Abstract
Protein-protein interfaces are often stabilized by a small number of dominant contacts, exemplified by the overrepresentation of arginine residues at oligomerization interfaces. Positively charged arginines are most commonly involved in ion pairs of opposite charge; however, previous work of Scheraga and coworkers described the stable, close range interaction between guanidinium pairs in a solvated environment. To extend this work, we searched over 70 thousand protein structures and complexes for unusual formations of arginine residues supported by the electron density. Symmetry transformations were used to generate full assemblies. Clusters of four to eight arginine residues with Cζ-Cζ distances < 5 Å, organized as rings with 4 to 8 members, stacks of two arginines, and strings of stacked arginines, are commonly located at the interfaces of oligomeric proteins. The positive charge is properly balanced by negatively charged counter ions in about 90% of the cases. We also observed planar stacking of guanidinium groups, bridged by hydrogen bonds and interactions with water molecules. The guanidinium groups are commonly involved in 5 hydrogen bonds with water molecules and acceptor groups from surrounding amino acids. Water molecules have a bridging effect on the arginine pairs, but in some cases small molecular weight chemicals in the crystallization buffer may be misinterpreted as water molecules. In summary, despite electrostatic repulsion, arginines do form various clusters that are exposed to interact with and potentially be controlled or switched by charged metabolites, membrane lipids, nucleic acids or side chains of other proteins. Control of the stability of arginine clusters may play an important role in protein-protein oligomerization, molecular recognition and ligand binding.
Keywords: arginine, guanidinium, ion pair interactions, solvation, electrostatic, protein structure, protein-protein oligomerization, protein complexes
INTRODUCTION
Arginine residues are important mediators of protein-protein and protein-nucleic acid interactions in a wide range of biological process.1,2 Unique physicochemical properties of arginine residues are related to the positively charged guanidinium with five hydrogen bond donors and a special type of aromatic system.3 Charged residues such as arginine are preferentially localized on the surface of proteins, whereas hydrophobic residues are predominantly buried in the protein core.4 The extended flexibility of the arginine side chain and the extremely high pKa value of the guanidinium group, facilitate the interaction with negatively charged groups. About 40% of the salt-bridges in proteins involve ion pairs between arginine and carboxylate groups of acidic amino acids.5 The positive charge is also important for planar stacking and polar interactions with aromatic residues.6,7 In addition, arginine residues can establish multiple hydrogen bonds with nucleic acids and play an important role in the catalytic mechanism of a number of enzymes.8 Moreover, arginines make dominant energetic contributions to the stabilization of protein-protein complexes and are one of the most common residues of “hot spot” protein interfaces.9, 10 While protein interfaces are commonly depleted of charged residues such as lysine, aspartic acid and glutamic acid, the number of arginines at protein interfaces is larger than expected from random distribution.11 In addition, guanidinium groups are significantly overrepresented at protein-protein interfaces, as opposed to other polar and charged groups, such as the terminal nitrogen in lysine, aspartate and glutamate carboxylates, and polar amide groups of asparagine and glutamine.12
Ion-pairs in protein structures are formed mostly between residues of opposite charge. However, previous work by Scheraga and co-workers identified an unusually large number of arginine-arginine pairs in protein structures from the PDB database.13 Because most of the negatively charged pairs were found at the protein surface, semi-empirical calculations using the AM1 molecular orbital method were used to study the interaction between guanidinium moieties in the solvated environment. The authors concluded that water molecules stabilize the positively charged guanidinium groups and have a bridging effect on the arginines pairs. Recently, we found unusual clusters of 5 and 6 arginine residues in pentameric and hexameric oligomers of the HIV CA protein, respectively.14 During CA protein oligomerization into the basic building blocks for capsid formation, one Arg18 from each subunit comes into close contact at the center of the rings, suggesting a mechanism of electrostatic regulation.15,16 This prompted us to examine the PDB database for patterns of arginine clusters. Our analysis showed that short-range interactions between arginine pairs and higher-order formations are commonly found at the interface of protein-protein oligomerization. Stabilizing interactions are discussed using a test set of high quality structures and representative examples from the PDB database.
EXPERIMENTAL METHODS
Clusters of positively charged arginines were identified using Molsoft Internal Coordinate Mechanics (ICM) software.17,18 among 67,520 X-ray crystal structures with resolution < 3 Å deposited in the PDB database19 The initial analysis was focused on arginine clusters with at least 4 positively charged residues. A script (available upon request) was written in the ICM language and was used to download the PDB entries and generate symmetry-related neighbors within a cutoff distance of 20 Å. Symmetry-related residues were generated by replication of the unit cell according to the crystallographic space group and the unit cell parameters provided in the PDB files. For each arginine within the unit cell, close proximity arginines with Cζ-Cζ distances < 5 Å were collected. This procedure was repeated until all arginines in close proximity were clustered together. The identified clusters at protein-protein interfaces were further confirmed to be biologically relevant using the available literature.
The arginine set was further filtered using electron density. A subset of high-quality clusters were identified by unambiguous electron density around arginine side chains to assure their direct interaction. The maps were downloaded from the Uppsala Electron Density server20 and fitted to the arginine side chains using a previously described algorithm.21 Arginine residues with low occupancy atoms or with side chain conformations not supported by the electron density were excluded from the analysis. Acidic residues, hydrogen bond acceptors and water molecules were counted, and the electron density for potentially misassigned or unassigned anions was analyzed (see below). Negatively charged hetero groups were counted after assigning MMFF atom types.22
RESULTS AND DISCUSSION
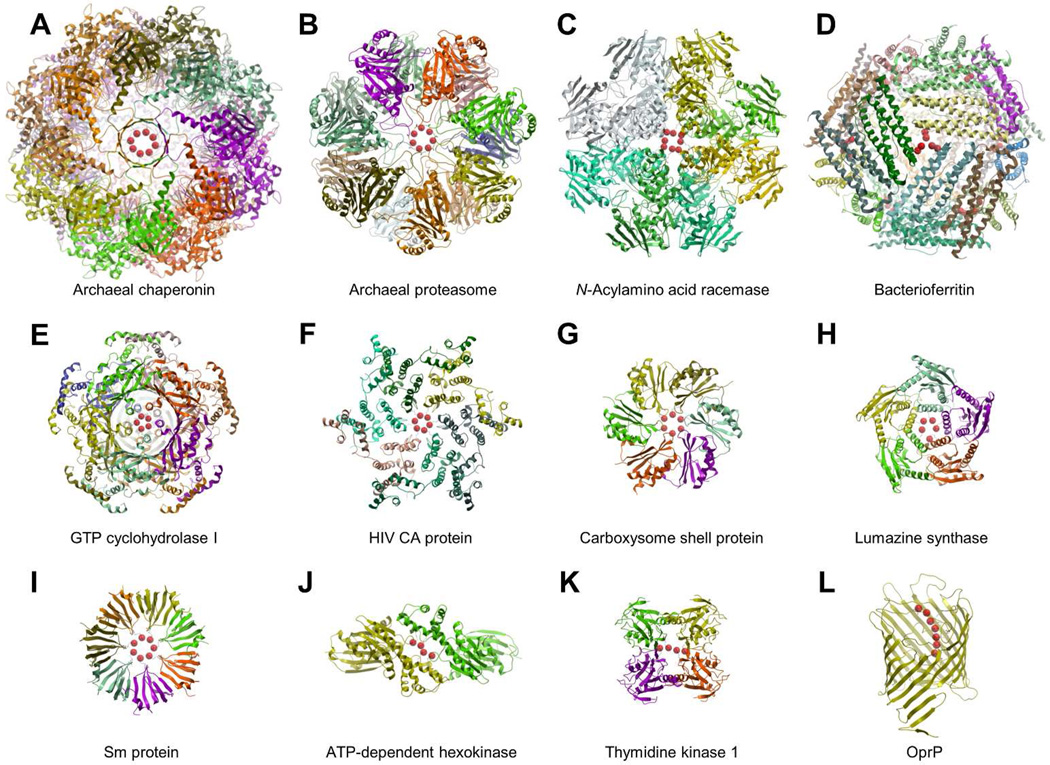
As anticipated by Scheraga and coworkers, we found a large number of short range interactions between positively charged arginine side chains in the PBD database. While most of the clusters were arginine pairs, a number of unusual geometrical formations with up to eight residues was found. Representative examples are shown in Figure 1 and listed in Table 1. Clusters of four to eight arginines with Cζ-Cζ distances < 5 Å, organized as rings with 4 to 8 members, stacks of two arginines, and strings of stacked arginines, are commonly located at the interface of oligomeric proteins. Furthermore, these arginine formations are usually exposed to interact with other external partners not necessarily present in the analyzed structures. These potential interaction partners may include charged metabolites, membranes, nucleic acids, and other proteins (possibly depending on their phosphorylation state).
Figure 1.
Representative examples of arginine clusters with Cζ-Cζ distances < 5 Å obtained from the PDB database. A) Archaeal chaperonin (PDB entry 3kfb); B) Archaeal proteasome (PDB entry 1j2q); C) N-Acylamino acid racemase (PDB entry 1r0m); D) Bacterioferritin (PDB entry 1nf4); E) GTP cyclohydrolase I (PDB entry 1wpl); F) HIV CA protein (PDB entry 3h4e); G) Carboxysome shell protein, (PDB entry 2a10); H) Lumazine synthase (PDB entry 2f59); I) Sm protein (PDB entry 1i8f); J) ATP-dependent hexokinase (PDB entry 2e2q); K) Thymidine kinase 1 (PDB entry 2wvj); L) Outer membrane protein (PDB entry 2o4v). The protein structures are represented with a ribbon model and colored by subunit. Cζ atoms are represented in red with space filling spheres.
Table 1.
Clusters of 4 to 8 arginines collected from the PDB database. Common types of arginine clusters include rings, stacks and strings of stacked residues.
| Protein | Number of arginines and type |
PDB entry | Description |
|---|---|---|---|
| Archaeal chaperonin | 8 – ring | 3kfb | Cylindrical protein complex with two stacking rings of 8 subunits each.34 |
| N-Acylamino acid racemase | 8 – stack | 1r0m | Homooctameric protein with a 4-fold symmetry axis.25,26 |
| Sm protein | 7 – ring | 1i8f | Heptameric protein in a ring.41,53 |
| Archaeal proteasome | 7 – ring | 1j2q | Four heptameric proteins in a barrel-shaped structure.35 |
| Outer membrane protein | 7 – string | 2o4v | Transporter protein.42 |
| HIV CA protein | 6 – ring | 3h4e | Protein hexamer. Assembles into hexamers and pentamers to form closed shell capsids.16 |
| Bacterioferritin | 6 – ring | 1nf4 | Spherical shell protein with 24 subunits.40 |
| Carboxysome shell protein | 6 – ring | 2a10 | Protein hexamer. Assembles into hexamers and pentamers to form icosahedral compartments.37 |
| GTP cyclohydrolase I | 5 – ring | 1wpl | Two rings of 5 subunits each.27 |
| Lumazine synthase | 5 – ring | 2f59 | Pentameric protein in a ring.54 |
| ATP-dependent hexokinase | 4 – string | 2e2q | Protein dimer.30 |
| Thymidine kinase 1 | 4 – string | 2wvj | Protein tetramer.31 |
Protein-protein oligomerization
The hydrophobic and aromatic interaction between non-polar residues, a major driving force for protein folding and protein-protein assembly, plays an important role at defining the architecture of oligomeric interfaces. However, subunit interface areas are more hydrophilic than a typical protein core, and have a larger number of polar and aromatic residues.23 Early studies showed that arginine residues are commonly involved in hydrogen bonding contacts at the interface of oligomeric proteins.24 Moreover, the Protein IntErface Recognition method (PIER) demonstrated that guanidinium groups are significantly overrepresented at protein interfaces.12 Our results show that quaternary interactions between clusters of arginines, negatively charged counter-ions, hydrogen bonding residues and water molecules, are involved in protein oligomer stabilization.
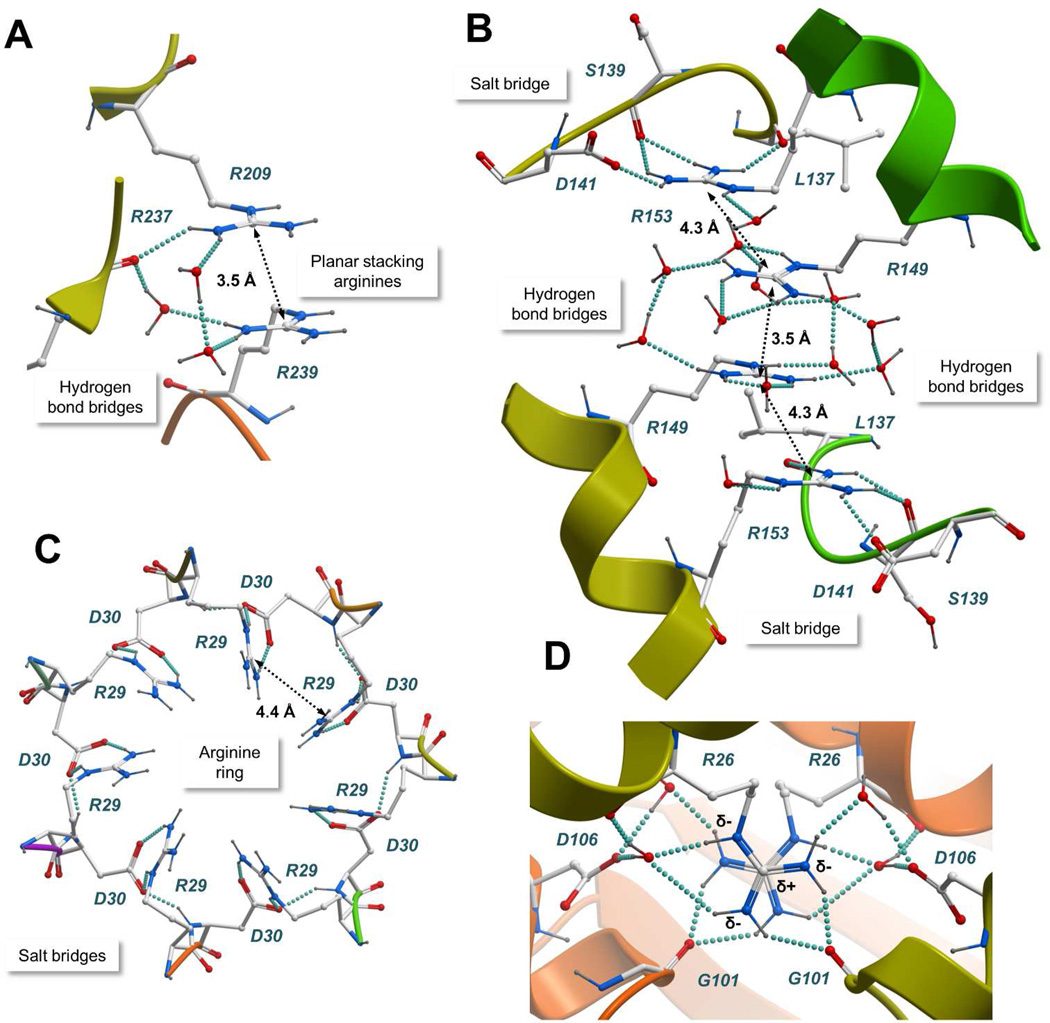
Figure 1 displays representative protein complexes with arginine clusters. N-Acylamino acid racemase catalyzes the interconversion of N-acylamino acid enantiomers. The 1.3 Å X-ray crystal structure of the enzyme reveals a homo-octameric architecture with individual subunits packed around a 4-fold symmetry axis.25,26 As shown in Figure 1C, two arginines from each of the 4 centrally located subunits form a large cluster of 8 positive charges. This unusually large cluster of arginines is exposed to the solvent and stabilized by a network of hydrogen bonds between the guanidinium groups, water molecules and the carbonyl oxygen of Arg237. Figure 2A illustrates the hydrogen bonding network between a pair of adjacent arginines with Cζ-Cζ separated by 3.5 Å. The guanidinium groups adopt a planar stacking conformation bridged by hydrogen bonds, involving either a pair of water molecules, or one water molecule and the carbonyl oxygen of Arg237. Arginine pairs in the same subunit are stabilized by a salt bridge with Asp212 and separated by a Cζ-Cζ distance of 4.6 Å.
Figure 2.
Atomic details of representative arginine clusters. A) N-Acylamino acid racemase (PDB entry 1r0m); B) ATP-dependent hexokinase (PDB entry 2e2q); C) Sm protein (PDB entry 1i8f); D) Nucleoside diphosphate kinase (PDB entry 2hur). The protein structures are represented with a ribbon model and colored by subunit. Interacting residues and water molecules are shown with a “balls and sticks” model. Hydrogen bonds are represented with cyan spheres and the Cζ-Cζ distances between arginine pairs are shown. Common stabilizing interactions such as planar stacking of arginines, counterions, and bridges of hydrogen bonds are shown.
GTP cyclohydrolase I is involved in the synthesis of the tetrahydrobiopterin cofactor. The protein assembles as two rings of homopentamers with a cluster of 5 arginines located at the central symmetry axis (Figure 1E). Individual arginines establish a hydrogen bond with the hydroxyl group of Ser241 located in the adjacent subunit.27 The guanidinium groups form a solvent accessible ring.
Figure 1K shows thymidine kinase, a key enzyme in the synthesis of thymidine monophosphate, and Figure 1J shows hexokinase, an enzyme that catalyzes the phosphorylation of glucose into glucose- 6-phosphate. Thymidine kinase has been reported to assemble as a low activity dimer and as a highly active tetramer.28 Hexokinase forms dimers in solution.29 Interestingly, both X-ray crystal structures show a typical string of stacked arginines with Cζ-Cζ distances < 5 Å.30,31 As shown in Figure 2B, the arginine cluster in a 2.0 Å structure of hexokinase is stabilized by salt bridges with Asp141 and by a network of hydrogen bonds. Arginines establish the maximum possible number of hydrogen bonding contacts, using either the solvent or hydrogen bond acceptors from surrounding residues. Bridges of 2 to 4 water molecules can be found at the geometrical center of the cluster (Arg149-Arg149 pair).
Highly dynamic oligomeric protein domains
We found a number of arginine clusters at highly dynamic domains of large protein complexes, such as chaperonins and the proteasome (Figures 1A and B). Molecular chaperones mediate protein folding, whereas the proteasome is involved in protein degradation.32,33 In both cases the interacting proteins are processed in an inner chamber, accessible through an axial channel that adopts distinct conformations in the open and closed state. Figure 1A illustrates a cluster of 8 arginines stabilizing the closed state of a built-in lid of the Methanococcus maripaludis archaeal chaperonin.34 The X-ray crystal structure shows a ring conformation with adjacent arginine pairs separated by an average Cζ-Cζ distance of 4.1 Å. This unusually large number of positive charges is stabilized by salt bridge interactions with an adjacent ring of negatively charged residues. Lid residues are highly conserved and play an important role in the chaperonin mechanism of protein folding. The arginine ring is disrupted in the open conformation in order to facilitate access to the folding chamber.
Archaeal proteasomes are another example of arginine rings in flexible protein domains. A highly conserved and essential YDR motif is involved in the gating mechanism that allows protein substrates to reach the inner chamber of the Archaeoglobus fulgidus 20 S proteasome.35 As shown in Figure 1B, seven arginines with an average Cζ-Cζ distance of 4.5 Å interact at the top of the channel entrance. The arginine side chains point towards the outside of the proteasome chamber suggesting that hydrogenbonding contacts with the solvent, as well as negatively charged counter ions, might be the major stabilizing interactions.
Closed shell assembly
The building blocks for HIV-1 capsid formation, a large closed shell structure enclosing the viral genome, are quasi-equivalent pentamers and hexamers of the viral CA protein.14 The capsid can be modeled as a fullerene-like cone assembled from ~250 hexamers and exactly 12 pentamers that are required to close the capsid. As shown in Figure 1F, 6 arginines with Cζ- Cζ distances < 5 Å are found in the X-ray crystal structure of the hexamer.16 The arginine side chains are exposed to the solvent and could be stabilized by water molecules and/or charge-compensated by negatively-charged counter ions from either a permanent charge of Glu or Asp or a phosphorylated Ser, Thr or Tyr. Other compensatory anions are also possible. An analogous ring of arginines resides at the center of the CA protein pentamers; however, in this case the residues pack in a very small area, suggesting that at least some electrostatic repulsion between individual subunits compared with the hexamer.15 Indeed, biochemical studies confirm that the oligomeric assembly of hexamers is preferred over the pentamers.36 Because the arginine residue is highly conserved in the HIV CA proteins from multiple strains, it has been proposed to function as an electrostatic switch, favoring the assembly of hexamers over pentamers.
Our analysis of the protein structure database identified other cases of arginine rings at the oligomerization interface of proteins involved in closed shell formation. Figure 1G shows the hexameric assembly of the carboxysome shell protein CcmK4, a basic building block for the construction of bacterial subcellular compartments involved in carbon fixation. A ring of 6 arginines with Cζ-Cζ distances < 5 Å is found at the center of the hexamer.37 Moreover, carboxysome shell proteins assemble as hexamers and pentamers, and the final carboxysome compartments are approximately icosahedral in shape.38 The negative charge at the center of the ring is conserved among different family members, suggesting a similar electrostatic switch mechanism for assembly of hexamers and pentamers.
Another example found in the PDB database is lumazine synthase (Figure 1H), an enzyme involved in the biosynthesis of riboflavin. In contrast to the CA protein, pentamers of lumazine synthase are stable in solution, and the enzyme oligomerizes into typical pentameric rings that exist either in the free state or as different quaternary associations such as decamers (dimers of pentamers), icosahedrons (dodecamers of pentamers) or large closed shell capsids.39
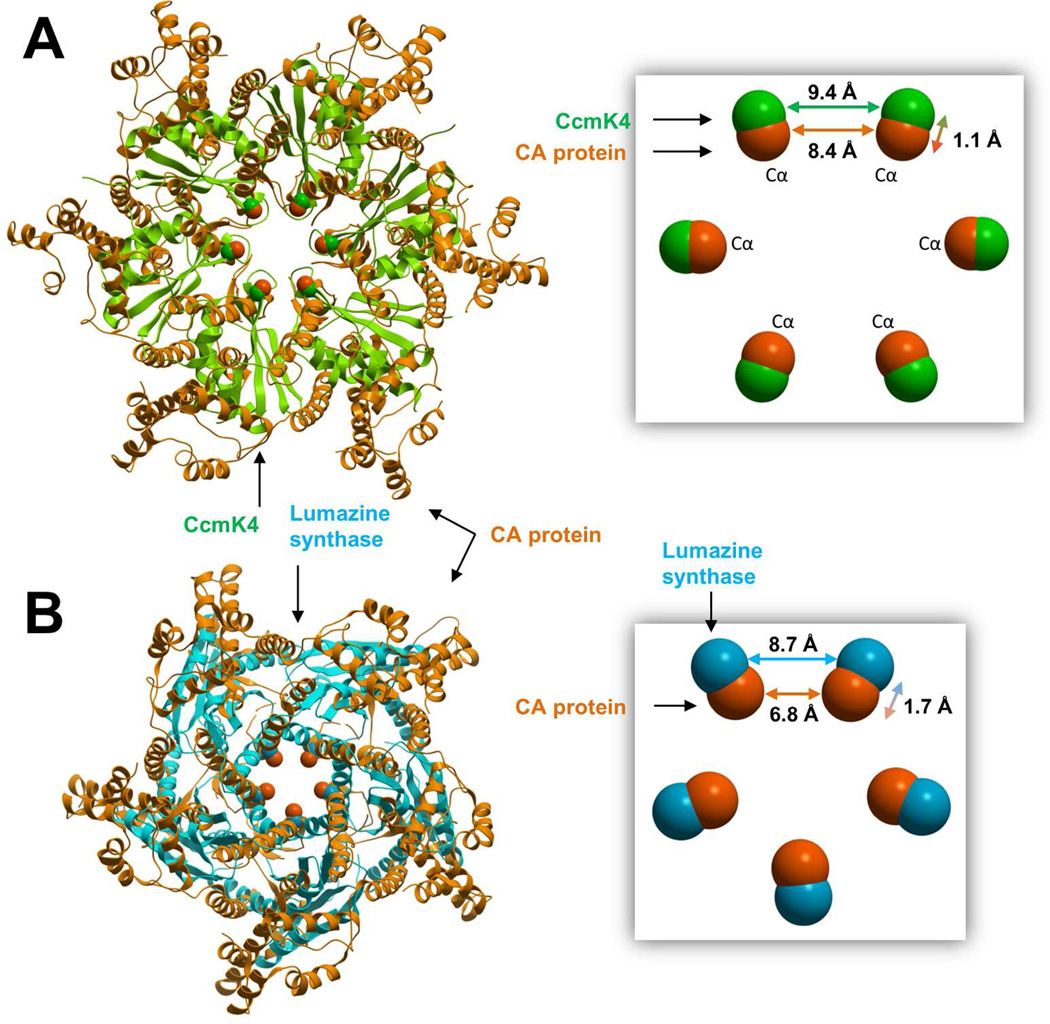
Hexamers of the CA protein were compared with similar types of rings assembled from the carboxysome shell protein, and the structures were superimposed by minimizing the root mean square distance between equivalent alpha carbons in the arginine rings. In a similar way, pentamers of the CA protein were compared with pentamers of lumazine synthase. As shown in Figure 3, the Cα-Cα distance between adjacent arginines in the CA proteins hexamers (8.4 Å) is similar to the equivalent distances in carboxysome shell protein (9.4 Å) and lumazine synthase (8.7 Å). By comparison, the Cα-Cα distance calculated from arginines pairs in pentamers of the CA protein is significantly lower (6.8 Å), suggesting that stronger electrostatic repulsion is involved.
Figure 3.
Three-dimensional superimposition of hexamers (A) and pentamers (B) assembled from the HIV CA protein (orange ribbon, PDB entries 3h4e and 3p05), carboxysome shell protein (green ribbon, PDB entry 2a10) and lumazine synthase (cyan ribbon, PDB entry 2f59). The proteins assemble into large closed shell structures and share a cluster of positively charged arginines at the pentamer/hexamer ring centers. Hexamers and pentamers were superimposed using alpha carbons from the arginine rings, represented with a space filling model. Cα-Cα distances between adjacent arginines are shown.
A large cluster of arginine residues was also found in ferritin, an intracellular iron storage protein present in most living organisms. The protein assembles into spherical shells composed of 24 subunits with two-, three- and four-fold symmetry axes.40 As shown in Figure 1D, large clusters of 6 arginine residues divided into two rings, are found at the three-fold symmetry axes stabilized by layers of negatively charged residues.
Arginine formations with specific known biological functions
Our analysis of the PDB database identified a number of cases where the positively charged arginine residues are involved in the molecular recognition of specific biological partners. Figure 1I illustrates the 3D crystal structure of a small nuclear ribonucleoprotein component (Sm protein) involved in pre-mRNA processing.41 A central ring of 7 arginines forms a cationic pore that interacts specifically with single-stranded RNA. The ring is stabilized by salt bridges with an adjacent ring of negatively charged residues (Figure 2C).
Figure 1L shows a string of 7 stacked arginines involved in the transport of phosphate anions.42 The arginines are stabilized by salt bridges with acidic residues and hydrogen bonds between the guanidinium groups, neighboring residues and water molecules.
Stabilizing interactions derived from a subset of high quality structures
A subset of high resolution arginine clusters with unambiguous fit to the electron density was further analyzed, and the following criteria were used for inclusion: i) the arginine side chain conformations were unambiguously supported by the electron density, ii) arginine residues with low occupancy atoms were excluded, and iii) clusters with arginines from crystallographic neighbors were removed. Using these filters, 13,218 high quality clusters from 6,427 PDB entries were found. The sequences were grouped into 2,978 unique clusters with < 90% identity between supporting protein sequences.
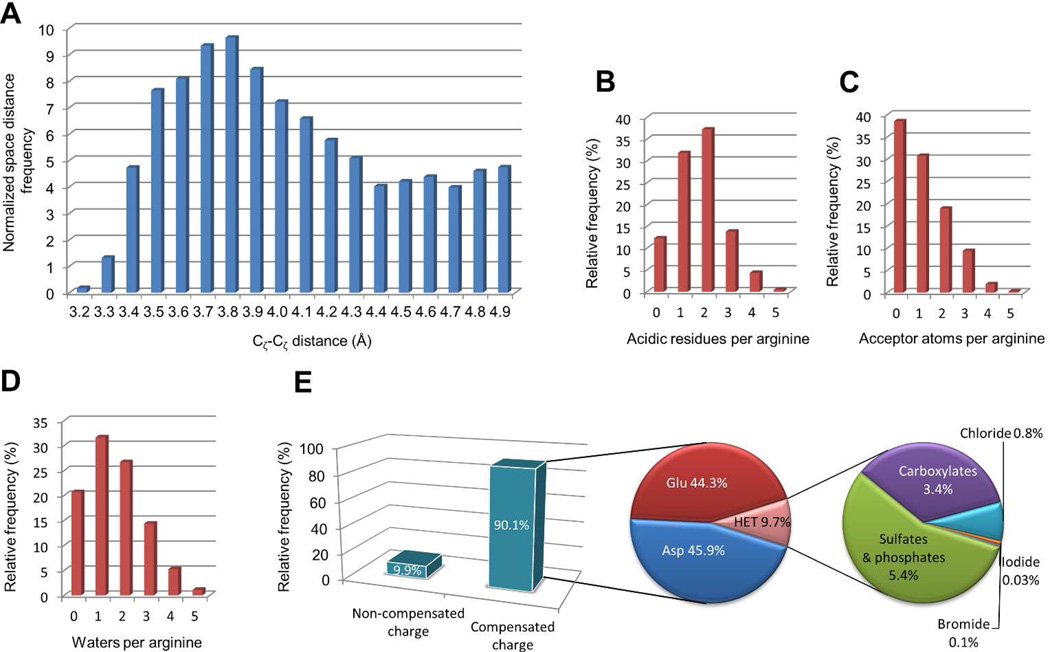
As shown in Figure 4A, the minimum distance between adjacent Cζ-Cζ pairs is 3.2 Å. This value is in good agreement with the stable 3.0 Å ion pair distance described by Scheraga and coworkers.13 Visual inspection revealed a consistent planar stacking of staggered guanidinium groups, exemplified in Figure 2D with an arginine pair at the interface of a nucleoside diphosphate kinase dimer (Cζ-Cζ distance= 3.2 Å).43 Planar stacking avoids steric clashes between the guanidinium groups. Close to 60- degree torsion angles along the N-Cζ-Cζ-N axis minimize the electrostatic repulsion by maximizing the distance between δ-nitrogens. Moreover, the electrostatic repulsion between positively charged arginines is perfectly balanced by the surrounding environment. As shown in Figure 2D, there are two counter ions from aspartic acid residues, and all guanidinium donors are involved in hydrogen bonds with water molecules or surrounding residues. Planar stacking of guanidinium groups, previously described for lumazine synthase44 and glutathione-S-transferase,45 is commonly found in high quality structures from the PDB database. The Cζ-Cζ distance distribution shows a clear preference for arginine pairs separated by approximately 3.8 Å. Because there is less electrostatic repulsion at this distance, the relative orientation of stacked guanidinium groups is determined primarily by interactions with counter ions and hydrogen bonds with the surrounding environment.
Figure 4.
Property distribution of a subset of 13,218 high quality, density-supported, arginine clusters from the PDB database. A) For each arginine, the Cζ-Cζ distance to the closest arginine residue was calculated, and the pairs were grouped into distance bins. The frequency was normalized to account for the cubic growth of spherical volume. The number of arginines that fall into each bin was divided by the square of the distance. Counts for the number of aspartic and glutamic acid residues within a 6 Å distance cutoff (B), the number of hydrogen bond acceptor atoms (C) and crystallographic water molecules (D) within a 3.35 Å distance cutoff are provided. The inset E shows the fraction of arginines surrounded by negatively charged counter ions. Common counter ions are indicated.
Statistical analysis revealed that 90.1% of the arginines are surrounded by negatively charged counter-ions (Figure 4E). As expected, ion pairs with carboxylate groups from aspartate and glutamate are the most frequently found (87.7% of the arginines have at least one acidic amino acid within a 6 Å cutoff distance, Figure 4B). On the other hand, hetero molecules account for 9.7% of the ion-pairs (phosphates and sulfates 5.4%, carboxylates 3.4%, chlorides 0.8% and other halogenated ions 0.1%, Figure 4E). Arginine clusters are involved in binding of phosphate-containing molecules such as nucleotides, NADP, FMN, FAD and acetyl-CoA.46–48 Moreover, arginine clusters play a crucial role in stabilizing the propionic acid side chains of heme groups and bind frequently to organic acids and negatively charged bioactive compunds.49,50 Finally, we found a number of arginine clusters stabilized by chlorides, sulfates, phosphates, acids and other molecules commonly present in the buffers used for protein purification and crystallization.
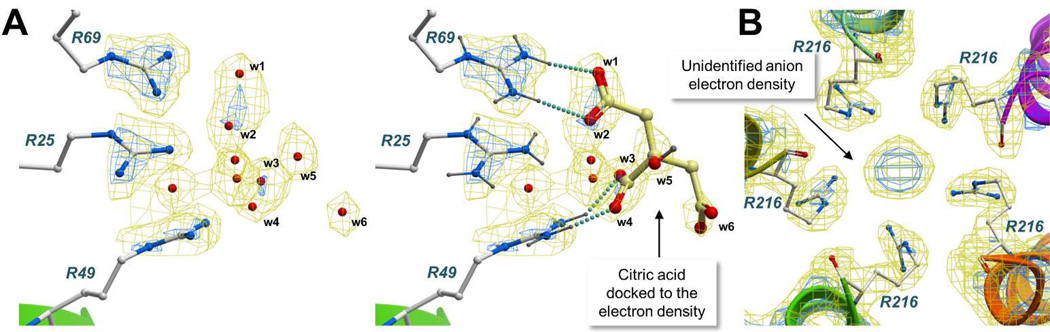
Non-compensated positive charge is found in 9.1% of the arginines in a cluster (Figure 4E). Guanidinium groups tend to be solvent exposed in these cases, or fit into a polar pocket with highly complementary physicochemical properties. The guanidinium cation acts as a strong hydrogen bond donor via the guanidine NH2 and NH groups, with complementary hydrogen bond acceptor counterparts, such as the amide carbonyl of glutamine and aspargine, the hydroxyl groups of serine, threonine and tyrosine, and the backbone carbonyl (Figure 4C). Crystallographic water molecules have an important stabilizing effect and are involved in hydrogen bonds or dipole-dipole interactions with 79% of the arginines (Figure 4D). However, because the electron density for placement of water oxygen atoms is often an ambiguous sphere of unknown origin, small anions such as chloride might be misinterpreted as water molecules or left uninterpreted. Figure 5A shows a cluster of 3 arginines surrounded by 9 crystallographic water molecules in PDB entry 1d3b.51 While the electron density is unambiguous for the arginine side chains, water molecules are placed along a continuous duct of electron density, indicating that they could be replaced by a small molecular weight chemical used in the crystallization buffer. Docking results confirmed that citric acid fits into the electron density surrounding the arginine triplet, replacing 6 water molecules and establishing two stabilizing saltbridges. Figure 5B shows a cluster of 5 arginines with a central lobe of unidentified density.52 Anions used in the crystallization media, such as sulfate, could easily fit into the electron density. Taken together, these results suggest that water molecules are important for the stability of positively charged arginines, but they might be overestimated in some cases due to ambiguous electron density and incorrect placement.
Figure 5.
Ambiguous water molecule fit and unidentified anion electron density. In both cases, negatively charged molecules from the buffers used for protein purification and crystallization are likely involved in stabilizing salt bridge interactions. A) A molecule of citric acid docked to an arginine triplet fits the electron density better than the original water molecules (PDB entry 1d3b). B) The unidentified electron density could easily accommodate anions used in the buffer, such as for example a sulfate molecule (PDB entry 1wuq).
CONCLUSION
In this work, unusual formations of positively charged arginine residues were found in the PDB database, suggesting their important role in protein-protein oligomerization and higher-order assembly, protein-ligand binding and nucleic acid recognition. Controlling a countercharge to these clusters may be used as a biological regulation mechanism. Despite the prevalence of short-range interactions between arginine pairs, higher-order clusters with up to eight guanidinium groups were found at protein oligomerization interfaces, as well as at sites involved in molecular recognition. The positive charge is properly balanced by negatively charged counter ions in about 90% of the cases. Arginine clusters stabilize enzymatic cofactors and bind to nucleotides, glutamates, aspartates and phosphorylated residues, and bioactive compounds bearing carboxylate, sulfate and phosphate moieties. The arginines form rings, stacks and strings of stacked side-chains. Ring formations are often located between layers of acidic amino acids, or at highly exposed sites easily accessible to water molecules and counter ions. Arginine pairs adopt a common planar stacking conformation in part to avoid steric clashes. At the lower Cζ-Cζ distance limit, staggering of guanidinium groups helps minimize the electrostatic repulsion between negatively polarized nitrogens. The guanidinium groups are commonly involved in five hydrogen bonds with water molecules and acceptor groups from surrounding amino acids. Water molecules have a bridging effect on the arginine pairs, but in some cases their electron density might be misinterpreted.
Taken together, our results suggest that clusters of interacting arginine residues play an important role in protein structure and function. They can be stabilized and/or regulated by multiple mechanisms such as stacking, hydrogen bonds and electrostatics, as well as complementary side chains, backbone carbonyls, water molecules, anions or ligands with negatively charged groups or hydrogen bond acceptors. Our results may be particularly relevant for the development of new scoring functions for protein-protein docking, identification of druggable protein sites and for the prediction of oligomerization interfaces.
ACKNOWLEDGMENTS
We thank Irina Kufareva for valuable advice and some tools used in this study. The authors thank Owen Pornillos, Barbie Ganser-Pornillos, Fung-Yi Chan, Manuel Rueda, Winston Chen, Chayan Acharya, Fiona McRobb, Falgun Shah, Chris Edwards for useful discussions and comments. Supported by postdoctoral grant SFRH/BPD/64216/2009 from the Fundação para a Ciência e a Tecnologia (FCT), Portugal (M.N.) and NIH grants R01 HL48908 (M.Y.), R01GM066087 (M.Y.), R01 GM084545 (M.Y.), P50 GM082545 (Wesley Sundquist and M.Y.), R01 GM071872 (R.A.), U01 GM094612 (R.A.) and U54 GM094618 (R.A.).
REFERENCES
- 1.Calnan BJ, Tidor B, Biancalana S, Hudson D, Frankel AD. Science. 1991;252:1167. doi: 10.1126/science.252.5009.1167. [DOI] [PubMed] [Google Scholar]
- 2.Matsuo M, Huang CH, Huang LC. Biochemical Journal. 1978;173:441. doi: 10.1042/bj1730441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams ML, Gready JE. Journal of Computational Chemistry. 1989;10:35. [Google Scholar]
- 4.Tsai CJ, Lin SL, Wolfson HJ, Nussinov R. Protein Science. 1997;6:53. doi: 10.1002/pro.5560060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barlow DJ, Thornton JM. Journal of Molecular Biology. 1983;168:867. doi: 10.1016/s0022-2836(83)80079-5. [DOI] [PubMed] [Google Scholar]
- 6.Burley SK, Petsko GA. FEBS Letters. 1986;203:139. doi: 10.1016/0014-5793(86)80730-x. [DOI] [PubMed] [Google Scholar]
- 7.Flocco MM, Mowbray SL. Journal of Molecular Biology. 1994;235:709. doi: 10.1006/jmbi.1994.1022. [DOI] [PubMed] [Google Scholar]
- 8.Guillén Schlippe YV, Hedstrom L. Archives of Biochemistry and Biophysics. 2005;433:266. doi: 10.1016/j.abb.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 9.Bogan AA, Thorn KS. Journal of Molecular Biology. 1998;280:1. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 10.Ben-Shimon A, Eisenstein M. Journal of Molecular Biology. 2010;402:259. doi: 10.1016/j.jmb.2010.07.021. [DOI] [PubMed] [Google Scholar]
- 11.Bordner AJ, Abagyan R. Proteins-Structure Function and Bioinformatics. 2005;60:353. doi: 10.1002/prot.20433. [DOI] [PubMed] [Google Scholar]
- 12.Kufareva I, Budagyan L, Raush E, Totrov M, Abagyan R. Proteins-Structure Function and Bioinformatics. 2007;67:400. doi: 10.1002/prot.21233. [DOI] [PubMed] [Google Scholar]
- 13.Magalhaes A, Maigret B, Hoflack J, Gomes JNF, Scheraga HA. Journal of Protein Chemistry. 1994;13:195. doi: 10.1007/BF01891978. [DOI] [PubMed] [Google Scholar]
- 14.Ganser-Pornillos BK, Yeager M, Sundquist WI. Current Opinion in Structural Biology. 2008;18:203. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pornillos O, Ganser-Pornillos BK, Yeager M. Nature. 2011;469:424. doi: 10.1038/nature09640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pornillos O, Ganser-Pornillos BK, Kelly BN, Hua Y, Whitby FG, Stout CD, Sundquist WI, Hill CP, Yeager M. Cell. 2009;137:1282. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abagyan R, Totrov M, Kuznetsov D. Journal of Computational Chemistry. 1994;15:488. [Google Scholar]
- 18.Neves MAC, Totrov M, Abagyan R. 2011 Submitted. [Google Scholar]
- 19.Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlić A, Quesada M, Quinn GB, Westbrook JD, et al. Nucleic Acids Research. 2011;39:D392. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleywegt GJ, Harris MR, Zou JY, Taylor TC, Wählby A, Jones TA. Acta Crystallographica Section D-Biological Crystallography. 2004;D60:2240. doi: 10.1107/S0907444904013253. [DOI] [PubMed] [Google Scholar]
- 21.Bottegoni G, Kufareva I, Totrov M, Abagyan R. Journal of Medicinal Chemistry. 2009;52:397. doi: 10.1021/jm8009958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halgren TA. Journal of Computational Chemistry. 1996;17:490. [Google Scholar]
- 23.Argos P. Protein Engineering. 1988;2:101. doi: 10.1093/protein/2.2.101. [DOI] [PubMed] [Google Scholar]
- 24.Janin J, Miller S, Chothia C. Journal of Molecular Biology. 1988;204:155. doi: 10.1016/0022-2836(88)90606-7. [DOI] [PubMed] [Google Scholar]
- 25.Wang WC, Chiu WC, Hsu SK, Wu CL, Chen CY, Liu JS, Hsu WH. Journal of Molecular Biology. 2004;342:155. doi: 10.1016/j.jmb.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 26.Chiu WC, You JY, Liu JS, Hsu SK, Hsu WH, Shih CH, Hwang JK, Wang WC. Journal of Molecular Biology. 2006;359:741. doi: 10.1016/j.jmb.2006.03.063. [DOI] [PubMed] [Google Scholar]
- 27.Maita N, Hatakeyama K, Okada K, Hakoshima T. Journal of Biological Chemistry. 2004;279:51534. doi: 10.1074/jbc.M409440200. [DOI] [PubMed] [Google Scholar]
- 28.Munch-Petersen B, Tyrsted G, Cloos L. Journal of Biological Chemistry. 1993;268:15621. [PubMed] [Google Scholar]
- 29.Nishimasu H, Fushinobu S, Shoun H, Wakagi T. Journal of Bacteriology. 2006;188:2014. doi: 10.1128/JB.188.5.2014-2019.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishimasu H, Fushinobu S, Shoun H, Wakagi T. Journal of Biological Chemistry. 2007;282:9923. doi: 10.1074/jbc.M610678200. [DOI] [PubMed] [Google Scholar]
- 31.Welin M, Kosinska U, Mikkelsen NE, Carnrot C, Zhu C, Wang L, Eriksson S, Munch-Petersen B, Eklund H. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:17970. doi: 10.1073/pnas.0406332102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baumeister W, Walz J, Zühl F, Seemüller E. Cell. 1998;92:367. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 33.Saibil H. Current Opinion in Structural Biology. 2000;10:251. doi: 10.1016/s0959-440x(00)00074-9. [DOI] [PubMed] [Google Scholar]
- 34.Pereira JH, Ralston CY, Douglas NR, Meyer D, Knee KM, Goulet DR, King JA, Frydman J, Adams PD. Journal of Biological Chemistry. 2010;285:27958. doi: 10.1074/jbc.M110.125344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Groll M, Brandstetter H, Bartunik H, Bourenkow G, Huber R. Journal of Molecular Biology. 2003;327:75. doi: 10.1016/s0022-2836(03)00080-9. [DOI] [PubMed] [Google Scholar]
- 36.Ganser-Pornillos BK, von Schwedler UK, Stray KM, Aiken C, Sundquist WI. Journal of Virology. 2004;78:2545. doi: 10.1128/JVI.78.5.2545-2552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO. Science. 2005;309:936. doi: 10.1126/science.1113397. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka S, Kerfeld CA, Sawaya MR, Cai F, Heinhorst S, Cannon GC, Yeates TO. Science. 2008;319:1083. doi: 10.1126/science.1151458. [DOI] [PubMed] [Google Scholar]
- 39.Ritsert K, Huber R, Turk D, Ladenstein R, Schmidt-Bäse K, Bacher A. Journal of Molecular Biology. 1995;253:151. doi: 10.1006/jmbi.1995.0542. [DOI] [PubMed] [Google Scholar]
- 40.Macedo S, Romão CV, Mitchell E, Matias PM, Liu MY, Xavier AV, LeGall J, Teixeira M, Lindley P, Carrondo MA. Nature Structural Biology. 2003;10:285. doi: 10.1038/nsb909. [DOI] [PubMed] [Google Scholar]
- 41.Mura C, Cascio D, Sawaya MR, Eisenberg DS. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5532. doi: 10.1073/pnas.091102298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moraes TF, Bains M, Hancock REW, Strynadka NCJ. Nature Structural & Molecular Biology. 2007;14:85. doi: 10.1038/nsmb1189. [DOI] [PubMed] [Google Scholar]
- 43.Moynié L, Giraud MF, Georgescauld F, Lascu I, Dautant A. Proteins-Structure Function and Bioinformatics. 2007;67:755. doi: 10.1002/prot.21316. [DOI] [PubMed] [Google Scholar]
- 44.Zhang X, Meining W, Fischer M, Bacher A, Ladenstein R. Journal of Molecular Biology. 2001;306:1099. doi: 10.1006/jmbi.2000.4435. [DOI] [PubMed] [Google Scholar]
- 45.Xiao B, Singh SP, Nanduri B, Awasthi YC, Zimniak P, Ji X. Biochemistry. 1999;38:11887. doi: 10.1021/bi990468i. [DOI] [PubMed] [Google Scholar]
- 46.Meissner B, Schleicher E, Weber S, Essen LO. Journal of Biological Chemistry. 2007;282:33142. doi: 10.1074/jbc.M704951200. [DOI] [PubMed] [Google Scholar]
- 47.Pitcher RS, Brissett NC, Picher AJ, Andrade P, Juarez R, Thompson D, Fox GC, Blanco L, Doherty AJ. Journal of Molecular Biology. 2007;366:391. doi: 10.1016/j.jmb.2006.10.046. [DOI] [PubMed] [Google Scholar]
- 48.Dall'Aglio P, Arthur CJ, Williams C, Vasilakis K, Maple HJ, Crosby J, Crump MP, Hadfield AT. Biochemistry. 2011;50:5704. doi: 10.1021/bi2003668. [DOI] [PubMed] [Google Scholar]
- 49.Erlanson DA, Lam JW, Wiesmann C, Luong TN, Simmons RL, DeLano WL, Choong IC, Burdett MT, Flanagan WM, Lee D, Gordon EM, O'Brien T. Nature Biotechnology. 2003;21:308. doi: 10.1038/nbt786. [DOI] [PubMed] [Google Scholar]
- 50.Riise EK, Lorentzen MS, Helland R, Smalås AO, Leiros H-KS, Willassen NP. Acta Crystallographica Section D-Biological Crystallography. 2007;D63:135. doi: 10.1107/S0907444906043812. [DOI] [PubMed] [Google Scholar]
- 51.Kambach C, Walke S, Young R, Avis JM, de la Fortelle E, Raker VA, Lührmann R, Li J, Nagai K. Cell. 1999;96:375. doi: 10.1016/s0092-8674(00)80550-4. [DOI] [PubMed] [Google Scholar]
- 52.Tanaka Y, Nakagawa N, Kuramitsu S, Yokoyama S, Masui R. Journal of Biochemistry. 2005;138:263. doi: 10.1093/jb/mvi120. [DOI] [PubMed] [Google Scholar]
- 53.Mura C, Kozhukhovsky A, Gingery M, Phillips M, Eisenberg D. Protein Science. 2003;12:832. doi: 10.1110/ps.0224703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klinke S, Zylberman V, Bonomi HR, Haase I, Guimarães BG, Braden BC, Bacher A, Fischer M, Goldbaum FA. Journal of Molecular Biology. 2007;373:664. doi: 10.1016/j.jmb.2007.08.021. [DOI] [PubMed] [Google Scholar]