Abstract
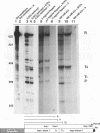
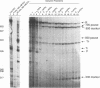
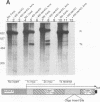
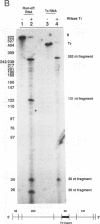
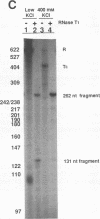
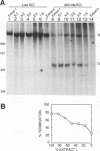
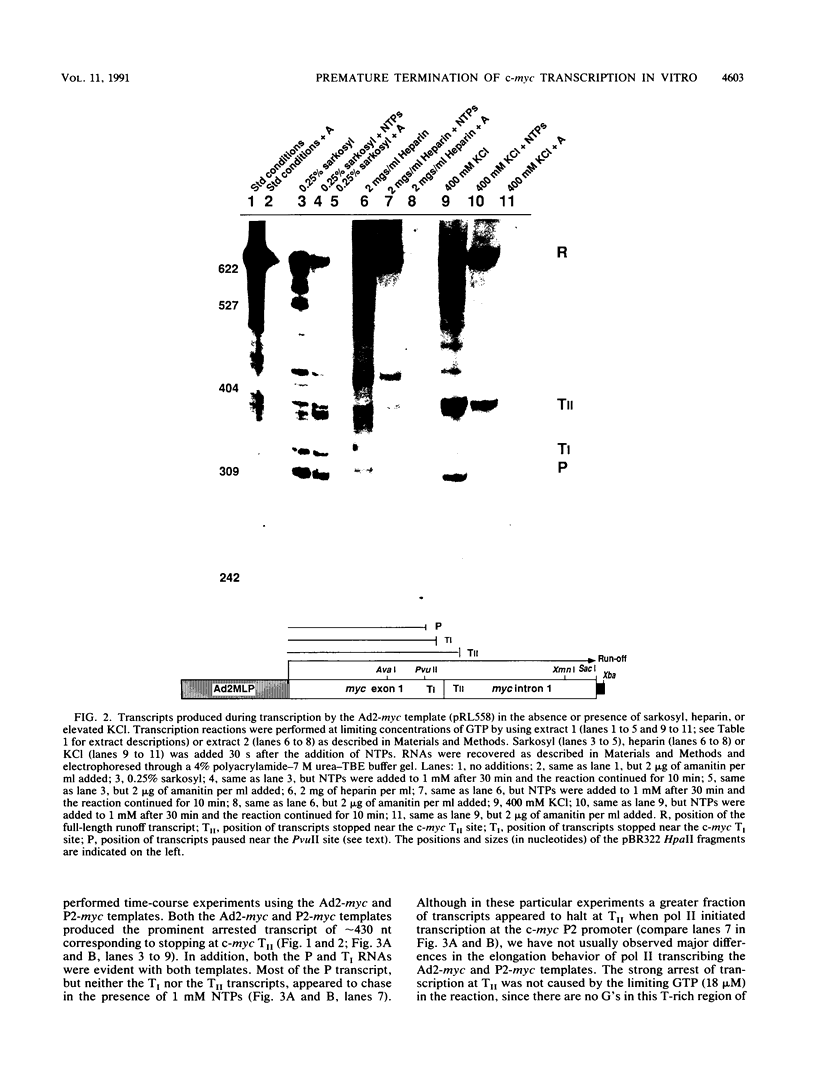
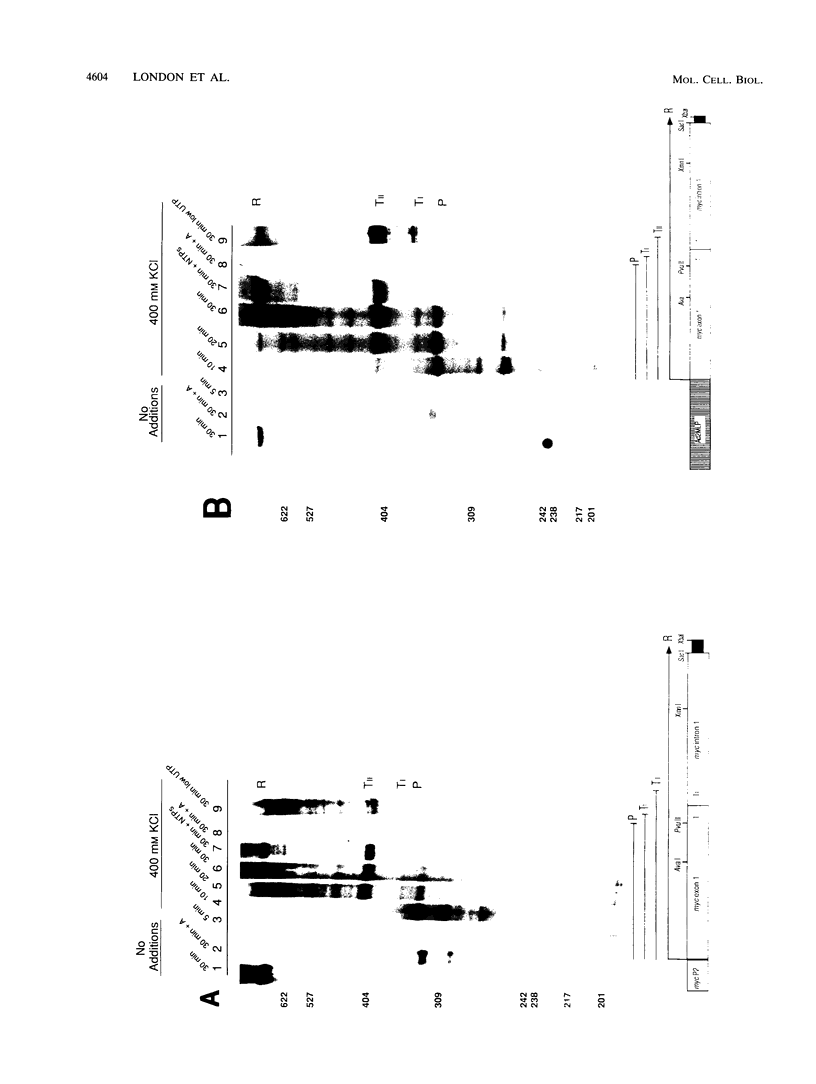
Transcriptional regulation of the human c-myc gene, an important aspect of cellular differentiation, occurs in part at the level of transcript elongation. In vivo, transcriptional arrest, due to either pausing or termination, occurs near the junction between the first exon and first intron and varies with the growth state of the cell. We have tested the transcription of c-myc templates in HeLa nuclear extracts. We did not observe significant arrest under standard conditions, but we found that a considerable fraction of transcription complexes stopped at the c-myc TII site (just past the first exon-intron junction) when the KCl concentration was raised to 400 mM during elongation. Transcriptional arrest at TII also was observed at KCl concentrations as low as 130 mM and when potassium acetate or potassium glutamate was substituted for KCl. Under these conditions, arrest occurred at the TII site when transcription was initiated at either the c-myc P2 promoter or the adenovirus 2 major late promoter. Further, the TII sequence itself, in forward but not reverse orientation, was sufficient to stop transcription in a HeLa nuclear extract. By separating the TII RNA from active transcription complexes by using gel filtration, we found that arrest at TII at 400 mM KCl resulted in transcript release and thus true transcriptional termination. The efficiency of termination at TII depended on the growth state of the cells from which the extracts were made, suggesting that some factor or factors control premature termination in c-myc.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alitalo K., Schwab M., Lin C. C., Varmus H. E., Bishop J. M. Homogeneously staining chromosomal regions contain amplified copies of an abundantly expressed cellular oncogene (c-myc) in malignant neuroendocrine cells from a human colon carcinoma. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1707–1711. doi: 10.1073/pnas.80.6.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amster-Choder O., Wright A. Regulation of activity of a transcriptional anti-terminator in E. coli by phosphorylation in vivo. Science. 1990 Aug 3;249(4968):540–542. doi: 10.1126/science.2200123. [DOI] [PubMed] [Google Scholar]
- Bender T. P., Thompson C. B., Kuehl W. M. Differential expression of c-myb mRNA in murine B lymphomas by a block to transcription elongation. Science. 1987 Sep 18;237(4821):1473–1476. doi: 10.1126/science.3498214. [DOI] [PubMed] [Google Scholar]
- Bengal E., Aloni Y. A block of transcription elongation by RNA polymerase II at synthetic sites in vitro. J Biol Chem. 1989 Jun 15;264(17):9791–9798. [PubMed] [Google Scholar]
- Bengal E., Flores O., Krauskopf A., Reinberg D., Aloni Y. Role of the mammalian transcription factors IIF, IIS, and IIX during elongation by RNA polymerase II. Mol Cell Biol. 1991 Mar;11(3):1195–1206. doi: 10.1128/mcb.11.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengal E., Goldring A., Aloni Y. Transcription complexes synthesizing attenuated RNA can serve as a model system for analyzing elongation factors. J Biol Chem. 1989 Nov 15;264(32):18926–18932. [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986 Jun 12;321(6071):702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. Sequence requirements for premature termination of transcription in the human c-myc gene. Cell. 1988 Apr 22;53(2):245–256. doi: 10.1016/0092-8674(88)90386-8. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Brown D. D. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981 Apr;24(1):261–270. doi: 10.1016/0092-8674(81)90522-5. [DOI] [PubMed] [Google Scholar]
- Briggs D., Jackson D., Whitelaw E., Proudfoot N. J. Direct demonstration of termination signals for RNA polymerase II from the sea urchin H2A histone gene. Nucleic Acids Res. 1989 Oct 25;17(20):8061–8071. doi: 10.1093/nar/17.20.8061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzone F. J., Britten R. J., Davidson E. H. Mapping of gene transcripts by nuclease protection assays and cDNA primer extension. Methods Enzymol. 1987;152:611–632. doi: 10.1016/0076-6879(87)52069-9. [DOI] [PubMed] [Google Scholar]
- Chen Z., Harless M. L., Wright D. A., Kellems R. E. Identification and characterization of transcriptional arrest sites in exon 1 of the human adenosine deaminase gene. Mol Cell Biol. 1990 Sep;10(9):4555–4564. doi: 10.1128/mcb.10.9.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodchoy N., Pandey N. B., Marzluff W. F. An intact histone 3'-processing site is required for transcription termination in a mouse histone H2a gene. Mol Cell Biol. 1991 Jan;11(1):497–509. doi: 10.1128/mcb.11.1.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson T. W., Clayton D. A. In vitro transcription of human mitochondrial DNA: accurate termination requires a region of DNA sequence that can function bidirectionally. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6277–6281. doi: 10.1073/pnas.83.17.6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J., Sussman D. J., Zeller R., Leder P. The c-myc gene encodes superimposed RNA polymerase II and III promoters. Cell. 1987 Dec 24;51(6):1001–1008. doi: 10.1016/0092-8674(87)90586-1. [DOI] [PubMed] [Google Scholar]
- Collart M. A., Tourkine N., Belin D., Vassalli P., Jeanteur P., Blanchard J. M. c-fos gene transcription in murine macrophages is modulated by a calcium-dependent block to elongation in intron 1. Mol Cell Biol. 1991 May;11(5):2826–2831. doi: 10.1128/mcb.11.5.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick D., Berger R., Polack A., Bornkamm G. W. Transcription of c-myc in human mononuclear cells is regulated by an elongation block. Oncogene. 1987;2(1):61–65. [PubMed] [Google Scholar]
- Eick D., Bornkamm G. W. Transcriptional arrest within the first exon is a fast control mechanism in c-myc gene expression. Nucleic Acids Res. 1986 Nov 11;14(21):8331–8346. doi: 10.1093/nar/14.21.8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores O., Maldonado E., Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II. Factors IIE and IIF independently interact with RNA polymerase II. J Biol Chem. 1989 May 25;264(15):8913–8921. [PubMed] [Google Scholar]
- Friedman D. I., Imperiale M. J., Adhya S. L. RNA 3' end formation in the control of gene expression. Annu Rev Genet. 1987;21:453–488. doi: 10.1146/annurev.ge.21.120187.002321. [DOI] [PubMed] [Google Scholar]
- Gollnick P., Ishino S., Kuroda M. I., Henner D. J., Yanofsky C. The mtr locus is a two-gene operon required for transcription attenuation in the trp operon of Bacillus subtilis. Proc Natl Acad Sci U S A. 1990 Nov;87(22):8726–8730. doi: 10.1073/pnas.87.22.8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D. J. Regulation of c-myc transcription in vitro: dependence on the guanine-rich promoter element ME1a1. Oncogene. 1990 Jan;5(1):47–54. [PubMed] [Google Scholar]
- Hawley D. K., Roeder R. G. Functional steps in transcription initiation and reinitiation from the major late promoter in a HeLa nuclear extract. J Biol Chem. 1987 Mar 15;262(8):3452–3461. [PubMed] [Google Scholar]
- Hawley D. K., Roeder R. G. Separation and partial characterization of three functional steps in transcription initiation by human RNA polymerase II. J Biol Chem. 1985 Jul 5;260(13):8163–8172. [PubMed] [Google Scholar]
- Hernandez N., Weiner A. M. Formation of the 3' end of U1 snRNA requires compatible snRNA promoter elements. Cell. 1986 Oct 24;47(2):249–258. doi: 10.1016/0092-8674(86)90447-2. [DOI] [PubMed] [Google Scholar]
- Horwitz R. J., Li J., Greenblatt J. An elongation control particle containing the N gene transcriptional antitermination protein of bacteriophage lambda. Cell. 1987 Nov 20;51(4):631–641. doi: 10.1016/0092-8674(87)90132-2. [DOI] [PubMed] [Google Scholar]
- Kao S. Y., Calman A. F., Luciw P. A., Peterlin B. M. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987 Dec 3;330(6147):489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- Kelly K., Siebenlist U. The regulation and expression of c-myc in normal and malignant cells. Annu Rev Immunol. 1986;4:317–338. doi: 10.1146/annurev.iy.04.040186.001533. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K., Kane C. M. Analysis of the signals for transcription termination by purified RNA polymerase II. Biochemistry. 1990 Jan 9;29(1):269–278. doi: 10.1021/bi00453a037. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K., Kane C. M. Intrinsic sites of transcription termination and pausing in the c-myc gene. Mol Cell Biol. 1988 Oct;8(10):4389–4394. doi: 10.1128/mcb.8.10.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler M., Ben-Asher E., Aloni Y. Elements modulating the block of transcription elongation at the adenovirus 2 attenuation site. J Biol Chem. 1989 Jun 15;264(17):9785–9790. [PubMed] [Google Scholar]
- Krystal G., Birrer M., Way J., Nau M., Sausville E., Thompson C., Minna J., Battey J. Multiple mechanisms for transcriptional regulation of the myc gene family in small-cell lung cancer. Mol Cell Biol. 1988 Aug;8(8):3373–3381. doi: 10.1128/mcb.8.8.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A., Bartsch I., Grummt I. Specific interaction of the murine transcription termination factor TTF I with class-I RNA polymerases. Nature. 1990 Apr 5;344(6266):559–562. doi: 10.1038/344559a0. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachman H. M., Skoultchi A. I. Expression of c-myc changes during differentiation of mouse erythroleukaemia cells. Nature. 1984 Aug 16;310(5978):592–594. doi: 10.1038/310592a0. [DOI] [PubMed] [Google Scholar]
- Leder P., Battey J., Lenoir G., Moulding C., Murphy W., Potter H., Stewart T., Taub R. Translocations among antibody genes in human cancer. Science. 1983 Nov 18;222(4625):765–771. doi: 10.1126/science.6356357. [DOI] [PubMed] [Google Scholar]
- Leirmo S., Harrison C., Cayley D. S., Burgess R. R., Record M. T., Jr Replacement of potassium chloride by potassium glutamate dramatically enhances protein-DNA interactions in vitro. Biochemistry. 1987 Apr 21;26(8):2095–2101. doi: 10.1021/bi00382a006. [DOI] [PubMed] [Google Scholar]
- Lindsten T., June C. H., Thompson C. B. Multiple mechanisms regulate c-myc gene expression during normal T cell activation. EMBO J. 1988 Sep;7(9):2787–2794. doi: 10.1002/j.1460-2075.1988.tb03133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher B., Eisenman R. N. New light on Myc and Myb. Part I. Myc. Genes Dev. 1990 Dec;4(12A):2025–2035. doi: 10.1101/gad.4.12a.2025. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McGeady M. L., Wood T. G., Maizel J. V., Vande Woude G. F. Sequences upstream from the mouse c-mos oncogene may function as a transcription termination signal. DNA. 1986 Aug;5(4):289–298. doi: 10.1089/dna.1986.5.289. [DOI] [PubMed] [Google Scholar]
- McStay B., Reeder R. H. A DNA-binding protein is required for termination of transcription by RNA polymerase I in Xenopus laevis. Mol Cell Biol. 1990 Jun;10(6):2793–2800. doi: 10.1128/mcb.10.6.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechti N., Piechaczyk M., Blanchard J. M., Jeanteur P., Lebleu B. Sequence requirements for premature transcription arrest within the first intron of the mouse c-fos gene. Mol Cell Biol. 1991 May;11(5):2832–2841. doi: 10.1128/mcb.11.5.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechti N., Piechaczyk M., Blanchard J. M., Marty L., Bonnieu A., Jeanteur P., Lebleu B. Transcriptional and post-transcriptional regulation of c-myc expression during the differentiation of murine erythroleukemia Friend cells. Nucleic Acids Res. 1986 Dec 22;14(24):9653–9666. doi: 10.1093/nar/14.24.9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller H., Asselin C., Dufort D., Yang J. Q., Gupta K., Marcu K. B., Nepveu A. A cis-acting element in the promoter region of the murine c-myc gene is necessary for transcriptional block. Mol Cell Biol. 1989 Dec;9(12):5340–5349. doi: 10.1128/mcb.9.12.5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepveu A., Levine R. A., Campisi J., Greenberg M. E., Ziff E. B., Marcu K. B. Alternative modes of c-myc regulation in growth factor-stimulated and differentiating cells. Oncogene. 1987;1(3):243–250. [PubMed] [Google Scholar]
- Neuman de Vegvar H. E., Dahlberg J. E. Initiation and termination of human U1 RNA transcription requires the concerted action of multiple flanking elements. Nucleic Acids Res. 1989 Nov 25;17(22):9305–9318. doi: 10.1093/nar/17.22.9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Postel E. H., Mango S. E., Flint S. J. A nuclease-hypersensitive element of the human c-myc promoter interacts with a transcription initiation factor. Mol Cell Biol. 1989 Nov;9(11):5123–5133. doi: 10.1128/mcb.9.11.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price D. H., Sluder A. E., Greenleaf A. L. Dynamic interaction between a Drosophila transcription factor and RNA polymerase II. Mol Cell Biol. 1989 Apr;9(4):1465–1475. doi: 10.1128/mcb.9.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J. How RNA polymerase II terminates transcription in higher eukaryotes. Trends Biochem Sci. 1989 Mar;14(3):105–110. doi: 10.1016/0968-0004(89)90132-1. [DOI] [PubMed] [Google Scholar]
- Ratnasabapathy R., Sheldon M., Johal L., Hernandez N. The HIV-1 long terminal repeat contains an unusual element that induces the synthesis of short RNAs from various mRNA and snRNA promoters. Genes Dev. 1990 Dec;4(12A):2061–2074. doi: 10.1101/gad.4.12a.2061. [DOI] [PubMed] [Google Scholar]
- Re G. G., Antoun G. R., Zipf T. F. Modulation of a constitutive transcriptional block at exon-1 controls human c-myc oncogene expression. Oncogene. 1990 Aug;5(8):1247–1250. [PubMed] [Google Scholar]
- Reines D., Chamberlin M. J., Kane C. M. Transcription elongation factor SII (TFIIS) enables RNA polymerase II to elongate through a block to transcription in a human gene in vitro. J Biol Chem. 1989 Jun 25;264(18):10799–10809. [PubMed] [Google Scholar]
- Resnekov O., Kessler M., Aloni Y. RNA secondary structure is an integral part of the in vitro mechanism of attenuation in simian virus 40. J Biol Chem. 1989 Jun 15;264(17):9953–9959. [PubMed] [Google Scholar]
- Robledo R., Gottesman M. E., Weisberg R. A. Lambda nutR mutations convert HK022 Nun protein from a transcription termination factor to a suppressor of termination. J Mol Biol. 1990 Apr 20;212(4):635–643. doi: 10.1016/0022-2836(90)90226-c. [DOI] [PubMed] [Google Scholar]
- Saltzman A. G., Weinmann R. Promoter specificity and modulation of RNA polymerase II transcription. FASEB J. 1989 Apr;3(6):1723–1733. doi: 10.1096/fasebj.3.6.2649403. [DOI] [PubMed] [Google Scholar]
- Samuels M., Fire A., Sharp P. A. Separation and characterization of factors mediating accurate transcription by RNA polymerase II. J Biol Chem. 1982 Dec 10;257(23):14419–14427. [PubMed] [Google Scholar]
- Sato K., Ito R., Baek K. H., Agarwal K. A specific DNA sequence controls termination of transcription in the gastrin gene. Mol Cell Biol. 1986 Apr;6(4):1032–1043. doi: 10.1128/mcb.6.4.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawadogo M., Roeder R. G. Factors involved in specific transcription by human RNA polymerase II: analysis by a rapid and quantitative in vitro assay. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4394–4398. doi: 10.1073/pnas.82.13.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby M. J., Bain E. S., Luciw P. A., Peterlin B. M. Structure, sequence, and position of the stem-loop in tar determine transcriptional elongation by tat through the HIV-1 long terminal repeat. Genes Dev. 1989 Apr;3(4):547–558. doi: 10.1101/gad.3.4.547. [DOI] [PubMed] [Google Scholar]
- Shapiro D. J., Sharp P. A., Wahli W. W., Keller M. J. A high-efficiency HeLa cell nuclear transcription extract. DNA. 1988 Jan-Feb;7(1):47–55. doi: 10.1089/dna.1988.7.47. [DOI] [PubMed] [Google Scholar]
- SivaRaman L., Reines D., Kane C. M. Purified elongation factor SII is sufficient to promote read-through by purified RNA polymerase II at specific termination sites in the human histone H3.3 gene. J Biol Chem. 1990 Aug 25;265(24):14554–14560. [PubMed] [Google Scholar]
- Sluder A. E., Greenleaf A. L., Price D. H. Properties of a Drosophila RNA polymerase II elongation factor. J Biol Chem. 1989 May 25;264(15):8963–8969. [PubMed] [Google Scholar]
- Sopta M., Burton Z. F., Greenblatt J. Structure and associated DNA-helicase activity of a general transcription initiation factor that binds to RNA polymerase II. Nature. 1989 Oct 5;341(6241):410–414. doi: 10.1038/341410a0. [DOI] [PubMed] [Google Scholar]
- Spencer C. A., Groudine M. Transcription elongation and eukaryotic gene regulation. Oncogene. 1990 Jun;5(6):777–785. [PubMed] [Google Scholar]
- Spencer C. A., LeStrange R. C., Novak U., Hayward W. S., Groudine M. The block to transcription elongation is promoter dependent in normal and Burkitt's lymphoma c-myc alleles. Genes Dev. 1990 Jan;4(1):75–88. doi: 10.1101/gad.4.1.75. [DOI] [PubMed] [Google Scholar]
- Tomizawa J., Masukata H. Factor-independent termination of transcription in a stretch of deoxyadenosine residues in the template DNA. Cell. 1987 Nov 20;51(4):623–630. doi: 10.1016/0092-8674(87)90131-0. [DOI] [PubMed] [Google Scholar]
- Toohey M. G., Jones K. A. In vitro formation of short RNA polymerase II transcripts that terminate within the HIV-1 and HIV-2 promoter-proximal downstream regions. Genes Dev. 1989 Mar;3(3):265–282. doi: 10.1101/gad.3.3.265. [DOI] [PubMed] [Google Scholar]
- Wiest D. K., Hawley D. K. In vitro analysis of a transcription termination site for RNA polymerase II. Mol Cell Biol. 1990 Nov;10(11):5782–5795. doi: 10.1128/mcb.10.11.5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., Bishop J. M. DNA sequences that mediate attenuation of transcription from the mouse protooncogene myc. Proc Natl Acad Sci U S A. 1989 Jan;86(2):505–509. doi: 10.1073/pnas.86.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]