of 3-methoxytyramine in patients with head and neck paragangliomas. J Clin Endocrinol Metab. 2010;95:209–214. doi: 10.1210/jc.2009-1632. [DOI] [PubMed] [Google Scholar]
- 27.Eisenhofer G, Lenders JW, Timmers H, Mannelli M, Grebe SK, Hofbauer LC, Bornstein SR, Tiebel O, Adams K, Bratslavsky G, et al. Measurements of plasma methoxytyramine, normetanephrine, and metanephrine as discriminators of different hereditary forms of pheochromocytoma. Clin Chem. 2011;57:411–420. doi: 10.1373/clinchem.2010.153320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fliedner SM, Kaludercic N, Jiang XS, Hansikova H, Hajkova Z, Sladkova J, Limpuangthip A, Backlund PS, Wesley R, Martiniova L, et al. Warburg effect's manifestation in aggressive pheochromocytomas and paragangliomas: insights from a mouse cell model applied to human tumor tissue. PLoS One. 2012;7:e40949. doi: 10.1371/journal.pone.0040949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Semenza GL. HIF-1 mediates the Warburg effect in clear cell renal carcinoma. J Bioenerg Biomembr. 2007;39:231–234. doi: 10.1007/s10863-007-9081-2. [DOI] [PubMed] [Google Scholar]
- 30.Qu A, Taylor M, Xue X, Matsubara T, Metzger D, Chambon P, Gonzalez FJ, Shah YM. Hypoxia-inducible transcription factor 2α promotes steatohepatitis through augmenting lipid accumulation, inflammation, and fibrosis. Hepatology. 2011;54:472–483. doi: 10.1002/hep.24400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schietke RE, Hackenbeck T, Tran M, Gunther R, Klanke B, Warnecke CL, Knaup KX, Shukla D, Rosenberger C, Koesters R, et al. Renal tubular HIF-2α expression requires VHL inactivation and causes fibrosis and cysts. PLoS One. 2012;7:e31034. doi: 10.1371/journal.pone.0031034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grosfeld A, Stolze IP, Cockman ME, Pugh CW, Edelmann M, Kessler B, Bullock AN, Ratcliffe PJ, Masson N. Interaction of hydroxylated collagen IV with the von Hippel-Lindau tumor suppressor. J Biol Chem. 2007;282:13264–13269. doi: 10.1074/jbc.M611648200. [DOI] [PubMed] [Google Scholar]
- 33.Kurban G, Duplan E, Ramlal N, Hudon V, Sado Y, Ninomiya Y, Pause A. Collagen matrix assembly is driven by the interaction of von Hippel-Lindau tumor suppressor protein with hydroxylated collagen IV α 2. Oncogene. 2008;27:1004–1012. doi: 10.1038/sj.onc.1210709. [DOI] [PubMed] [Google Scholar]
- 34.Kurban G, Hudon V, Duplan E, Ohh M, Pause A. Characterization of a von Hippel Lindau pathway involved in extracellular matrix remodeling, cell invasion, and angiogenesis. Cancer Res. 2006;66:1313–1319. doi: 10.1158/0008-5472.CAN-05-2560. [DOI] [PubMed] [Google Scholar]
- 35.Pollard PJ, El-Bahrawy M, Poulsom R, Elia G, Killick P, Kelly G, Hunt T, Jeffery R, Seedhar P, Barwell J, et al. Expression of HIF-1α, HIF-2α (EPAS1), and their target genes in paraganglioma and pheochromocytoma with VHL and SDH mutations. J Clin Endocrinol Metab. 2006;91:4593–4598. doi: 10.1210/jc.2006-0920. [DOI] [PubMed] [Google Scholar]
- 36.Morais C, Gobe G, Johnson DW, Healy H. The emerging role of nuclear factor kappa B in renal cell carcinoma. Int J Biochem Cell Biol. 2011;43:1537–1549. doi: 10.1016/j.biocel.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 37.Pacak K, Sirova M, Giubellino A, Lencesova L, Csaderova L, Laukova M, Hudecova S, Krizanova O. NF-κB inhibition significantly upregulates the norepinephrine transporter system, causes apoptosis in pheochromocytoma cell lines and prevents metastasis in an animal model. Int J Cancer. 2012;131:2445–2455. doi: 10.1002/ijc.27524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sourbier C, Danilin S, Lindner V, Steger J, Rothhut S, Meyer N, Jacqmin D, Helwig JJ, Lang H, Massfelder T. Targeting the nuclear factor-κB rescue pathway has promising future in human renal cell carcinoma therapy. Cancer Res. 2007;67:11668–11676. doi: 10.1158/0008-5472.CAN-07-0632. [DOI] [PubMed] [Google Scholar]
- 39.Favier J, Igaz P, Burnichon N, Amar L, Libe R, Badoual C, Tissier F, Bertherat J, Plouin PF, Jeunemaitre X, et al. Rationale for anti-angiogenic therapy in pheochromocytoma and paraganglioma. Endocr Pathol. 2012;23:34–42. doi: 10.1007/s12022-011-9189-0. [DOI] [PubMed] [Google Scholar]
- 40.Takekoshi K, Isobe K, Yashiro T, Hara H, Ishii K, Kawakami Y, Nakai T, Okuda Y. Expression of vascular endothelial growth factor (VEGF) and its cognate receptors in human pheochromocytomas. Life Sci. 2004;74:863–871. doi: 10.1016/j.lfs.2003.07.036. [DOI] [PubMed] [Google Scholar]
- 41.Au PY, Martin N, Chau H, Moemeni B, Chia M, Liu FF, Minden M, Yeh WC. The oncogene PDGF-B provides a key switch from cell death to survival induced by TNF. Oncogene. 2005;24:3196–3205. doi: 10.1038/sj.onc.1208516. [DOI] [PubMed] [Google Scholar]
- 42.Jimenez C, Cabanillas ME, Santarpia L, Jonasch E, Kyle KL, Lano EA, Matin SF, Nunez RF, Perrier ND, Phan A, et al. Use of the tyrosine kinase inhibitor sunitinib in a patient with von Hippel-Lindau disease: targeting angiogenic factors in pheochromocytoma and other von Hippel-Lindau disease-related tumors. J Clin Endocrinol Metab. 2009;94:386–391. doi: 10.1210/jc.2008-1972. [DOI] [PubMed] [Google Scholar]
- 43.Fottner C, Minnemann T, Kalmbach S, Weber MM. Overexpression of the insulin-like growth factor I receptor in human pheochromocytomas. J Mol Endocrinol. 2006;36:279–287. doi: 10.1677/jme.1.01975. [DOI] [PubMed] [Google Scholar]
- 44.Yuen JS, Cockman ME, Sullivan M, Protheroe A, Turner GD, Roberts IS, Pugh CW, Werner H, Macaulay VM. The VHL tumor suppressor inhibits expression of the IGF1R and its loss induces IGF1R upregulation in human clear cell renal carcinoma. Oncogene. 2007;26:6499–6508. doi: 10.1038/sj.onc.1210474. [DOI] [PubMed] [Google Scholar]
- 45.Yuen JS, Akkaya E, Wang Y, Takiguchi M, Peak S, Sullivan M, Protheroe AS, Macaulay VM. Validation of the type 1 insulin-like growth factor receptor as a therapeutic target in renal cancer. Mol Cancer Ther. 2009;8:1448–1459. doi: 10.1158/1535-7163.MCT-09-0101. [DOI] [PubMed] [Google Scholar]
- 46.Ilvesmaki V, Liu J, Heikkila P, Kahri AI, Voutilainen R. Expression of insulin-like growth factor binding protein 1–6 genes in adrenocortical tumors and pheochromocytomas. Horm Metab Res. 1998;30:619–623. doi: 10.1055/s-2007-978945. [DOI] [PubMed] [Google Scholar]
- 47.Fernandez MC, Venara M, Nowicki S, Chemes HE, Barontini M, Pennisi PA. Igf-I regulates pheochromocytoma cell proliferation and survival in vitro and in vivo. Endocrinology. 2012;153:3724–3734. doi: 10.1210/en.2012-1107. [DOI] [PubMed] [Google Scholar]
- 48.Eisenhofer G, Huynh TT, Pacak K, Brouwers FM, Walther MM, Linehan WM, Munson PJ, Mannelli M, Goldstein DS, Elkahloun AG. Distinct gene expression profiles in norepinephrine- and epinephrine-producing hereditary and sporadic pheochromocytomas: activation of hypoxia-driven angiogenic pathways in von Hippel-Lindau syndrome. Endocr Relat Cancer. 2004;11:897–911. doi: 10.1677/erc.1.00838. [DOI] [PubMed] [Google Scholar]
- 49.Lee S, Nakamura E, Yang H, Wei W, Linggi MS, Sajan MP, Farese RV, Freeman RS, Carter BD, Kaelin WG, Jr, et al. Neuronal apoptosis linked to EglN3 prolyl hydroxylase and familial pheochromocytoma genes: developmental culling and cancer. Cancer Cell. 2005;8:155–167. doi: 10.1016/j.ccr.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 50.Aprelikova O, Chandramouli GV, Wood M, Vasselli JR, Riss J, Maranchie JK, Linehan WM, Barrett JC. Regulation of HIF prolyl hydroxylases by hypoxia-inducible factors. J Cell Biochem. 2004;92:491–501. doi: 10.1002/jcb.20067. [DOI] [PubMed] [Google Scholar]
- 51.Kolby L, Bernhardt P, Levin-Jakobsen AM, Johanson V, Wangberg B, Ahlman H, Forssell-Aronsson E, Nilsson O. Uptake of meta-iodobenzylguanidine in neuroendocrine tumours is mediated by vesicular monoamine transporters. Br J Cancer. 2003;89:1383–1388. doi: 10.1038/sj.bjc.6601276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saveanu A, Muresan M, De Micco C, Taieb D, Germanetti AL, Sebag F, Henry JF, Brunaud L, Enjalbert A, Weryha G, et al. Expression of somatostatin receptors, dopamine D2 receptors, noradrenaline transporters, and vesicular monoamine transporters in 52 pheochromocytomas and paragangliomas. Endocr Relat Cancer. 2011;18:287–300. doi: 10.1530/ERC-10-0175. [DOI] [PubMed] [Google Scholar]
- 53.Huynh TT, Pacak K, Brouwers FM, Abu-Asab MS, Worrell RA, Walther MM, Elkahloun AG, Goldstein DS, Cleary S, Eisenhofer G. Different expression of catecholamine transporters in phaeochromocytomas from patients with von Hippel-Lindau syndrome and multiple endocrine neoplasia type 2. Eur J Endocrinol. 2005;153:551–563. doi: 10.1530/eje.1.01987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nurse CA, Vollmer C. Role of basic FGF and oxygen in control of proliferation, survival, and neuronal differentiation in carotid body chromaffin cells. Dev Biol. 1997;184:197–206. doi: 10.1006/dbio.1997.8539. [DOI] [PubMed] [Google Scholar]
- 55.Hofsli E, Wheeler TE, Langaas M, Laegreid A, Thommesen L. Identification of novel neuroendocrine-specific tumour genes. Br J Cancer. 2008;99:1330–1339. doi: 10.1038/sj.bjc.6604565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 57.Tamura K, Makino A, Hullin-Matsuda F, Kobayashi T, Furihata M, Chung S, Ashida S, Miki T, Fujioka T, Shuin T, et al. Novel lipogenic enzyme ELOVL7 is involved in prostate cancer growth through saturated long-chain fatty acid metabolism. Cancer Res. 2009;69:8133–8140. doi: 10.1158/0008-5472.CAN-09-0775. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.