Summary
The nematode C. elegans consumes benign bacteria such as E. coli and is repelled by pathogens and toxins. Here we show that RNAi and toxin-mediated disruption of core cellular activities, including translation, respiration, and protein turnover, stimulates behavioral avoidance of attractive E. coli. RNAi of such essential processes also induces expression of detoxification and innate immune response genes in the absence of toxins or pathogens. Disruption of core processes in non-neuronal tissues can stimulate aversion behavior, revealing a neuroendocrine axis of control. Microbial avoidance requires serotonergic and Jnk kinase signaling. We propose that surveillance pathways oversee critical cellular activities to detect pathogens, many of which deploy toxins and virulence factors to disrupt these same host pathways. Variation in cellular surveillance and endocrine pathways controlling behavior, detoxification and immunity selected by past toxin or microbial interactions could underlie aberrant responses to foods, medicines, and microbes.
Introduction
Organisms compete for finite resources, driving the evolution of interspecies conflict. Host-pathogen relationships are an example of this conflict, where the volley of measures and countermeasures is complex and highly evolved. For example, microbial pathogens release an array of toxins or virulence factors aimed at crippling host functions and defenses. To counter pathogen or toxic attack, the host mobilizes immunity and detoxification responses. Chemical and protein toxins are wildly diverse, and operate by sabotaging critical cellular activities such as transcription, translation, or mitochondrial respiration. If the host is to survive such an assault, it must first detect the attack, then mobilize xenobiotic detoxification programs, compensatory mechanisms to rescue disabled cellular processes, and immunity-mediated eradication of the toxin’s microbial source. A major question is how hosts detect pathogens and xenobiotics. One possibility is that cellular receptors (like nuclear hormone receptors and GPCRs, or LRR proteins in plants) recognize specific toxins and virulence factors (Jones and Dangl, 2006). Such a solution suffers from the diversity of chemical and protein toxins and virulence factors that could escape specific receptors, and the predicted promiscuity of more generic receptors. Another solution is that hosts monitor the damage induced by toxins and virulence factors to initiate detoxification and immune defenses (Jones and Dangl, 2006).
C. elegans is a motile, soil-dwelling metazoan with astute sensory capabilities that allows them to escape a broad range of environmental hazards, including pathogens (Schulenburg and Ewbank, 2007) (Pujol et al., 2001)(Zhang et al., 2005). Here we show that surveillance pathways in C. elegans monitor core cellular activities and interpret disruption of core activities as a pathogenic attack, engaging behavioral, immune and detoxification responses. We demonstrate that C. elegans aversion behavior is a common response to RNAi or toxin-induced disruption of many vital cellular processes. RNAi of cellular components also stimulated detoxification and innate immune defenses, suggesting a generalizable mechanism enabling organisms to sense invading microbes. Aversion required an intact neurosensory system, but could be produced by disruption of cellular activities in non-neuronal tissues, revealing that animals integrate internal with sensory cues to coordinate behavioral defenses. A JNK kinase cascade is a key element in this surveillance system.
Results
Disruption of core cellular processes by RNAi stimulates microbial aversion behavior
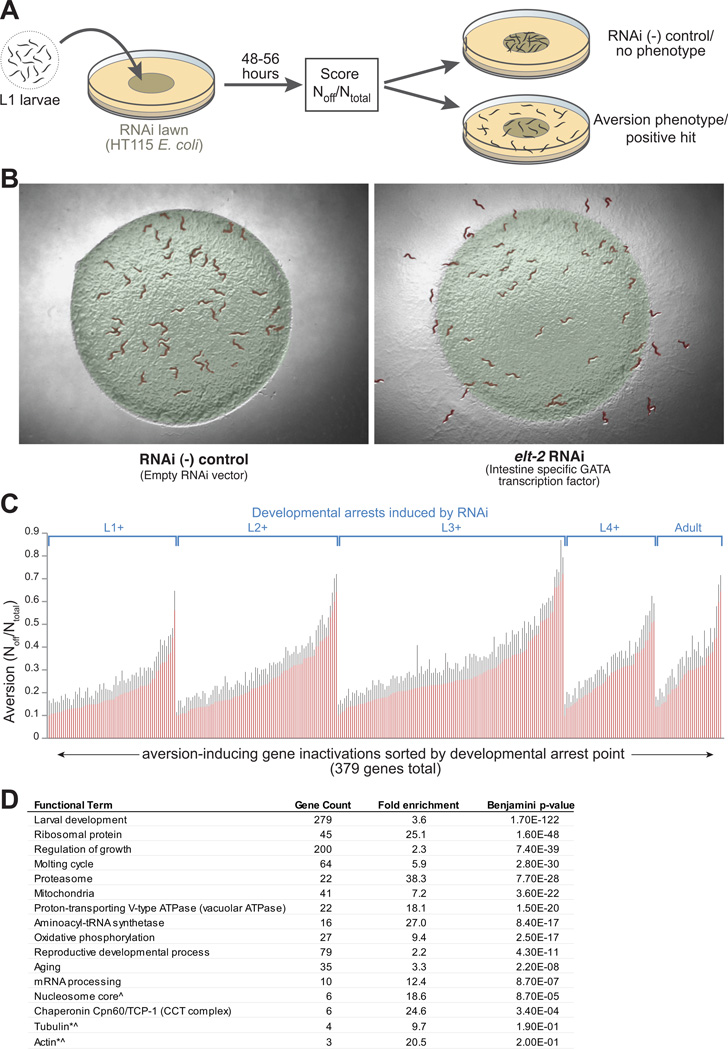
E. coli is a nutritive laboratory food source attractive to C. elegans. RNAi is performed by growing animals on E. coli expressing dsRNAs complementary to C. elegans target genes. RNAi libraries allow the inactivation of ~90% of the C. elegans genome (Kamath et al., 2003). We tested whether RNAi of a small set of C. elegans genes could alter the affinity of animals for an E. coli lawn, and found that inactivation of many genes essential for normal growth and development stimulated a behavioral avoidance phenotype (Figures 1A-B, Figures S1A-B and Movies S1–3), a response we refer to as “microbial aversion behavior.” The aversion phenotype could reflect a programmed behavioral response to inactivation of essential processes, or simply a reduction in the nutritive quality of dsRNA-expressing E. coli. To distinguish these possibilities, we tested whether RNAi of these essential genes produced the aversion phenotype in RNAi-defective animals. The RNAi defective C. elegans mutants sid-1(qt9) and rde-1(ne219) failed to exhibit the aversion response when grown on RNAi bacteria repulsive to wild type controls (Figure S1A). In the absence of dsRNA targeting essential C. elegans pathways, basal frequencies of food avoidance were very low. For animals raised on RNAi control bacteria, aversion was undetectable (0+/-0%) in the first 3 larval stages (L1, L2, L3), became weakly detectable in the L4 stage (0.5% +/− 0.5%), and increased to ~5% in adults (+/-3%) (Figure S1C).
Figure 1. Inactivation of essential cellular pathways stimulates microbial avoidance behavior.
(A) Schematic of RNAi microbial aversion assay.
(B) Example of control and aversion phenotypes after 48hr of growth on elt-2 or RNAi control bacteria. elt-2 encodes a transcription factor necessary for gut development and homeostasis.
(C) Developmental stage- and rank-ordered aversion levels +SEM for 379 gene inactivations (of 4,062 screened) exhibiting aversion ≥10% off bacteria at 48-58hr of growth on RNAi bacteria.
(D) DAVID bioinformatic analysis of aversion genes showing enrichment for specific functional categories. *enriched gene classes that were not statistically significant due to small N. ^high intra-class homology that could produce an elevated false positive rate due to off-target RNAi effects.
To identify disruptions capable of evoking an aversion response, we inactivated 4,062 essential and metabolic genes. Essential genes were defined as those required for first- or second-generation viability in whole genome screens of wild type C. elegans (N2) or enhanced RNAi strains. Metabolic genes were selected based on assembled GO term annotations. We found that 379 of the 4,062 gene inactivations assayed stimulated bacterial aversion >10% (Table S1A). Many of the genes identified also caused larval delay or arrest phenotypes (Figure 1C, S1D-E; Table S1A). The frequency of developmental phenotypes in the aversion gene set was significantly higher than in the set of 4,062 genes tested (85% vs. 21% of genes, p<.0001, chi-square test) (Figure S1D-E). There was a ~10-fold increased frequency of aversion phenotypes in the essential gene set compared to a set of 192 randomly selected genes (p<.0001, chi-square test) (data not shown).
DAVID gene enrichment analysis (Huang et al., 2009) revealed that inactivation of genes from particular functional classes preferentially induced the aversion response (Figure 1D). Enriched functional classes were comprised of genes involved in protein translation (25- and 27-fold enrichment of ribosome components and tRNA synthetases, respectively), the molting cycle (6-fold enriched), proteasome machinery (38-fold enriched), TCP chaperonins (25-fold enriched), mitochondria (7-fold enriched), vacuolar ATPases (18-fold enriched), mRNA processing (12-fold enriched), and histones (19-fold enriched). While not statistically significant (possibly due to small sample size), cytoskeleton components were also enriched in the aversion gene set (~10-fold and 20-fold enrichment for tubulin and actins, respectively). Moreover, we found that aversion-associated genes exhibited 2-fold higher conservation between C. elegans and Homo sapiens than the screen set, and 4-fold higher conservation than the entire C. elegans genome, suggesting that ultra-conserved cellular processes are more likely to be subject to a form of surveillance that controls adaptive behaviors (Table S1B). Human homologs of many aversion genes have also been implicated in human disease (Table S1C).
Chemicals and bacterial toxins stimulate microbial aversion
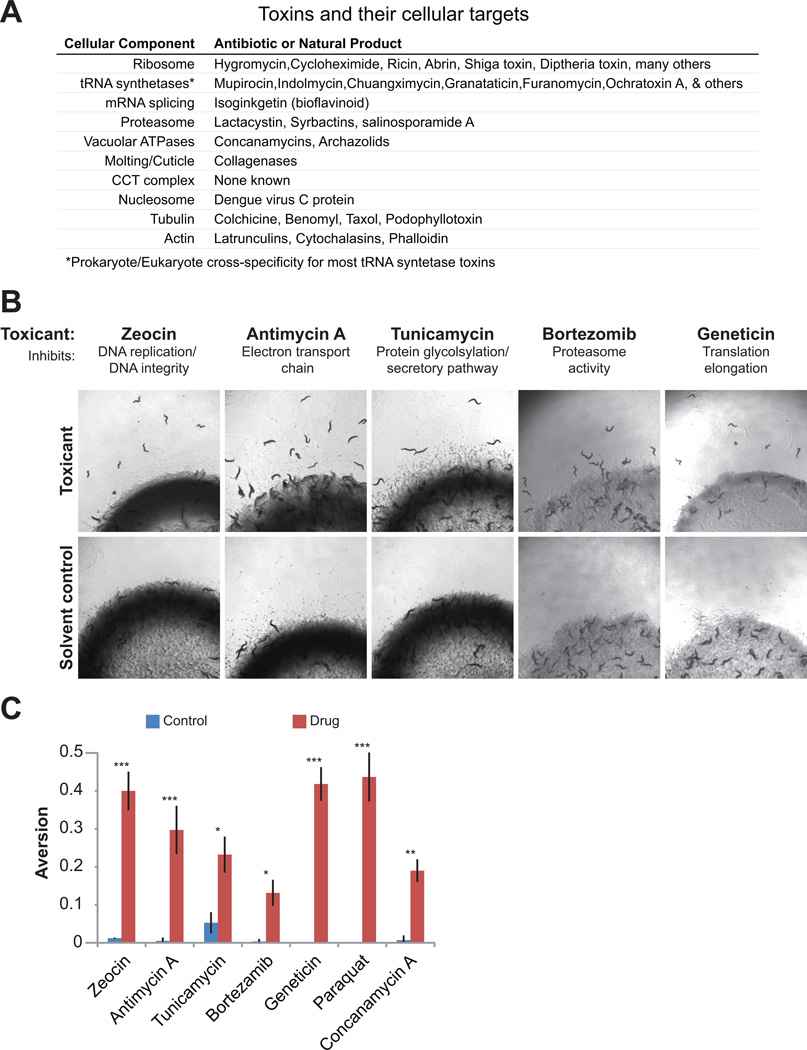
Many of the cellular processes identified by our RNAi screen are known targets of microbial toxins (Figure 2A). For example, the eukaryotic ribosome is inhibited by dozens of pathogen-derived toxins, including hygromycin, cycloheximide, and diptheria toxins. The electron transport chain (ETC), the proteasome, tRNA synthetases, mRNA splicing, vacuolar ATPases, tubulins and actins were identified by our screen and are also targets of microbial toxins. Because wild C. elegans species live in microbe-rich environments, we hypothesized that RNAi of critical cellular pathways mimics the impact of microbial toxins on cellular physiology, and that surveillance pathways within the animal oversee cellular activities as a broadly adaptable means of toxin detection. Under these conditions, activation of an aversive behavioral program represents an adaptive response.
Figure 2. Toxins stimulate the microbial aversion behavior.
(A) Toxin-induced microbial aversion phenotypes. Drug concentrations are provided in Supplemental Materials.
(B) Aversion levels +/−SEM for each drug at peak time points: Zeocin (16 hr); Paraquat (8 hr); Antimycin A (6 hr); Tunicamycin (24 hr); Geneticin (3 hr); Bortezomib (6 hr); Concanamycin A (8 hr).
*p<0.01; *p<0.001; ***p<0.0001 by t-test.
To test the hypothesis that aversion behavior is a generalized response to toxin exposure, we supplemented benign E. coli lawns with various poisons, many of which target essential activities identified in our screen, to determine if animals developed aversion towards an otherwise attractive E. coli strain (OP50). Geneticin/G418 was used to inhibit translation, antimycin A to inhibit the mitochondrial ETC, Bortezomib to inhibit proteasome activity, concanamycin A to inhibit vacuolar ATPAse activity, zeocin to induce genotoxic stress, and paraquat to induce oxidative stress. Notably, antimycin and concanamycin are toxins produced by Streptomyces species resident to soil and compost, both natural habitats for C. elegans. L4 larvae/young adults were placed on E. coli OP50 lawns supplemented with one of these compounds, and assayed for aversion behavior over time. Exposure to any of these toxins stimulated avoidance of the microbial lawn, usually within 4-16 hrs of exposure (Figure 2B-C). Treatment of larval stage animals produced similar results (data not shown). Aversion behaviors developed more rapidly in response to poisons than RNAi, probably because drugs directly inhibit protein activities while RNAi degrades mRNAs to prevent new protein translation but does not inhibit pre-existing cellular proteins.
Inactivation of essential cellular activities stimulates pathogen and detoxification responses
Because bacterial pathogens are known to stimulate food-avoidance behavior in C. elegans, the observation that inactivation of cellular processes using toxins or RNAi produced a similar behavioral response suggests an adaptable mechanism for toxin and pathogen detection mediated by cellular surveillance pathways in the host. We reasoned that if animals use internal physiologic cues to recognize the presence of pathogens, then perhaps RNAi of essential cellular components would trigger physiologic defenses associated with pathogen infection or toxin exposure – in the absence of pathogens or toxins.
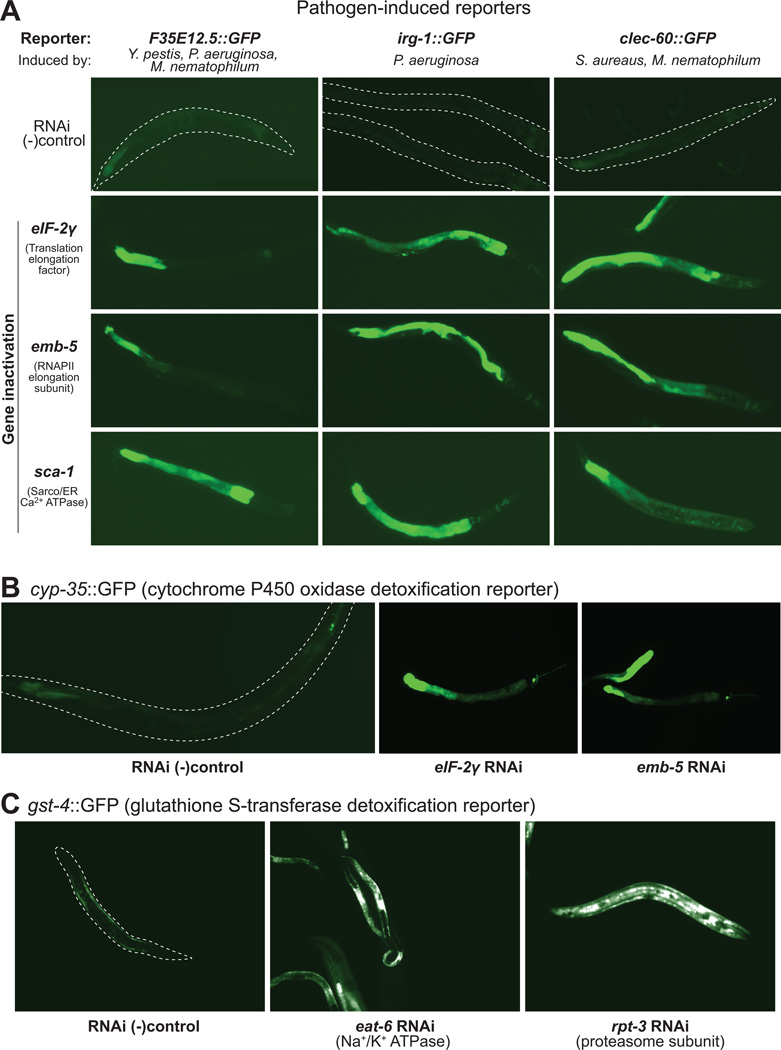
To address whether aversion-inducing gene inactivations can engage transcriptional responses to pathogen exposure in C. elegans, we examined the expression of several GFP reporters previously shown to represent activation of innate immune programs. For each reporter, GFP was fused to the promoter of a gene induced by a microbial pathogen but not by benign E. coli or attenuated pathogenic strains. Three of these genes have proposed functions in innate immunity because they contain conserved domains associated with innate immune responses in C. elegans and other species: clec-60 (a C-type lectin/CUB domain protein induced by S. aureus and M. nematophilum) (O'Rourke et al., 2006), F35E12.5 (a CUB domain protein induced by Y. pestis, M.nematophilum and P. aeruginosa) (Troemel et al., 2006)(O'Rourke et al., 2006)(Bolz et al., 2010), and nlp-29 (a conserved glycine/tyrosine-rich antimicrobial peptide induced by D. coniospora, S. marcescens and wounding) (Pujol et al., 2008). Finally, irg-1::GFP is a reporter induced by P. aeruginosa and cadmium poisoning whose expression in response to pathogen exposure requires ZIP-2, a bZIP transcription factor (Estes et al., 2010). clec-60::GFP, F35H12.5::GFP and irg-1::GFP are all primarily induced in the intestine, while nlp-29::GFP induction is generally hypodermal.
Induction of each pathogen reporter was analyzed against a panel of ~100 gene inactivations representing the major functional classes identified in our aversion screen and a random panel of genes from the whole genome RNAi library. For all 4 pathogen-response genes, we observed a significantly elevated frequency of activation by the aversion gene set relative to the control set (p<0.0001, chi-squared test) (Figure 3, S2C and Table S2A). The clec-60::GFP reporter was activated by 59% of aversion gene inactivations tested, F35H12.5::GFP by 24%, irg-1::GFP by 45%, and nlp-29::GFP by 28%, representing a 2 to 9 fold increase in induction frequencies relative to the random set (Figure S2C). In general, the three intestinal reporters were induced by RNAi against most functional classes, with most potent induction following disruption of protein synthesis, metabolic and mitochondrial functions (Table S2A). In contrast, the hypodermal nlp-29::GFP reporter was most consistently activated by disruption of the molting program. Starvation also induced the clec-60::GFP and nlp-29::GFP reporters.
Figure 3. Activation of pathogen-associated and detoxification responses.
(A) Innate immunity-associated reporter induction in response to growth on RNAi lawns inactivating the C. elegans eIF2γ homolog Y39G10AR.8, emb-5 and sca-1 genes.
(B) RNAi of eIF-2γ and emb-5 stimulates cyp-35B1::GFP, a reporter for cytochrome P450 enzymes involved in phase I drug detoxification.
(C) RNAi of eat-6 and rpt-3 stimulates a gst-4::GFP reporter for induction of glutathione S-transferase, a phase II detoxification enzyme.
If host surveillance pathways detect the presence of chemical toxins by monitoring core cellular activities, then RNAi-mediated disruption of these activities might also stimulate drug detoxification responses, such as cytochrome P450 and glutathione S-transferase genes. To test this hypothesis, we assayed GFP transcriptional reporters for induction of xenobiotic detoxification enzymes, cyp-35B1 (an intestinally-expressed cytochrome P450 oxidase) (Iser et al., 2011) and gst-4 (a glutathione S-transferase induced by drugs and toxins) (Figure 3B,C). Each of these reporters was induced at a frequency ~4-fold above background (~40% of aversion-inducing essential gene inactivations tested induced cyp-35B1::GFP and ~20% induced gst-4::GFP, p<0.0001, chi-squared test) (Figure S2C). The pattern of gene inactivations that stimulated expression of xenobiotic detoxification responses was overlapping but distinct (Table S2A). Both reporters were robustly induced by inactivation of genes involved in protein synthesis, mitochondrial function and metabolism. gst-4::GFP alone was induced by RNAi of proteasome subunits and cyp-35B1::GFP alone was induced by RNAi of splicing and vesicular trafficking components. The cyp-35B1::GFP reporter was also induced by inactivation of many molting and cuticle-specific genes, despite an expression pattern that was limited to the intestine.
General reporters of cellular stress, such as the mitochondrial unfolded protein response (UPRmito), and the endoplasmic reticulum unfolded protein response (ERUPR), assayed using hsp-6::GFP (Yoneda et al., 2004) and hsp-4::GFP (Calfon et al., 2002; Urano et al., 2002) were also triggered by many essential gene inactivations, as was a sod-3::GFP oxidative stress reporter (Libina et al., 2003) and a gpdh-1::GFP osmotic stress reporter (Lamitina et al., 2006). The majority of gene inactivations resulted in activation of at least one cellular stress reporter and many gene inactivations stimulated more than one reporter (Figure S2, Table S2B).
Microbial aversion behavior is controlled by a neuroendocrine circuit
Neurosensory inputs are necessary for pathogen avoidance behavior (Styer et al., 2008) but the contribution of sensory signals originating within an animal’s internal tissues has been difficult to determine. Three lines of evidence suggest that aversion behavior induced by essential gene inactivations is not exclusively a neuronal response but is also mediated by endocrine signaling between the C. elegans nervous system and other tissues: (1) both drugs and RNAi targeting the same process can stimulate aversion, suggesting that drug detection by neuronal chemoreceptors is not essential, (2) RNAi works poorly in C. elegans neurons, and almost always requires an enhanced RNAi mutant strain to elicit detectable phenotypes (Calixto et al., 2010) and (3) a subset of genes identified in the aversion screen exhibit post-embryonic expression patterns that exclude the nervous system but include the intestine, hypodermis, gonad, pharynx, vulva, body wall muscle and/or the excretory cell, suggesting that any or all of these tissues may be capable of triggering a neuroendocrine-mediated aversion response (Table S3). For example, the ELT-2 GATA transcription factor acts exclusively in the intestine to control gut development (Fukushige et al., 1998), and the QUA-1 hedgehog homolog acts in the hypodermis to control molting (Hao et al., 2006). Thus the aversion response to inactivation of essential genes is likely to operate through an endocrine-mediated relay of stress signals between non-neuronal tissues and the neurons that generate aversive behavior.
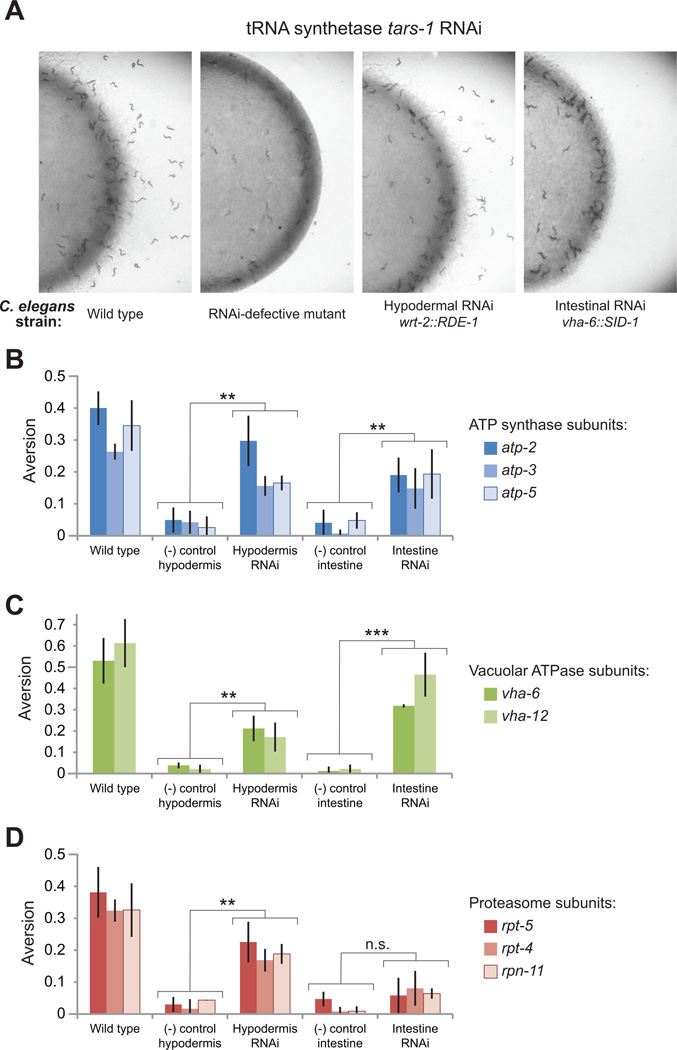
To test directly whether non-neuronal cues can trigger the aversion response, we generated C. elegans strains in which RNAi operates only in specified tissues by tissue-restricted restoration of RNAi in RNAi-defective mutants (either rde-1(ne219) or sid-1(qt9)). Tissue-specific rescue of rde-1 or sid-1 null mutants can restrict RNA interference-based gene inactivation to select tissues (Qadota et al., 2007)(Jose et al., 2009). rde-1 expression from the hypodermal wrt-2 promoter was used to restore RNAi activity to hypodermal and seam cells in rde-1(ne219) animals. A gut-specific vha-6 promoter driving expression of sid-1 restored RNAi to intestinal cells in a sid-1(qt9) mutant. Tissuespecificity for RNAi was confirmed by introducing a ubiquitously expressed GFP reporter (sur-5::GFPNLS) into hypodermal and intestinal RNAi lines and performing RNAi against GFP (Figure S3A-D). In wrt-2::RDE-1 animals, GFP silencing was observed in the hypodermis, but not in other major tissues (the muscle, intestine, neurons or pharynx). In the vha-6::GFP line, GFP was silenced in the intestine but not elsewhere. As further reassurance of the tissue-specificity of these lines, we inactivated several genes with known restricted patterns of expression (Table S3), and found that aversion was stimulated only in the tissue-specific RNAi strain where those genes are expressed (Figure S4A-B).
We tested whether tissue-restricted RNAi of essential functional classes identified in the aversion screen could stimulate microbial aversion. We found that most essential gene classes could stimulate aversion from either hypodermis or intestine. Genes encoding three ATP synthase subunits (atp-2, atp-3 or atp-5) or two vacuolar ATPase subunits (vha-6 and vha-12) could induce aversion when inactivated in either the intestine or the hypodermis (Figure 4B-C). Although aversion phenotypes were milder than in wild type animals, tissue-restricted RNAi animals exhibited significantly higher aversion than RNAi-defective strains. Not all essential functional classes tested stimulated aversive behavior from both tissues. For instance, hypodermal inactivation of proteasome subunits or tRNA synthetases stimulated aversion, while intestinal inactivations did not (Figures 4D, S4C-D). Inactivation of protein translation by RNAi of ribosomal components in either tissue stimulated aversion (Figure S4C-D).
Figure 4. Neuroendocrine control of aversion behavior.
(A) Aversion is stimulated by inactivation of the threonyl tRNA synthetase tars-1 in the hypodermis but not the intestine.
(B-D) Aversion +/−S.D. is induced by RNAi of (B) mitochondrial ATP synthase subunits and (C) vacuolar ATPase subunits in the hypodermis or intestine, and (D) proteasome subunits in the hypodermis only. Representative experiments +/−S.D. shown. *p<0.01; **p<0.001; ***p<0.0001; n.s. not significant.
(See also Figures S3–S4, Table S3.)
Aversion behavior requires a Jnk-like MAP kinase pathway
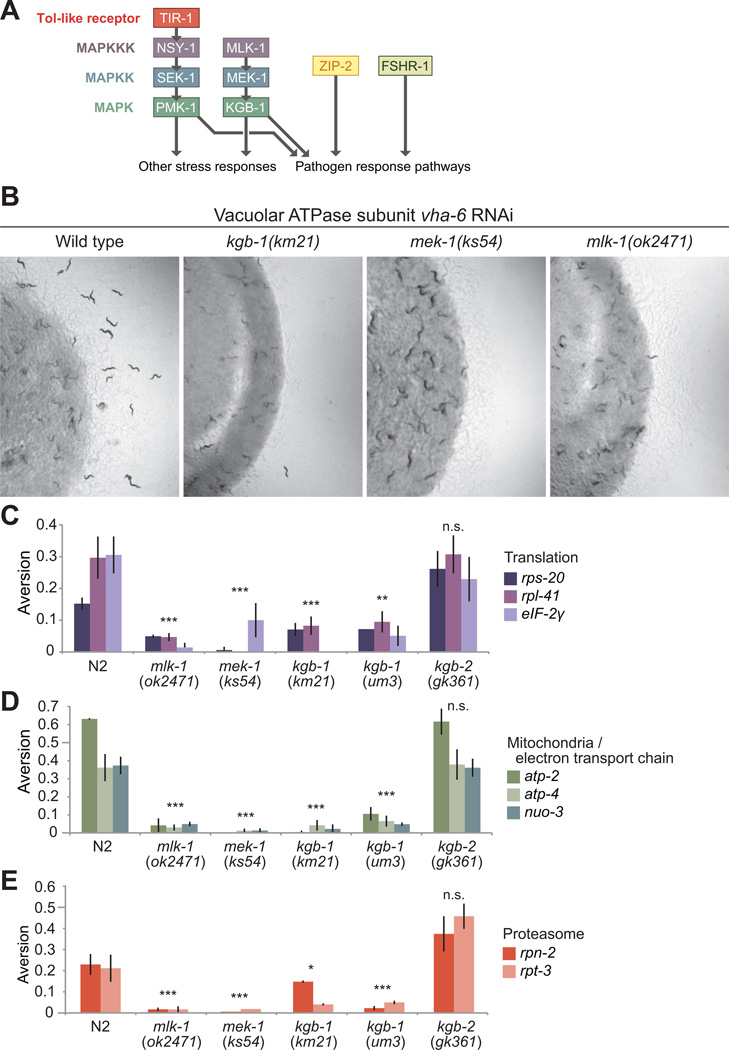
Multiple pathways contribute to pathogen resistance in C. elegans. Immune responses to P. aeruginosa may be the most intensively characterized. Animals are protected from P. aeruginosa infection by at least 3 genetic pathways, represented by: (1) FSHR-1, a G protein-coupled receptor specifically required in the intestine (Powell et al., 2009), (2) ZIP-2, a bZip family transcription factor controlling the induction of many pathogen-responsive genes (Estes et al., 2010), and (3) the NSY-1/SEK-1/PMK-1 p38 family MAP kinase cassette downstream of the TIR-1 Toll-like receptor necessary for innate immunity in many systems (Figure 5A). The p38 MAPK pathway was an especially attractive candidate because VHP-1, an inhibitory phosphatase of PMK-1 (Kim et al., 2004), stimulates microbial aversion when inactivated by RNAi (Table S1A). We examined mutants in these pathways for a role in aversion behavior. At least two mutant alleles were tested for all genes except fshr-1 (Table S4). Mutants in none of these pathways attenuated microbial aversion behavior (Figure S5A-B, and data not shown), although both sek-1 mutant alleles exhibited a partial reduction in aversion in response to RNAi of vacuolar ATPase, ATP synthase and tRNA synthetase genes (Fig S5A and data not shown), without an accompanying requirement for tir-1, nsy-1 or pmk-1.
Figure 5. The Jnk MAP kinase pathway is required for aversion.
(A) Four candidate pathogen and stress response pathways in C. elegans.
(B) Aversion phenotypes for Jnk pathway mutants following inactivation of vha-6 (vacuolar ATPase subunit).
(C-E) The Jnk pathway is required for aversion induced by RNAi of (C) translation components, (D) mitochondrial ETC components and (E) proteasome subunits. Representative experiment +/− SD shown. * p<0.01, ** p<0.001 and *** p<0.0001. n.s. not significant.
(See also Figure S5.)
However, a Jnk-like MAP kinase pathway was required for the aversion response. MLK-1/MEK-1/KGB-1 comprise a MAP kinase cassette required for resistance to the ER toxin tunicamycin (Mizuno et al., 2008), transcriptional responses to pore-forming toxins (Kao et al., 2011) and S. cerevisiae infection (Yun and Politz, 2011). The VHP-1 phosphatase antagonizes KGB-1 (as well as PMK-1) (Mizuno et al., 2004), so it seemed possible that vhp-1 RNAi-induced aversion behavior might be caused by hyperactivation of KGB-1. We also tested the kgb-1 homolog kgb-2, for which no function has yet been reported. We found that the mlk-1, mek-1 and kgb-1 genes were fully required for microbial aversion behavior in response to RNAi of all functional gene classes tested (Figure 5B-E), although kgb-1 was only partially required when tRNA synthetases or vacuolar ATPases were inactivated (Figure S5C). kgb-2 was not required for aversion in any gene category. For kgb-1, mek-1 and mlk-1 mutants, inactivation of electron transport chain components (atp-2, atp-4 or nuo-3), protein translation factors (rps-20, rpl-41 or Y39G10AR.8, the eIF-2γ homolog), or proteasomal subunits (rpn-2 or rpt-3) did not elicit aversion significantly above basal levels (Figure 5C-E). kgb-1 and sek-1 were each partially required for aversion induced by RNAi of vacuolar ATPases and tRNA synthetases (Figure S5A,C), suggesting that for some subtypes of cellular stress, these two kinases operate collectively to control the microbial aversion response.
Essential gene RNAi and pathogens exhibit similar neurobiological traits
C. elegans develops an avoidance of pathogenic bacteria it initially finds attractive, possibly by pairing internal with chemosensory cues of infection (Schulenburg and Ewbank, 2007). Pathogen avoidance requires an intact chemosensory system (Styer et al., 2008) and operates via a serotonergic circuit (Zhang et al., 2005). Reduced ingestion of pathogenic microbes has also been reported (Schulenburg and Muller, 2004; Sicard et al., 2007). We examined the behavioral profiles of essential gene RNAitreated animals to determine if aversion behaviors stimulated by disruption of core cellular processes share similar neurological characteristics with pathogen avoidance behavior.
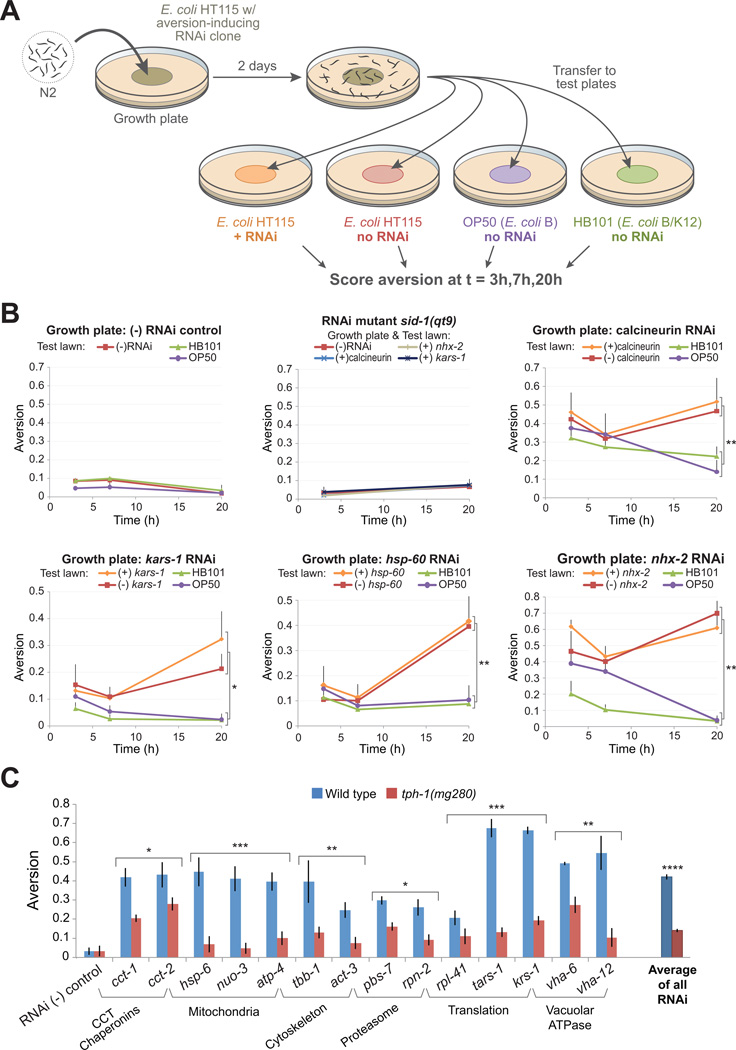
We examined whether animals with prior experience of aversion-inducing ”noxious” RNAi bacteria were capable of associating internal with chemosensory cues to guide future feeding decisions. RNAi and control RNAi (-) bacteria are the same E. coli K12 strain (HT115), differing only in whether they express dsRNA targeting a C. elegans gene. It seemed likely that expression of dsRNAs corresponding to a C. elegans gene would not significantly impact the repertoire of gustatory and olfactory signals produced by HT115. We predicted that if experience-dependent neural associations reinforce aversion behavior stimulated by disruption of cellular processes, animals raised on a noxious RNAi lawn should develop behavioral avoidance of both deleterious and benign HT115 - but perhaps not against less-related E. coli or other bacterial strains. To test this prediction, animals were raised for two days on noxious RNAi “growth plates” and transferred to “test plates” containing one of four microbial options: (1) the same noxious RNAi bacteria upon which they were grown, (2) the benign RNA (-) control, (3) OP50 E. coli or (4) HB101 E. coli (Figure 6A). Options (3) and (4) represent genetically distinct E. coli isolates from HT115. OP50 is an E. coli B strain and HB101 is an E. coli B/K12 hybrid strain. Animals were assayed for aversion at 3, 7 and 20hr after transfer to test plates. Four aversion-inducing gene inactivations were selected to disrupt a range of biological functions: kars-1 (tRNA synthetase), Y71H2AL.1 (calcineurin B subunit), nhx-2 (Na+/H+ transporter), and hsp-60 (mitochondrial chaperone). Animals raised on all four noxious RNAi lawns exhibited similar levels of microbial aversion for all 4 tester strains in the first 7hr after transfer. Between 7-20hr, animals on the (-) control and noxious RNAi test plates exhibited sustained or increasing levels of aversion. In contrast, by 20hr animals transferred to OP50 or HB101 test plates had undergone a decline in aversion, often to near-basal levels for that bacterial strain (Figure 6B, top left panel). Animals on the four test plates were of the same developmental stage and size, indicating that the reduction in lawn aversion on OP50 and HB101 plates was not attributable to attenuation of RNAi activity (Figure S6A). Animals raised on benign HT115 plates exhibited no aversion after transfer to tester plates. Similarly, RNAi-defective mutant animals displayed no aversion despite continuous growth on noxious RNAi bacteria (Figure 6B, top middle panel). We also tested several bacterial species that may represent more ecologically relevant food choices for C. elegans than laboratory E. coli. After 2dy of growth on kars-1 RNAi plates, animals were transferred to test plates containing microbial species known to reside in soil environments: B. subtilis (a gift from R. Losick), B. simplex, and a Comamonas sp. soil isolate (gifts from L. Avery). At 2 and 5hr, animals did not display significantly different aversion phenotypes on any tester plate, but by 14 and 22hr animals transferred to any of the three soilassociated species exhibited lower aversion than those transferred to kars-1 RNAi and control HT115 plates (Figure S6B).
Figure 6. Learned avoidance and serotonergic signaling mediate aversion behavior.
(A) Schematic outline of learning experiment.
(B) Aversion time courses for animals grown on RNAi bacteria inactivating the kars-1 tRNA synthetase, hsp-60 mitochondrial chaperone, nhx-2 Na+/K+ potassium pump, or Y71H2AL.1 calcineurin B. Control experiments: Top left, animals grown on HT115 RNAi control and tested on HT115, HB101 or OP50 E. coli. Top middle, sid-1 RNAi defective animals were grown and tested on the same bacterial type (e.g. grown and tested on nhx-2 RNAi). For graphical clarity, only +S.D. bars shown.
(C) Serotonin deficient tph-1(mg280) animals exhibited a partial aversion defect (~35% of wild type) in response to gene inactivations representing major functional classes.*p<0.01; **p<0.001; ***p<0.0001, ****p<10−19.
(See also Figures S6–S7.)
The observation that animals grown on noxious RNAi HT115 E. coli caused them to avoid benign HT115 E. coli suggested that neurosensory associations reinforce and sustain aversion behavior. We assayed aversion responsiveness in mutants defective for distinct aspects of neurosensation: chemosensation (che-2(e1033) and che-3(e1124)), mechanosensation (mec-3(e1388), mec-4(u45) and mec-7(e1527)), thermotaxis (ttx-1(p767) and ttx-4(nj3)), and aerotaxis (gcy-35(ok769)). We also tested the tax-4(p678) mutant, which inactivates a broadly expressed nucleotide-gated channel required for multiple neurosensory responses, including chemotaxis (Bargmann, 2006), chemorepulsion, aerotaxis, and pathogen avoidance (Styer et al., 2008; Yook and Hodgkin, 2007). We expected that sensory-defective mutants might exhibit aversion phenotypes on benign bacteria, possibly due to deficits in chemical, mechanical or oxygen-related aspects of food recognition. Similarly, we thought sensory mutants might fail to exhibit aversion behavior in response to noxious RNAi due to their inability to detect environmental cues necessary to form learned associations with internal signals. We measured baseline aversion levels of sensory mutants over a developmental time course on RNAi (-) control bacteria. While several mutants exhibited slightly elevated basal aversion behavior, che-2(1033) was the only mutant exhibiting significantly higher aversion than wild type by 48hr (0.18+/−.05 vs 0.02+/−.01, p<0.0001,student’s t-test) (Figure S6C). Because sensory mutant defects might be masked in an aversion assay by their naturally higher aversion rates, we plotted raw and baseline-subtracted aversion levels for all sensory mutants. Mutants were tested for aversion responses after 48hr of growth on RNAi bacteria targeting the vha-6, kars-1, cco-1 or elt-2 genes. Only tax-4, which is required for multiple sensory modalities, was strictly required for the aversion response in all four cases (Figure S6D-G). che-2 and che-3 mutants, which lack functional olfactory neurons, exhibited no significant defect in response to aversion induced by vha-6 or kars-1 RNAi (Figure S6D-E) but were strongly defective for aversion behavior in response to RNAi of cco-1 or elt-2 (Figure S6F-G). None of the mutants impaired in mechanosensation, thermotaxis or aerotaxis caused significant differences in aversion compared to wild type.
We also tested how animals in “aversion” states respond to new sensory cues. Our learning data showing that growth on noxious RNAi bacteria causes animals to avoid all bacterial foods for up to 7hrs suggested that animals raised on noxious RNAi bacteria are not as initially attracted to positive stimuli as naïve animals. To explore this possibility, we examined the stimulus-responsiveness of RNAi-treated animals in a panel of olfactory, gustatory and tactile assays that report on both attractive and repulsive behavioral responses. We first examined chemotaxis to the attractive odorant isoamyl alcohol in animals that were previously grown on noxious RNAi bacteria disrupting a range of cellular activities. Most gene inactivations did not interfere with chemoattraction but several caused mild deficits (qua-1, atp-3, ptc-1, pqn-47, vha-6 RNAi) (Figure S7A). Chemotaxis to the soluble attractant NH4Cl was used to evaluate animal attraction to gustatory cues after growth on noxious RNAi bacteria (Figure S7B). One gene inactivation (cmd-1 RNAi) showed significant defects in chemotaxis; several others were slightly less attracted to NH4Cl than the RNAi control but these reductions were not statistically significant. Five RNAi treatments were tested for avoidance of the volatile repellant, octanol, but no deficit in behavioral repulsion was detected (Figure S7C). A light touch assay was used to assess mechanosensory reactions and none of the gene inactivations tested exhibited detectable defects in tactile avoidance (Figure S7D).
Pathogen avoidance behavior requires serotonin signaling (Zhang, Y. et al, 2005). To determine if noxious RNAi avoidance also engages a serotonergic circuit, we analyzed aversion responses in the serotonin biosynthetic mutant, tph-1(mg280). We found that serotonin-deficient animals exhibited a consistent ~65% reduction in aversion behavior relative to wild type controls for every functional category tested (Figure 6C) (p<10−9, student’s t-test).
In addition to locomotory avoidance of pathogens, some studies have reported a reduction in microbial ingestion (Yook, K. and J. Hodgkin, 2007). We measured pharyngeal pumping rates for a panel of gene inactivations yielding fertile adults (so all samples are stage-matched with controls) (Figure S7E). Nine of ten RNAi treatments yielded a mild to strong reduction in pumping rate that was statistically significant (p<10−4, student’s t-test with Bonferroni correction).
DISCUSSION
Food avoidance is a behavioral response to disruption of essential cellular activities
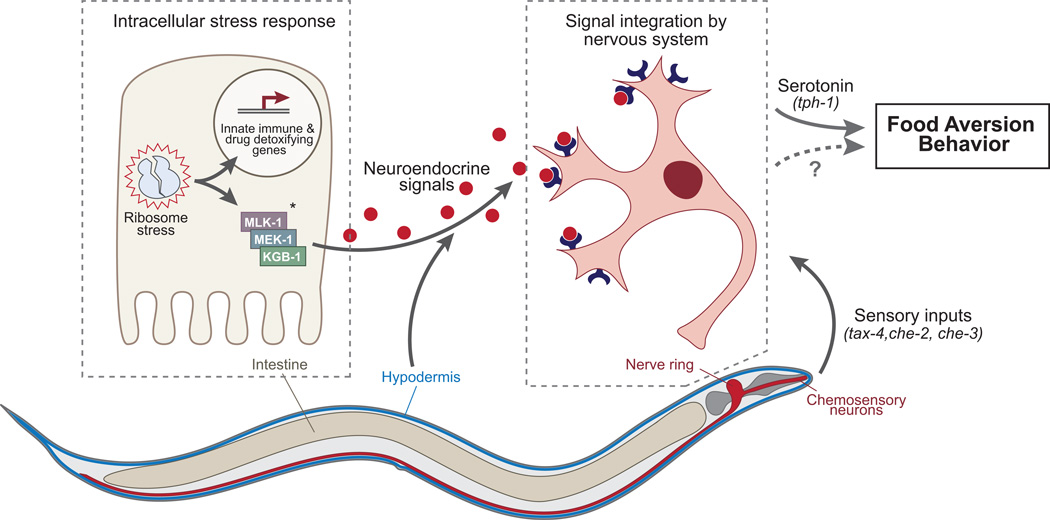
We have shown that C. elegans develops a behavioral revulsion to otherwise nutritive, non-pathogenic E. coli when those bacteria produce dsRNA that inactivates C. elegans genes required for fundamental cellular activities - such as protein translation or oxidative respiration. RNAi-induced disruption of these highly conserved and fundamental processes may mimic the inactivation of these same processes by toxins or virulence factors produced by noxious microorganisms in the natural habitats of C. elegans. The generality of aversion behavior as a response to functionally diverse gene inactivations allowed us to conduct an RNAi screen to identify the range of physiologic processes that may be subject to surveillance. From this screen, we discovered that aversion was induced by disruption of protein translation, mitochondrial functions, the proteasome, vacuolar ATPases, the tubulin and actin cytoskeletons, mRNA processing, chromatin packaging, central metabolism, and the molting program. Disruption of essential genes implicated in aversion often also reduced food ingestion. Multiple cellular surveillance pathways are already well-known, including the ER unfolded protein response, DNA damage and cytoskeleton checkpoints. Ribotoxic stress and mitochondrial stress response pathways have also been reported (Iordanov et al., 1997)(Haynes and Ron, 2010). However, many of the cellular processes and molecules identified in our screen are not associated with previously reported surveillance pathways and may therefore reveal new cellular defense programs (Figure 7).
Figure 7. Model of aversion behavior.
Distress signals arising in internal tissues are integrated with sensory inputs to control the microbial aversion response. *Although Jnk pathway components are depicted as acting in the intestine, the site(s) of action have not yet been determined.
Surveillance of cellular functions provides a new mechanism for toxin and pathogen detection
C. elegans are repelled by pathogenic bacteria or fungi, exhibiting both locomotory avoidance and reduced feeding (Schulenburg and Ewbank, 2007). Multiple tissues, including the intestine and hypodermis are susceptible to pathogen invasion, and many microbial signals and host pathways are likely to be involved in mounting an effective immune response. Our results with innate immune and xenobiotic detoxification reporters suggest that inactivation of a single cellular process may be sufficient to mount both immune and detoxification responses. RNAi-mediated disruption of translation, mitochondrial functions, vacuolar ATPases, and basic metabolic pathways were all potent inducers of the clec-60::GFP, irg-1::GFP and F35H12.5::GFP pathogen-responsive reporters – despite the absence of a pathogen or a toxin in these experiments. Pathogen-associated reporters were weakly stimulated by inactivation of proteasomal and secretory pathway components. For the engagement of innate immune responses, mechanisms of pathogen recognition are thought to involve both direct sensation of pathogen-intrinsic signals (PAMPs, or pathogen-associated molecular patterns) such as cell wall components and sensation of pathogen-extrinsic signals (DAMPs, or damage-associated molecular patterns) caused by toxin or virulence factor-induced damage in the host (Jones and Dangl, 2006; Vance et al., 2009). DAMPs are generally believed to be small molecules or cellular proteins released into the extracellular environment when host cells die. Our findings suggest that cellular surveillance pathways detect invading pathogens by sensing disruption to cellular processes (like protein translation or oxidative respiration) caused by secreted toxins and virulence factors. Upon detection of cellular disturbances, such surveillance mechanisms engage host defenses by stimulating xenobiotic detoxification genes to neutralize toxins and immune defenses to eradicate microbes, activating compensatory programs to restore function to pathways disabled by pathogens and toxins, and enabling escape from pathogens by locomotory avoidance and suppression of pathogen ingestion. One advantage of such a system is that it can detect decreases in core cellular functions long before cell death, when DAMP signals are released. In contrast to natural pathogens, which transfer to hosts a suite of virulence factors and chemical toxins with pathogenic and perhaps anti-surveillance functions that may be difficult to isolate, our system of induction of pathogen response pathways by inactivation of individual host genes allows the removal of many layers of evolved measure/countermeasure complexity in pathogen/host interactions. This advantage will lend itself to dissection of individual surveillance pathways, and the elucidation of corrective mechanisms for disruptions to particular processes.
Many of the essential cellular functions identified in our screen are known targets of microbial toxins. We found that supplementation of innocuous E. coli food with microbial toxins (antimycin A, tunicamycin, and concanamycin A) targeting the same cellular processes identified in our screen (the mitochondrial ETC, the secretory pathway, and vacuolar ATPases) also trigger the aversion response. Analysis of the gst-4::GFP and cyp-35B::GFP detoxification reporters revealed that many essential gene inactivations engaged xenobiotic detoxification responses. These data suggest that many drugs are detected via the surveillance of the core cellular processes targeted by drugs rather than by detection of the drug itself. Thus the detection of toxins and bacterial pathogens may use common surveillance elements and may couple to induction of cytochrome P450 and other detoxification systems as well as induction of anti-bacterial immunity functions. Because bacteria and fungi produce a large array of xenobiotic secondary metabolites, the products of polyketide synthase genes or non-ribosomal peptide synthesis genes which constitute 5-10% of many microbial genomes, the logic that any toxin is likely to come from a bacteria, fungus, or plant on which the animal was feeding is reasonable. While toxins are diverse, they are biased toward the disruption of highly conserved cellular machinery, such as ribosomal and mitochondrial components, perhaps to be pathogenic to the widest range of hosts. Significantly, the gene inactivations that most potently induce antibacterial behaviors and gene expression are among the most conserved C. elegans genes, suggesting the “core proteome” is targeted by microbial toxins and virulence factors. Direct surveillance of these systems may enable the host to defend itself effectively from pathogens with which it has no evolutionary history. Other emerging studies support this cellular surveillance model of pathogen detection. Exposure of C. elegans to the Pseudomonas aeruginosa virulence factor ToxA, which disables translation elongation factor EF2 (one of the most highly conserved eukaryotic genes and an aversion hit in our screen), and hygromycin, which inhibits ribosome translocation, induces pathogenresponse genes (McEwan et al., 2012) and inactivation of translation using RNAi or the translation inhibitor cycloheximide stimulates the pathogen response genes (Dunbar et al., 2012).
Control of aversion through MAP kinase and endocrine pathways
Cellular stress responses are coupled to endocrine pathways in C. elegans. Insulin signaling regulates dauer arrest in response to high population density or starvation. Disruption of the ETC in neurons activates the mitochondrial UPR in intestinal cells (Durieux et al., 2011) and neuroendocrine signals control proteotoxicity (Prahlad and Morimoto, 2011) and heat shock responses (Prahlad et al., 2008) in non-neuronal tissues. Using tissue-restricted RNAi, we have shown that the hypodermis and the intestine are each capable of stimulating the aversion response for many essential gene inactivations. Given that both tissues contact the external environment, they make attractive candidates as extraneuronal sensory organs for microbial toxins and virulence factors. However the expression patterns of some aversion genes suggest additional tissues may be competent to trigger the aversion response, including the gonad, vulva, pharynx, body wall muscle and excretory cell. Regardless of the range of tissues involved in sensation of cellular stressors, the finding that aversion can be triggered by disruption of core processes in non-neuronal tissues argues that neuroendocrine signals are relayed between peripheral tissues and the neurons that mediate aversion behavior.
We tested if known pathogen response pathways also mediate the aversion response. We found no evidence that three major pathways controlling P. aeruginosa resistance, the ZIP-2, FSHR-1 and p38 MAPK pathway were involved in aversion behavior. However a Jnk-family MAP kinase pathway involved in C. elegans responses to tunicamycin and pore-forming toxins, comprised of MLK-1 (MAPKKK), MEK-1 (MAPKK) and KGB-1 (MAPK), was clearly required for microbial aversion behavior. All three kinases were essential for aversion in response to most gene inactivations, including translation, mitochondrial ETC and proteasome genes. This kinase cascade could act in the intestine or hypodermis to couple distress signals from cellular surveillance pathways to the release of neuroendocrine signals, or it could function in the neural circuits that generate aversion behavior. The mammalian homolog of KGB-1, Jun kinase, is activated when cells are exposed to toxins that inhibit elongation of protein translation (Iordanov et al., 1997), suggesting that C. elegans and mammals may respond to toxin-mediated disruption of cellular functions via a common signaling pathway.
Similarities of pathogen avoidance behavior and RNAi-induced food aversion
Although aversion can be stimulated by cues emerging from non-neuronal tissues, our analysis of neurosensory mutants demonstrated that aversion behavior also requires an intact neurosensory system. tax-4, which encodes a cyclic nucleotide gated channel, is required for many sensory responses and for pathogen avoidance behavior. tax-4 was fully required for induction of aversion by inactivation of essential cellular pathways. However, mutants in individual sensory modalities (touch, heat, oxygen-sensation) had little or no effect on aversion responses. che-2 and che-3 were required for aversion induced by cco-1 and elt-2 RNAi, but not vha-6 or kars-1 RNAi, suggesting that chemosensation may be required for aversion behavior in some instances. Together these findings indicate that the sensory system as a whole is necessary for the aversion response, but that aversion is likely redundantly reinforced by multiple sensory modalities. We also found that, like pathogen avoidance behavior, noxious RNAi-induced aversion behavior exhibits a significant requirement for serotonin. On average, ~65% of the aversion response relies on serotonergic circuit(s), indicating that other forms of neurotransmission must provide the residual aversion observed in serotonin mutants.
The ability of C. elegans to integrate sensory with undefined internal cues may be the basis for learned pathogen avoidance behavior. We have shown that animals reared on aversion-inducing RNAi E. coli (the HT115 strain) develop and maintain an avoidance of HT115, even when shifted to an HT115 lawn that does not produce noxious dsRNAs, consistent with pathogen avoidance association studies. Not only does previous experience with noxious RNAi bacteria cause animals to avoid the HT115 strain, it causes them to avoid entirely different bacterial species at levels comparable to the noxious RNAi strain for up to 7 hours. After 7hr, aversion behavior on non-HT115 bacteria subsided so that by 20hr aversion was barely detectable. This result suggests that once entrained on noxious bacteria, animals may adopt an “aversive state” that is resistant to modulation by new sensory cues, even attractive ones. This proposition is supported by our chemosensory data showing that prior growth on noxious RNAi bacteria can cause a reduction in chemotaxis to attractive olfactory or gustatory cues, but has no observable effect on behavioral responses to aversive stimuli.
A surveillance system for pathogens and xenobiotics
Our screen for microbial aversion behavior has shown that a wide range of critical cellular processes are monitored by surveillance pathways, many of which are not associated with known cellular surveillance systems. Many of the cellular processes identified are targets of toxins produced by the expansive diversity of bacteria in the natural ecosystem of any animal or plant. Given that aversion-associated genes are among the most conserved across eukaryotic phylogeny, host surveillance pathways are likely to be ancient and shared by many or all eukaryotes. Consistent with this view, we found that cellular surveillance signals require a conserved JNK-family MAP kinase cascade to produce the aversion response. We would also suggest that cellular surveillance systems detect xenobiotics and medicinal drugs via their inhibition of core cellular pathways and may likewise couple (through JNK or related MAP kinase pathways) to behavioral modifications and detoxification responses. Because inactivation of a cellular pathway can act as a signal of bacterial infection, toxicity caused by medicinal drugs may in some cases be due to inappropriate induction of antibacterial behaviors or engagement of innate immune defenses. For example, if nausea represents the human analog of “microbial aversion behavior”, then the nausea produced by chemotherapeutic treatments may indicate the misapplication of a normally adaptive response to the ingestion of tainted foods.
The physiological diversity of virulence factors and bacterial toxins represents the billion year arms race with eukaryotic surveillance and detoxification systems: as the host evolves surveillance, detoxification, and immunity functions, bacteria will evolve countermeasures that may neutralize the surveillance pathways required for immune defense. There may be corresponding complexity and genetic variation in the surveillance pathways marshaled by eukaryotes, perhaps based on their varied toxin and pathogen histories. Such variation in humans may underlie distinct responses to drugs, therapeutic or addictive, or to normally benign chemicals or foods. The molecular components of these surveillance pathways, such as the conserved JNK kinase cascade, may constitute new drug targets to alleviate the nausea and appetite suppression caused by many therapeutic drugs, or the toxic side effects that derail many drugs in development. Inactivation of these surveillance pathways may also suppress aberrant induction of xenobiotic and pathogen response pathways in a variety of diseases.
Experimental Procedures
Microbial avoidance assays
Saturated overnight cultures were concentrated 30-40X for HT115 RNAi bacteria or ~20X for OP50 bacteria (used in drug assays) in S Basal, 50ul aliquots were dropped to 6-well RNAi plates and left on the benchtop overnight. For toxin avoidance, toxins were dropped to lawns ~1hr prior to use. Synchronized L1s (for RNAi) or L4/A stage (for drugs) were dropped directly onto lawns and wells were scored for aversion = Noff lawn/Ntotal at desired time points. Each RNAi or drug treatment was conducted in triplicate in each experiment, and was repeated in two or more independent trials. Drug concentrations used can be found in Supplemental Materials.
Neurosensory and Feeding Assays
Chemotaxis, mechanosensation and pharyngeal pumping experiments were performed using standard procedures (Hart, 2006).
Statistical Methods
The student’s t-test and chi-squared tests were used to validate behavioral results as stated in figure legends. DAVID bioinformatic analysis (http://david.abcc.ncifcrf.gov/home.jsp) was used to identify enriched gene classes from the RNAi screen. Multiple comparison correction was made for several experiments.
See Supplemental Experimental Procedures for detailed descriptions of behavioral assays, animal imaging, tissue-restricted RNAi experiments, statistical methods and see Table S4 for an inventory of C. elegans and bacterial strains used in this study.
Highlights.
-
-
Inactivation of vital cellular processes by RNAi stimulates microbial aversion
-
-
Drugs and toxins targeting the vital cell processes stimulates aversion behavior
-
-
Immunity and detoxification defenses are induced by RNAi against cellular processes
-
-
Jnk kinase and serotonergic signaling mediate the aversion response
Supplementary Material
Acknowledgements
We thank F. Ausubel, D. McEwen, R. Pukkila-Worley, E. Troemel, S.Curran, A.Frand, J.Kim, Y.Tabach, B.Cezairliyan, A.Jose, J.Gray, B.Shtonda, L.Avery, C.Bargmann, and D.Hopwood for helpful discussions, David Shore for participation in stress reporter assays, E.O’Rourke and B.Samuel for assistance with pharyngeal pumping assays and Rusty Howson for figure support. We thank the CGC, OMRF knockout consortium, National Bioresource Project of Japan, Wormbase, and F.Ausubel, D.Kim, E.Troemel, J.Irazoqui, J.Kaplan, J.Kim, A.Soukas, L.Avery and R.Losick for strains. For sharing unpublished results, we thank F.Ausubel, D.Kim, E.Troemel, Y.Tabach, L.Avery and S.Politz. Salary support for J.A.M. from the Jane Coffin Childs Memorial Fund, the Bank of America Charles King Trust and the MGH Fund for Medical Discovery. This work was funded by NIH grant DK070147.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bolz DD, Tenor JL, Aballay A. A conserved PMK-1/p38 MAPK is required in caenorhabditis elegans tissue-specific immune response to Yersinia pestis infection. J Biol Chem. 2010;285:10832–10840. doi: 10.1074/jbc.M109.091629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Calixto A, Chelur D, Topalidou I, Chen X, Chalfie M. Enhanced neuronal RNAi in C. elegans using SID-1. Nat Methods. 2010;7:554–559. doi: 10.1038/nmeth.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar TL, Yan Z, Balla KM, Troemel ER. C. elegans detects pathogen-induced translational inhibition to activate bZIP immune signaling. Cell Host Microbe. 2012 doi: 10.1016/j.chom.2012.02.008. IN PRESS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes KA, Dunbar TL, Powell JR, Ausubel FM, Troemel ER. bZIP transcription factor zip-2 mediates an early response to Pseudomonas aeruginosa infection in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2010;107:2153–2158. doi: 10.1073/pnas.0914643107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushige T, Hawkins MG, McGhee JD. The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev Biol. 1998;198:286–302. [PubMed] [Google Scholar]
- Hao L, Mukherjee K, Liegeois S, Baillie D, Labouesse M, Burglin TR. The hedgehog-related gene qua-1 is required for molting in Caenorhabditis elegans. Dev Dyn. 2006;235:1469–1481. doi: 10.1002/dvdy.20721. [DOI] [PubMed] [Google Scholar]
- Hart AC. In: Wormbook. Hart AC, Ambros V, editors. The C. elegans Research Community; 2006. [Google Scholar]
- Haynes CM, Ron D. The mitochondrial UPR - protecting organelle protein homeostasis. J Cell Sci. 2010;123:3849–3855. doi: 10.1242/jcs.075119. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Iordanov MS, Pribnow D, Magun JL, Dinh TH, Pearson JA, Chen SL, Magun BE. Ribotoxic stress response: activation of the stress-activated protein kinase JNK1 by inhibitors of the peptidyl transferase reaction and by sequence-specific RNA damage to the alpha-sarcin/ricin loop in the 28S rRNA. Mol Cell Biol. 1997;17:3373–3381. doi: 10.1128/mcb.17.6.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iser WB, Wilson MA, Wood WH, 3rd, Becker K, Wolkow CA. Co-regulation of the DAF-16 target gene, cyp-35B1/dod-13, by HSF-1 in C. elegans dauer larvae and daf-2 insulin pathway mutants. PLoS One. 2011;6:e17369. doi: 10.1371/journal.pone.0017369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Jose AM, Smith JJ, Hunter CP. Export of RNA silencing from C. elegans tissues does not require the RNA channel SID-1. Proc Natl Acad Sci U S A. 2009;106:2283–2288. doi: 10.1073/pnas.0809760106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kao CY, Los FC, Huffman DL, Wachi S, Kloft N, Husmann M, Karabrahimi V, Schwartz JL, Bellier A, Ha C, et al. Global functional analyses of cellular responses to pore-forming toxins. PLoS Pathog. 2011;7:e1001314. doi: 10.1371/journal.ppat.1001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Liberati NT, Mizuno T, Inoue H, Hisamoto N, Matsumoto K, Ausubel FM. Integration of Caenorhabditis elegans MAPK pathways mediating immunity and stress resistance by MEK-1 MAPK kinase and VHP-1 MAPK phosphatase. Proc Natl Acad Sci U S A. 2004;101:10990–10994. doi: 10.1073/pnas.0403546101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamitina T, Huang CG, Strange K. Genome-wide RNAi screening identifies protein damage as a regulator of osmoprotective gene expression. Proc Natl Acad Sci U S A. 103(32):12173–12178. doi: 10.1073/pnas.0602987103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 115(4):489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- McEwan DL, Kirienko NV, Ausubel FM. Pseudomonas aeruginosa Exotoxin A Triggers an Immune Response in Caenorhabditis elegans through Translational Inhibition. Cell Host Microbe. 2012 doi: 10.1016/j.chom.2012.02.007. IN PRESS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T, Fujiki K, Sasakawa A, Hisamoto N, Matsumoto K. Role of the Caenorhabditis elegans Shc adaptor protein in the c-Jun N-terminal kinase signaling pathway. Mol Cell Biol. 2008;28:7041–7049. doi: 10.1128/MCB.00938-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T, Hisamoto N, Terada T, Kondo T, Adachi M, Nishida E, Kim DH, Ausubel FM, Matsumoto K. The Caenorhabditis elegans MAPK phosphatase VHP-1 mediates a novel JNK-like signaling pathway in stress response. EMBO J. 2004;23:2226–2234. doi: 10.1038/sj.emboj.7600226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke D, Baban D, Demidova M, Mott R, Hodgkin J. Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of C. elegans with M. nematophilum. Genome Res. 2006;16:1005–1016. doi: 10.1101/gr.50823006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JR, Kim DH, Ausubel FM. The G protein-coupled receptor FSHR-1 is required for the Caenorhabditis elegans innate immune response. Proc Natl Acad Sci U S A. 2009;106:2782–2787. doi: 10.1073/pnas.0813048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahlad V, Cornelius T, Morimoto RI. Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science. 2008;320:811–814. doi: 10.1126/science.1156093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahlad V, Morimoto RI. Neuronal circuitry regulates the response of Caenorhabditis elegans to misfolded proteins. Proc Natl Acad Sci U S A. 2011;108:14204–14209. doi: 10.1073/pnas.1106557108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol N, Cypowyj S, Ziegler K, Millet A, Astrain A, Goncharov A, Jin Y, Chisholm AD, Ewbank JJ. Distinct innate immune responses to infection and wounding in the C. elegans epidermis. Curr Biol. 2008;18:481–489. doi: 10.1016/j.cub.2008.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol N, Link EM, Liu LX, Kurz CL, Alloing G, Tan MW, Ray KP, Solari R, Johnson CD, Ewbank JJ. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr Biol. 2001;11:809–821. doi: 10.1016/s0960-9822(01)00241-x. [DOI] [PubMed] [Google Scholar]
- Qadota H, Inoue M, Hikita T, Koppen M, Hardin JD, Amano M, Moerman DG, Kaibuchi K. Establishment of a tissue-specific RNAi system in C. elegans. Gene. 2007;400:166–173. doi: 10.1016/j.gene.2007.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulenburg H, Ewbank JJ. The genetics of pathogen avoidance in Caenorhabditis elegans. Mol Microbiol. 2007;66:563–570. doi: 10.1111/j.1365-2958.2007.05946.x. [DOI] [PubMed] [Google Scholar]
- Schulenburg H, Muller S. Natural variation in the response of Caenorhabditis elegans towards Bacillus thuringiensis. Parasitology. 2004;128:433–443. doi: 10.1017/s003118200300461x. [DOI] [PubMed] [Google Scholar]
- Sicard M, Hering S, Schulte R, Gaudriault S, Schulenburg H. The effect of Photorhabdus luminescens (Enterobacteriaceae) on the survival, development, reproduction and behaviour of Caenorhabditis elegans (Nematoda: Rhabditidae) Environ Microbiol. 2007;9:12–25. doi: 10.1111/j.1462-2920.2006.01099.x. [DOI] [PubMed] [Google Scholar]
- Styer KL, Singh V, Macosko E, Steele SE, Bargmann CI, Aballay A. Innate immunity in Caenorhabditis elegans is regulated by neurons expressing NPR-1/GPCR. Science. 2008;322:460–464. doi: 10.1126/science.1163673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, Kim DH. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2006;2:e183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano F, Calfon M, Yoneda T, Yun C, Kiraly M, Clark SG, Ron D. A survival pathway for Caenorhabditis elegans with a blocked unfolded protein response. J Cell Biol. 2002;158:639–646. doi: 10.1083/jcb.200203086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda T, Benedetti C, Urano F, Clark SG, Harding HP, Ron D. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J Cell Sci. 2004;117:4055–4066. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
- Yook K, Hodgkin J. Mos1 mutagenesis reveals a diversity of mechanisms affecting response of Caenorhabditis elegans to the bacterial pathogen Microbacterium nematophilum. Genetics. 2007;175:681–697. doi: 10.1534/genetics.106.060087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun M, Politz S. Personal communication: the mlk-1/mek-1/kgb-1 pathway is required for C. elegans resistance to killing by S cerevisiae. 2011 [Google Scholar]
- Zhang Y, Lu H, Bargmann CI. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature. 2005;438:179–184. doi: 10.1038/nature04216. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.