Phylogenetic and functional analysis of strigolactone pathway genes across the plant kingdom suggests considerable promiscuity in events upstream allowing for signal diversity and its later refinement.
Abstract
Strigolactones (SLs) are carotenoid-derived phytohormones with diverse roles. They are secreted from roots as attractants for arbuscular mycorrhizal fungi and have a wide range of endogenous functions, such as regulation of root and shoot system architecture. To date, six genes associated with SL synthesis and signaling have been molecularly identified using the shoot-branching mutants more axillary growth (max) of Arabidopsis (Arabidopsis thaliana) and dwarf (d) of rice (Oryza sativa). Here, we present a phylogenetic analysis of the MAX/D genes to clarify the relationships of each gene with its wider family and to allow the correlation of events in the evolution of the genes with the evolution of SL function. Our analysis suggests that the notion of a distinct SL pathway is inappropriate. Instead, there may be a diversity of SL-like compounds, the response to which requires a D14/D14-like protein. This ancestral system could have been refined toward distinct ligand-specific pathways channeled through MAX2, the most downstream known component of SL signaling. MAX2 is tightly conserved among land plants and is more diverged from its nearest sister clade than any other SL-related gene, suggesting a pivotal role in the evolution of SL signaling. By contrast, the evidence suggests much greater flexibility upstream of MAX2. The MAX1 gene is a particularly strong candidate for contributing to diversification of inputs upstream of MAX2. Our functional analysis of the MAX1 family demonstrates the early origin of its catalytic function and both redundancy and functional diversification associated with its duplication in angiosperm lineages.
Strigolactones (SLs) are carotenoid-derived terpenoid lactones, which have been identified as signaling molecules in several areas of plant biology. SLs were first identified as germination stimulants for seeds of plants in the genus Striga (Cook et al., 1966). Striga spp. and related Orobanchaceae are parasitic weeds that germinate in response to host plant root exudates and develop haustoria to penetrate the host tissue and draw nutrients. Striga spp. are major agricultural pests across much of tropical and subtropical Asia and are present in two-thirds of arable land in Africa, where they are the greatest biological cause of crop damage (Humphrey and Beale, 2006). The secretion of SLs by roots, despite its exploitation by Striga spp., has been preserved because it also serves to recruit arbuscular mycorrhizal (AM) fungi (Akiyama et al., 2005). AM fungi form symbiotic associations with most land plants, whereby the plant gains access to mineral nutrients, particularly phosphate, absorbed by the fungal hyphae, and in exchange the fungus gains fixed carbon from the plant. In several flowering plant species, SL production is correspondingly increased when phosphate availability is limiting, thereby presumably increasing fungal recruitment (Yoneyama et al., 2007, 2012).
AM symbioses can be traced back to the origin of land plants, between 360 to 450 million years ago, and are thought to have facilitated plant colonization of the terrestrial environment (Simon et al., 1993). Although AM symbiosis has been lost from some lineages, such as Brassicaceae, it is still widespread, with 80% of land plants able to form associations with AM fungi (Schüssler et al., 2001). In support of a similarly ancient origin for SL secretion, the liverwort Marchantia polymorpha and the moss Physcomitrella patens, both basal land plant groups, have been shown to produce SLs (Proust et al., 2011; Delaux et al., 2012). Furthermore, the presence of SLs in charophyte algae indicates that SL production may predate the emergence of land plants (Delaux et al., 2012), and Chara corallina responds to SL treatment by producing longer rhizoids (Delaux et al., 2012). In P. patens, SLs appear to act as intercolony coordination signals, regulating colony growth and competition by controlling flexible developmental processes such as protonemal branching (Proust et al., 2011; Delaux et al., 2012). In flowering plants, SLs have also been implicated in development, including several processes regulated in response to phosphate limitation (Kohlen et al., 2011; Ruyter-Spira et al., 2011). In particular, SLs play important roles in the regulation of shoot branching in higher plants (Gomez-Roldan et al., 2008; Umehara et al., 2008). It is through work on their effects on shoot branching that some of the genes in the SL pathway were first identified.
Arabidopsis (Arabidopsis thaliana) MORE AXILLARY GROWTH (MAX) mutants show increased branching and reduced stature relative to wild-type plants, and analogous phenotypes have been identified in pea (Pisum sativum; RAMOSUS [RMS]), petunia (Petunia hybrida; DECREASED APICAL DOMINANCE [DAD]), and rice (Oryza sativa; DWARF [D] or HIGH TILLERING DWARF) mutants. So far, six MAX/RMS/DAD/D genes have been identified, with roles in SL biosynthesis or signaling. MAX3/RMS5/HIGH TILLERING DWARF1/D17 (Booker et al., 2004; Johnson et al., 2006; Zou et al., 2006) and MAX4/RMS1/DAD1/D10 (Sorefan et al., 2003; Snowden et al., 2005; Arite et al., 2007) encode carotenoid cleavage dioxygenases (CCD7 and CCD8, respectively). These enzymes are capable of sequentially cleaving the carotenoid 9-cis-β-carotene to produce a novel compound, carlactone, a putative strigolactone intermediate (Alder et al., 2012). Another biosynthetic gene, D27, was originally mutationally defined in rice (Lin et al., 2009), and reverse genetic approaches in Arabidopsis indicate a similar function in this species (Waters et al., 2012a). D27 is an iron-containing protein with isomerase activity that can produce the 9-cis-β-carotene substrate for MAX3 from all-trans-β-carotene (Alder et al., 2012). The fourth gene known to be involved in SL biosynthesis, MAX1, encodes a cytochrome p450 monooxygenase belonging to the CYP711 clan (Booker et al., 2005). Mutant phenotypes associated with this gene have so far only been identified in one species, Arabidopsis, although the gene is present in all tracheophytes (Nelson et al., 2008). The excessive-branching phenotypes associated with mutations in all of these genes can be rescued by exogenous application of SL, while mutants in the two remaining genes in the pathway are SL insensitive. D14 encodes an α/β hydrolase, which is proposed to act in signaling or in the hydrolysis of SLs to an active compound and provides specificity to signaling via MAX2/RMS4/D3, an F-box protein that mediates both SL signaling and signaling of karrikins (Stirnberg et al., 2002, 2007; Ishikawa et al., 2005; Johnson et al., 2006; Arite et al., 2009; Hamiaux et al., 2012; Waters et al., 2012b). Karrikins are compounds structurally related to SLs that are found in smoke and act as germination stimulants for plants that colonize ground cleared by forest fires (Nelson et al., 2010; Waters et al., 2012b).
Homology searches described in the original publications for each of the MAX/D genes revealed two general patterns. MAX1, MAX3, and MAX4 are members of widespread gene families and are more closely related to nonplant sequences than to other plant genes (Sorefan et al., 2003; Booker et al., 2005). By contrast, MAX2, D14, and D27 are members of plant-specific gene families (Stirnberg et al., 2002; Arite et al., 2009; Lin et al., 2009). These contrasting patterns of SL pathway gene ancestry and the diverse biological roles of SLs present interesting evolutionary questions. The identification of SLs and SL responses in charophyte algae demonstrate their early evolution, but these species lack many of the genes required for SL synthesis and signaling in angiosperms. In an attempt to trace the evolution of the angiosperm SL pathway, we conducted a phylogenetic analysis of the known SL biosynthesis and signaling genes, allowing the correlation of events in the evolution of the genes with the evolution of SL function. Our analysis suggests that the notion of a distinct SL pathway is inappropriate. Instead, the angiosperm pathway seems to have been defined by the rapid evolution of MAX2 in early land plants. Upstream of MAX2, there appears to be much greater flexibility, especially in the requirements for the synthesis of SLs. We present evidence for the contribution of MAX1 to this flexibility. Our functional analysis of MAX1 orthologs from phylogenetically diverse species demonstrates the early origin of its catalytic activity and both redundancy and functional diversification associated with its duplication in angiosperm lineages.
RESULTS
Gene Families
Orthologs and paralogs of known SL genes were surveyed across published and prepublished plant genomes to establish the presence of the SL pathway throughout the plant kingdom. For 18 genomes, putative orthologous and paralogous sequences for the target genes were identified using InParanoid 4.0 (Berglund et al., 2008) and clustered to produce ortholog sets. Incorporating singleton (orphan) sequences that had not been assigned to any ortholog cluster by the InParanoid algorithm typically had no effect on the number of putative orthologs/paralogs; however, in approximately 16% of cases, additional paralogous sequences were inferred (Table I). For six additional genomes, orthologs/in-paralogs were identified using BLAST searches. As expected given the simpler approach, the number of false positives was higher; however, the ortholog sets identified in related genomes using InParanoid allowed the homology-based assignment to be checked during phylogenetic reconstruction, as the true orthologs among the additional sequences were expected to lie within the same monophyletic clades as the previously identified sequences. We identified orthologs of each of the six SL pathway genes described to date in the majority of species surveyed. Where previous studies identified MAX/D orthologs in species for which genome sequence data were not available, these sequences were also included in the gene trees.
Table I. Distribution of orthologs and paralogs of the MAX pathway genes across the surveyed species predicted using InParanoid 4 (Berglund et al., 2008).
Where sequences were inferred to be nonorthologous during phylogenetic reconstruction, the proportion of retained sequences is indicated. Values in parentheses are orphan sequences that were not assigned to any ortholog set but were more similar to a sequence within the ortholog set than any other sequence.
| Method | Species | MAX1 | MAX2 | MAX3 | MAX4 | D14 | D27 |
|---|---|---|---|---|---|---|---|
| InParanoid | |||||||
| P. patens | 0 | 1 | 1 | 1 | 0 | 0a | |
| S. moellendorffii | 1 (0/4) | 1 | 1 | 1 | 0 | 0 | |
| Brachypodium distachyon | 5 | 1 | 1 | 1 | 1 | 1 | |
| Rice | 5 (0/1) | 1 (0/1) | 1 | 2 | 1 | 0b | |
| Sorghum bicolor | 4 | 1 | 1 (0/1) | 2 (0/3) | 1 | 1 | |
| Maize | 1 (2) | 1 | 1 | 1 | 2 | 2 | |
| M. guttatus | 1/2 (0/5) | 1 | 1 (0/8) | 1 (0/5) | 2 | 1 | |
| Vitis vinifera | 1 (0/1) | 1 | 1 | 1 | 1 | 1 | |
| Carica papaya | 1 | 1 | 1 | 1 | 1 | 1 | |
| Arabidopsis | 1 | 1 | 1 | 1 | 1 | 1 | |
| Arabidopsis lyrata | 1 | 1 | 1 | 1 | 1 | 1 | |
| Cucumis sativus | 1 | 1 | 1 | 1 | 1 | 1 | |
| Soybean | 2/4 | 2 | 2 (0/1) | 2 | 2 | 2 | |
| M. truncatula | 2 | 1 | 1 | 1 | 1 | 2 | |
| L. japonicus | 1 | 1 (0/3) | 0 | 1 | 1 | 1 | |
| P. trichocarpa | 2 (0/3) | 2 (0/1) | 1 | 2 | 2 | 2 | |
| Manihot esculenta | 1 | 2 (0/1) | 1 | 1 | 1 | 1 | |
| Ricinus communis | 1 (0/1) | 1 (0/2) | 1 | 1 | 1 | 1 | |
| BLAST | |||||||
| P. dactylifera | 2 | 1 | 1 | 2 | 0 | 1 | |
| A. caerulea | 1 | 1 | 1 | 3 | 1 | 1 | |
| S. lycopersicum | 1 | 2 | 1 | 1 | 1 | 1 | |
| C. sinensis | 1 | 2 | 1 | 2 | 1 | 1 | |
| Eucalyptus grandis | 1 | 1 | 1 | 2 | 2 | 1 | |
| Strawberry | 2 | 1 | 1 | 1 | 1 | 1 |
Two putative orthologs were inferred during phylogenetic reconstruction of the D27 gene family tree. bThe known D27 sequence in rice (LOC_Os11g37650) is not annotated as a functional gene in the version of the rice genome used for ortholog prediction, but was included in subsequent analyses; no other orthologs were identified.
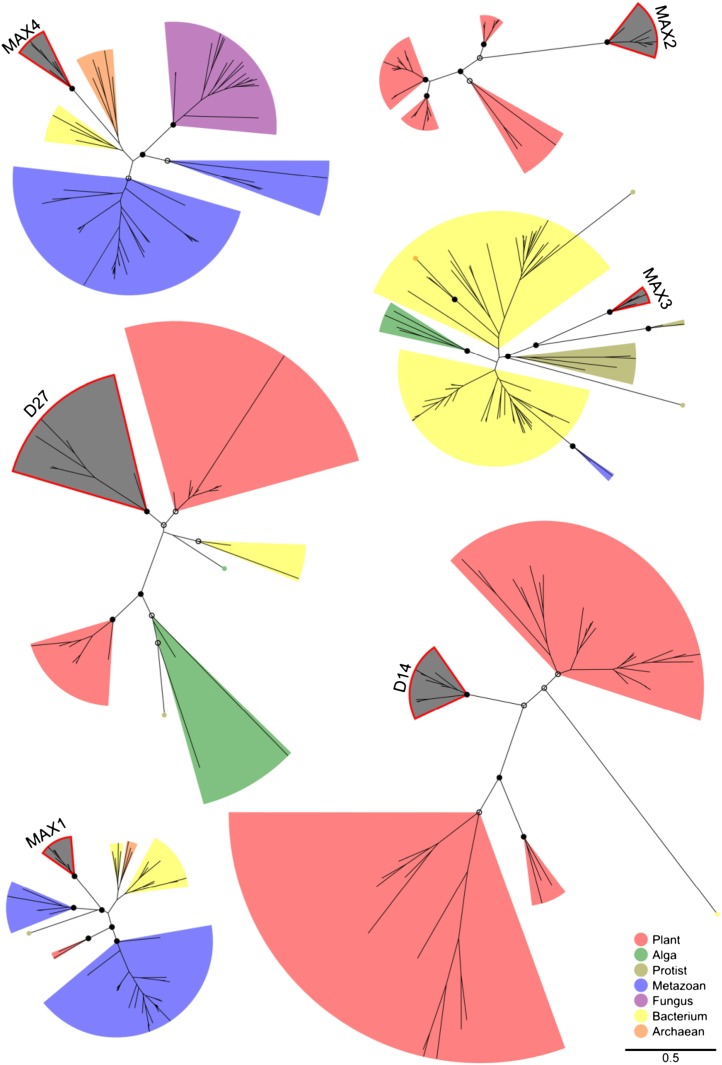
To determine relationships between the target genes and their wider gene families, BLAST searches using the target genes were performed against the Kyoto Encyclopedia of Genes and Genomes (KEGG; Kanehisa and Goto, 2000) database of nonredundant protein sequences from a wide taxonomic range. Up to 500 of the highest scoring hit sequences were analyzed, and after filtering to remove incomplete gene sequences, these files contained 460 (MAX1), 217 (MAX2), 377 (MAX3), 444 (MAX4), 477 (D14), and 29 (D27) homologs. The resulting very low number of homologous sequences for D27 was due to only 51 sequences being retrieved in the initial BLAST search rather than to excessive stringency of filtering. Following further filtering to remove sequences that could not be reliably aligned or that were inferred to be phylogenetically distant, 33 (MAX2), 82 (MAX3), 75 (MAX4), 101 (D14), and 29 (D27) sequences were used for phylogenetic reconstruction (see Supplemental Data Set S1). Maximum-likelihood gene family protein sequence phylogenies inferred using RAxML (for Randomized Axelerated Maximum Likelihood; Stamatakis, 2006) are presented in Figure 1 to the same scale for each of the target genes. Uniquely for MAX1, no suitable well-supported node could be identified in preliminary phylogenies for use as a phylogenetic distance cutoff, so a subtree was pruned at an arbitrary node in the maximum-likelihood phylogeny (with 21% bootstrap support), leaving 69 MAX1 sequences, approximately the mean number of sequences for the other genes. Because these phylogenies show only the most closely related sequences regardless of taxonomy, algal sequences have only been included in two cases (MAX3 and D27), and no nonplant sequences were inferred to be sufficiently closely related to MAX2 to justify inclusion.
Figure 1.
Phylogenetic trees depicting the relationship between each of the MAX genes and the most similar sequences in the KEGG database (Kanehisa and Goto, 2000). Protein sequences were aligned using MAFFT (Katoh et al., 2005), and phylogenies were reconstructed using RAxML (Stamatakis, 2006) under the JTT substitution matrix. Support for the maximum-likelihood phylogenies was assessed using 100 rapid bootstrap resamplings (Stamatakis et al., 2008). Colored areas indicate clades belonging to a single taxonomic group; MAX gene clades are shaded in gray with a red outline. Support for these clades is indicated using black (>90%) and white (>50%) circles.
Two general patterns are apparent from the gene family phylogenies (Fig. 1). MAX1, MAX3, and MAX4 are each more closely related to clades of nonplant sequences (Metazoa, Protists, and Bacteria/Archaea, respectively) than to other land plant sequences, while MAX2, D14, and D27 are each most closely related to clades of plant sequences. The phylogenetic distances between the target gene orthologs and the rest of each gene family phylogeny are summarized in Table II. Both D14 and D27 are closely related to sequences in neighboring clades of plant sequences, which contain the previously identified D14-like (Arite et al., 2009; Waters et al., 2012b) and D27-like (Lin et al., 2009) genes, respectively. MAX1, MAX3, and MAX4 are more distantly related to neighboring clades in Metazoa, Archaea, and Protists. The most distinct of the gene families analyzed is that of MAX2, for which the basal branch length indicates divergence at the rate of two substitutions per site since the last common ancestor of MAX2 and its sister clade.
Table II. Alignment and phylogeny statistics for the MAX pathway genes.
| Gene | Average Sequence Length | Ortholog Set Alignment Length | Conserved Sites in Ortholog Set Alignment | Uniquely Conserved Sites in Ortholog Set Alignment | Basal Branch Length in Gene Family Tree | Total Distance to Nearest Sister Sequence/Clade in Gene Family Tree |
|---|---|---|---|---|---|---|
| bp | substitutions per site | |||||
| MAX1 | 529.3 | 834 | 168 (20.1%) | 48 (5.8%) | 0.72 | 1.17 |
| MAX2 | 701.9 | 1049 | 213 (20.3%) | 113 (10.8%) | 2.06 | 2.29 |
| MAX3 | 581.7 | 817 | 216 (26.4%) | 83 (10.2%) | 1.34 | 1.99 |
| MAX4 | 547.9 | 1018 | 247 (24.3%) | 77 (7.6%) | 1.20 | 1.31 |
| D14 | 275.8 | 350 | 170 (48.6%) | 14 (4.0%) | 0.31 | 0.55 |
| D27 | 241.0 | 414 | 71 (17.2%) | 17 (4.1%) | 0.36 | 0.65 |
Ortholog Set Phylogenies
Relationships among the target gene ortholog sets were examined in more detail in arbitrarily rooted maximum-likelihood phylogenies of protein sequences (Fig. 2; Supplemental Fig. S2). Sequences inferred to be nonorthologous in preliminary phylogenies were excluded from the final ortholog set. Only two of the orphan sequences added to the InParanoid sets were inferred to be true orthologs, both maize (Zea mays) MAX1 sequences. Two additional sequences (for P. patens) were identified as putative orthologs of D27 following the gene family phylogenetic reconstruction; however, these sequences were shown to be only distantly related to the remaining orthologs during preliminary phylogenetic reconstruction and have been excluded from the presented set of orthologs. Protein sequences and alignments of the sequences included in the phylogenies are presented in Supplemental Data Set S1.
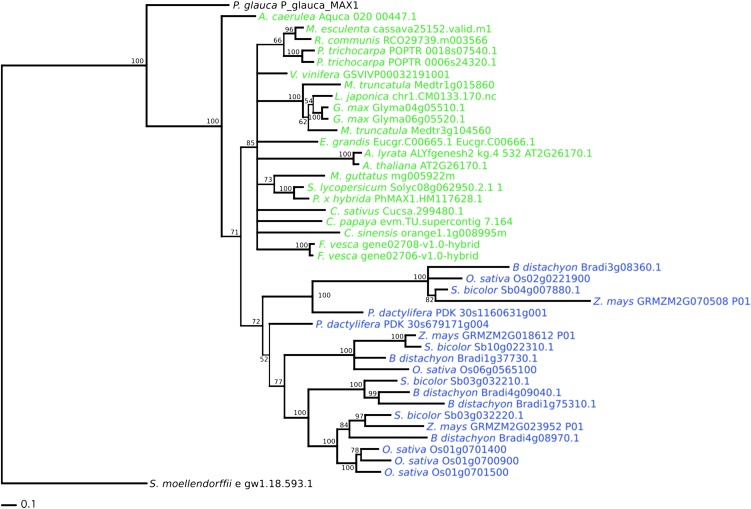
Figure 2.
Phylogenetic tree depicting the relationship between putative orthologs of MAX1 across 24 plant species. Additional sequences were included for some experimentally determined orthologs in species for which no genome sequence was available. Protein sequences were aligned using MAFFT (Katoh et al., 2005), and DNA sequences were aligned to the protein alignment using Transeq from the EMBOSS suite (Rice et al., 2000). Phylogenies were reconstructed from the DNA alignments under the general time-reversible plus γ model using RAxML (Stamatakis, 2006), and support was assessed using 100 rapid bootstrap resamplings (Stamatakis et al., 2008). Dicotyledon sequences are colored green, and monocotyledons are colored blue.
With the exception of MAX3, which was absent from the predicted gene models of Lotus japonicus, and D14, which was absent from Phoenix dactylifera, each of the target genes has an orthologous sequence in all monocot and dicot species surveyed (Table I), indicating a general conservation of the known SL pathway in angiosperms. In addition, most dicot species had only a single ortholog of each gene, with the exception of soybean (Glycine max) and Populus trichocarpa, recent paleopolyploids (Tuskan et al., 2006; Schmutz et al., 2010), which typically had a second paralogous copy. Monocots also most commonly possessed a single ortholog of each gene, with the exceptions of MAX1, for which multiple paralogous sequences were present, and MAX4, for which two copies were identified in both rice and P. dactylifera. However, no putative orthologs of canonical D14 or D27 were identified in the basal plant species Selaginella moellendorffii and P. patens, and MAX1 also was absent from P. patens. In each case, the phylogenetic position inferred was consistent with expectations, supporting the hypothesis that the remaining sequences are orthologous and are likely to perform similar functions in their respective species.
For each gene, the reconstructed phylogeny broadly reflects the major taxonomic divisions among the higher plants. The arbitrary rooting of the phylogenies on P. patens or S. moellendorffii sequences when present is supported by the wider gene family trees. Where the phylogenetic relationships are sufficiently well resolved, both monocots and dicots are typically monophyletic. The exceptions to this rule are Aquilegia caerulea MAX1, which is placed as a sister group to the monocot and the remaining dicot MAX1 sequences, with a bootstrap probability of 71%; Arabidopsis lyrata and Arabidopsis MAX4 and D27, which are placed as sister groups to the monocot and the remaining dicot sequences, with bootstrap probabilities of 59% and 100%, respectively; and P. dactylifera MAX3, which is placed as a sister group to the dicots rather than the remaining monocots, with a bootstrap probability of 75%.
Among the monocots, at least three separate clades of MAX1 are inferred and possibly two clades of MAX4, although the second clade is only represented by two species. For MAX2, Citrus sinensis and P. trichocarpa have paralogs in two separate clades, and two paralogous clades of petunia and Solanum lycopersicum apparently reflect a basal duplication in the Solanaceae. Where relationships are sufficiently resolved, the remaining duplications are either monophyletic or present as sister groups. However, in most phylogenies, the basal relationships among the dicotyledons are poorly resolved, so the apparent paraphyly in the remaining gene trees is likely to result from uncertainty in the true tree topology.
MAX1, MAX4, and D27 share a similar pattern in which a small number of sequences are inferred to form a sister group to the main angiosperm clade. While this is relatively weakly supported for MAX1 and MAX4 (71% and 59%, respectively), paraphyly of dicotyledon D27 genes received 100% bootstrap support. In each case, the sister sequences are from relatively basal dicotyledons, and thus, the paraphyly likely represents shared ancestral polymorphism.
Functional Conservation of MAX1
Uniquely among these genes, the phylogenetic analysis indicates that the MAX1 family possesses neither orthologs in the bryophyte P. patens nor any closely related genes in plants that would be expected to supply the same function. In addition, while most of the other genes are generally copy number conserved, MAX1 is present in multiple conserved clades in the monocots. This suggests that MAX1 might be a later addition to the SL pathway than the other genes and that its action may have diversified in the monocots. To test these hypotheses, the functional conservation of MAX1 action was assessed using a complementation approach. MAX1 orthologs were cloned, placed under the control of the strong Cauliflower mosaic virus 35S promoter, and introduced into the max1-1 mutant of the model angiosperm Arabidopsis. The ability of the orthologs to replace the native function of AtMAX1 was measured by the degree to which several shoot phenotypes conferred by max1-1 were restored to the wild type in the resultant transgenic lines. As only two nonangiosperm land plant genomes have been sequenced (P. patens, in which MAX1 is absent, and S. moellendorffii), to increase the taxonomic resolution for testing the incorporation of MAX1 into the SL pathway, BLAST searches of expressed sequence tags were used to identify a putative full-length gymnosperm ortholog from white spruce (Picea glauca). To compare the evolutionary fates of MAX1 paralogs in angiosperms, the two in-paralogs of model dicot Medicago truncatula and three of the five in-paralogs of model monocot rice (representing all three monocot MAX1 clades) were also included.
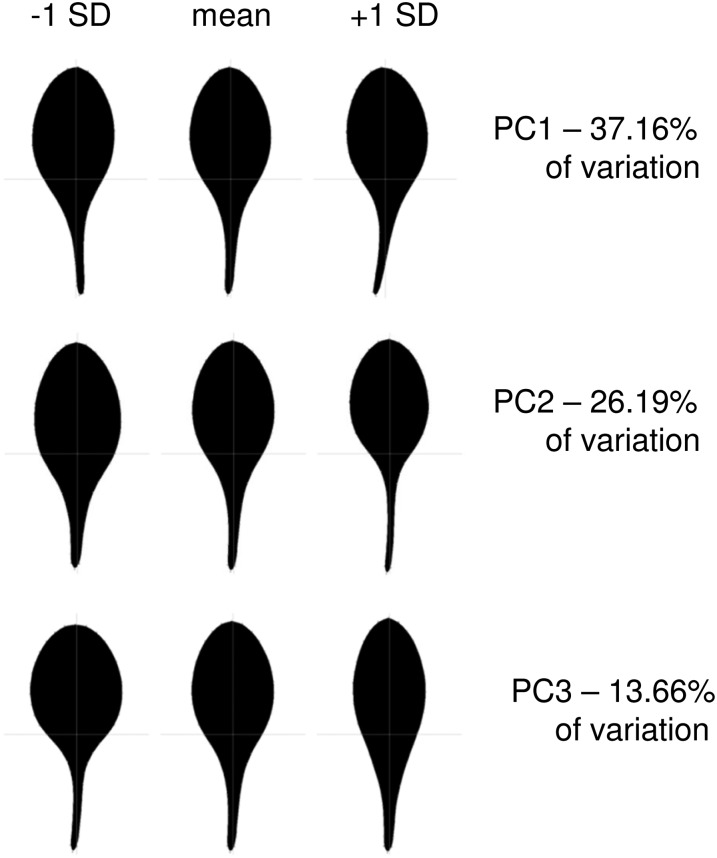
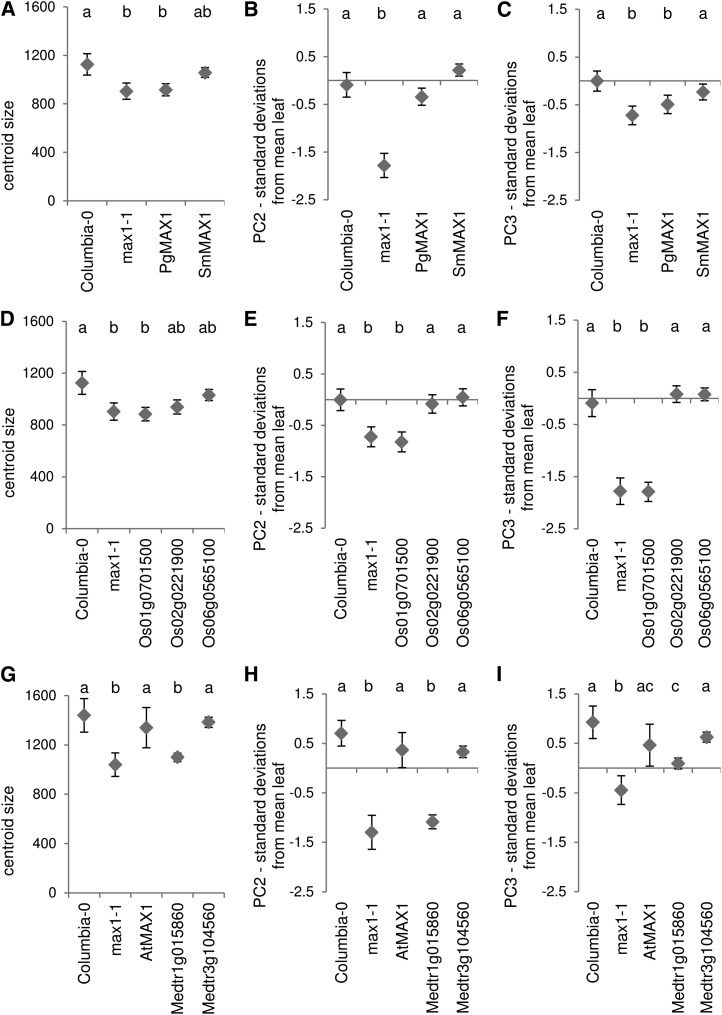
The degree of primary branching following decapitation and leaf size/shape phenotypes were used to assess the ability of each ortholog to rescue the Arabidopsis max1-1 mutant (Stirnberg et al., 2002). Morphometric analysis of max1-1 leaves with LeafAnalyser software (Weight et al., 2008) was used to quantify precisely the leaf phenotypes. This analysis captured leaf size as centroid size, and identified significant differences between the wild type and max1-1 in principal component 2 (PC2) and principal component 3 (PC3) of leaf shape variation identified among a library of leaves from natural accessions (Fig. 3; Danisman et al., 2012). PC2 represents variation in the width and length of the petiole and the position of the lamina on the petiole, which becomes more distal from negative to positive across the variation in the population and as leaves become more elongated. PC3 captures the leaf aspect ratio of lamina length to lamina width, which increases from the negative sd to the positive. Leaves of max1-1 plants have more negative scores in this analysis than the Columbia ecotype for all these principal components, reflecting their smaller size and shorter, wider shape.
Figure 3.
Leaf shape models for the first three principal components of leaf shape in 10 natural Arabidopsis accessions produced by LeafAnalyser (Danisman et al., 2012).
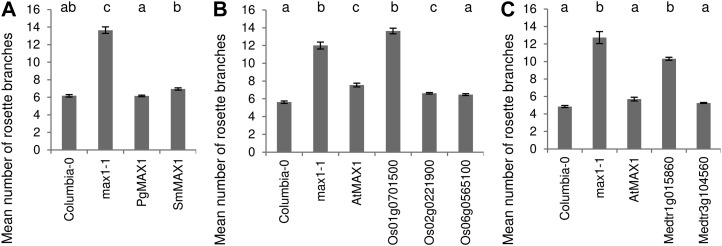
Orthologs from both the lycophyte S. moellendorffii and the gymnosperm white spruce were capable of rescuing the max1-1 high-branching phenotype completely (Fig. 4A). Both orthologs were also capable of complete rescue of leaf shape PC2, but unlike SmMAX1, PgMAX1 was not able to rescue PC3 or leaf centroid size, and the mean centroid size of the SmMAX1 plants was only intermediate between the wild type and max1-1 (Fig. 5, A–C).
Figure 4.
Rosette-branching rescue of Arabidopsis max1 by putative orthologs from nonangiosperm species white spruce and S. moellendorffii (A), in-paralogs from rice (B), and in-paralogs from M. truncatula (C), compared with the Columbia wild type and max1-1. The AtMAX1 line (S. Ward, J. Salmon, S. Hanley, A. Karp, and O. Leyser, unpublished data) is included as a control for the native gene function. n = 20 plants for each of eight to 11 independent transgenic lines, except for the single-line genotypes Columbia, max1-1, and 35S:AtMAX1 max1-1, for which n = 40. Error bars indicate se of the mean. Shared letters indicate no significant difference in a Kruskal-Wallis test at P ≤ 0.001.
Figure 5.
Leaf phenotype rescue of Arabidopsis max1 by putative orthologs from nonangiosperm species white spruce and S. moellendorffii (A–C), in-paralogs from rice (D–F), and in paralogs from M. truncatula (G–I), compared with the Columbia wild type and max1-1. The AtMAX1 line is included as a control for the native gene function. Phenotypes used to judge rescue, as produced by LeafAnalyser, are as follows: leaf size measured by centroid size (A, D, and G) and mean number of sds of leaves from the natural accession mean for PC2 (B, E, and H) and PC3 (C, F, and I). Six to 10 plants of each of two representative lines were used per ortholog, except for the M. truncatula orthologs, in which all eight to 10 lines are included. Error bars indicate se of the mean. Shared letters indicate no significant difference in Tamhane’s T2 post hoc test (P ≤ 0.001, centroid size and PC2) or Tukey’s honestly significant difference mean-separation test (P ≤ 0.05, PC3).
Of the three rice (O. sativa ssp. japonica) orthologs, Os01g0701500 is unable to function at all in rescue of shoot branching, leaf shape, or leaf size (Figs. 4B and 5, D–F). The other two orthologs are capable of rescuing the branching phenotype. Both of these also fully rescue the PC2 component of leaf shape; however, mean leaf centroid size in these lines was again intermediate between max1-1 and the wild type (Figs. 4B and 5, D–F). The two orthologs from the dicotyledon M. truncatula showed divergence in their ability to rescue. Medtr3g104560 rescued completely, while Medtr1g015860 showed no statistical difference to max1, except for PC3, which provided values intermediate between max1 and the wild type (Figs. 4C and 5, G–I).
The results of these phenotypic rescue experiments are summarized in Supplemental Table S3. Overall, Os01g0701500 and Medtr1g015860 showed little or no evidence of rescue of any of the phenotypes assessed. Most of the remaining orthologs showed weaker rescue of leaf size than the other phenotypes, while PgMAX1, which rescued branching and PC2 completely, did not rescue PC3. Together, these results suggest that the leaf size SL phenotype has a higher threshold for successful rescue than branching and PC2.
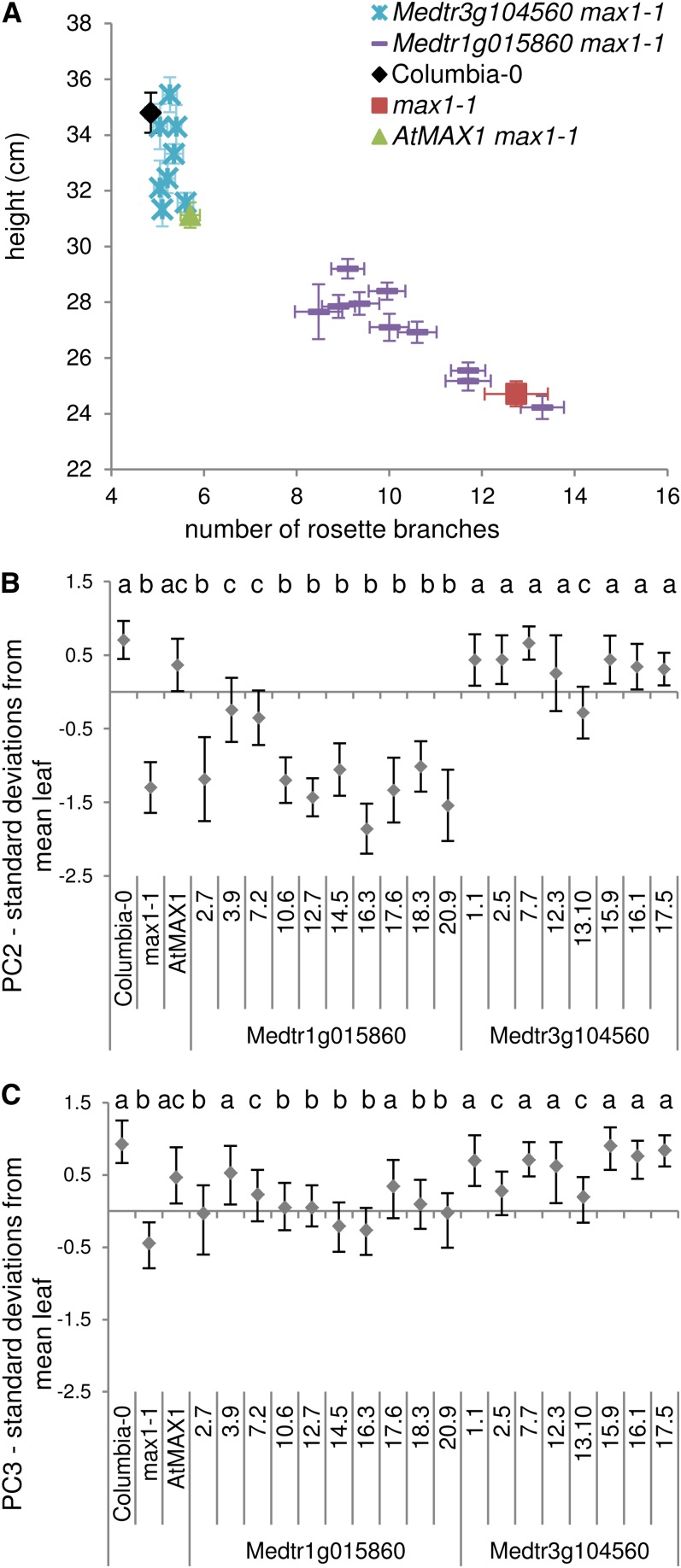
The poor rescue by Os01g0701500 is likely due to a premature stop codon 20 residues from the end of the consensus protein sequence. By contrast, there is no obvious reason for the poor rescue by Medtr1g015860. To investigate the differences in function between Medtr1g015860 and Medtr3g104560 in more detail, individual transgenic lines were compared using the leaf shape and branching phenotypes, as well as the reduced height phenotype observed in max1-1 mutants, which is suspected to be at least partially causally linked to the increased branching (Fig. 6A). This analysis revealed that a subpopulation of Medtr1g015860 overexpression lines were statistically different from max1-1 in branch number, increased height relative to the mutant, and in some cases, partial rescue of leaf shape PC2 and PC3 (Fig. 6, B and C). The degree of rescue in the individual transgenic lines does not correlate with overall transgene expression, suggesting that the low degree of rescue is not due to low expression of the transgene (Supplemental Fig. S3). This indicates that Medtr1g015860 retains some capacity to catalyze the reaction(s) lost in max1 mutants, but not to the same efficiency as the endogenous gene or its in-paralog.
Figure 6.
Phenotypic rescue of Arabidopsis max1-1 by individual transgenic lines of putative orthologs from M. truncatula. Rosette branching compared with final height of longest branch (A) mean number of sds of leaves from the natural accession mean for PC2 of leaf shape, as produced by LeafAnalyser (B), and mean number of sds of leaves from the natural accession mean for PC3 of leaf shape (C). For branching, n = 20, except for Columbia, max1-1, and AtMAX1 max1-1, for which n = 40, and for leaf shape analysis, n = 6–10. Error bars indicate se of the mean. Shared letters indicate no significant difference in Tamhane’s T2 post hoc test to Columbia (a), max1-1 (b), or AtMAX1 max1-1 (c) for lines not already labeled “a” (P ≤ 0.001, PC2; P ≤ 0.05, PC3). Col-0, Columbia. [See online article for color version of this figure.]
Conserved Sites
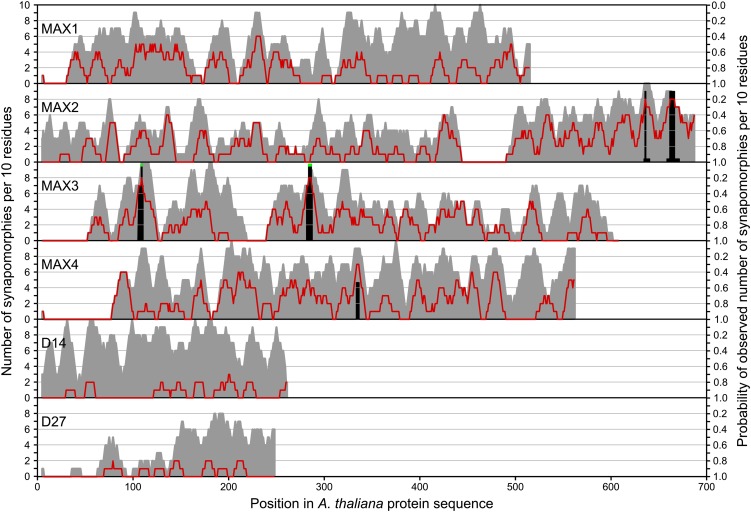
Conservation at the molecular level was then examined in more detail for all members of the SL pathway. A number of sites in each target gene are conserved across almost the entire ortholog set (defined as, at most, four mismatches at a given amino acid position in the ortholog set), representing between 17.2% (D27) and 48.6% (D14) of their respective alignments (Table II). A subset of these sites are synapomorphic (see “Materials and Methods”) between 4.0% (D14) and 10.8% (MAX2) of the alignment length. The synapomorphic sites include both insertions and nonsynonymous substitutions (see Supplemental Data Set S1) and are typically distributed throughout the gene (Fig. 7). Some regions have very low numbers of synapomorphic sites, particularly the N-terminal ends of all the target genes. Typically, this is due to low sequence conservation in these regions; however, for MAX2 and D14, the lack of N-terminal synapomorphies reflects similarity to the outgroup sequences within a well-conserved region. While there are peaks of at least six synapomorphic residues per 10 amino acids in each of MAX1, MAX2, MAX3, and MAX4, only two sites in MAX2, two sites in MAX3, and one site in MAX4 have a probability of less than one of occurring due to chance once a Bonferroni correction is applied (Table III). Of these, only two (both in MAX3) are significant at the 5% level.
Figure 7.
Sliding-window analysis (n = 10) highlighting the distribution of uniquely conserved residues across the ortholog set alignments for each of the MAX genes. For each gene, the gray area indicates the density of conserved residues, the red line indicates the density of synapomorphic residues that are conserved in the ingroup but differ from the wider gene family, and black bars show the probability of the observed density of synapomorphic residues occurring due to chance; regions with less than 5% probability are highlighted with green tips.
Table III. Sequence alignment features corresponding to peaks in Figure 7.
Positions are relative to the Arabidopsis ortholog sequence for each gene. Conserved positions in motifs are shown in uppercase. Predicted secondary structure elements were identified using PHYRE (Kelley and Sternberg, 2009). H, α-Helix; E, β-sheet; C, random coil. Uppercase letters indicate greater than 70% confidence in the predicted structure. Asterisks indicate significance at 5% level.
| Gene | Peak | P Value | Conserved Positions | Motif | Structure | Indels |
|---|---|---|---|---|---|---|
| MAX2 | 636 | 0.10 | 631–640 | RKLFIHGTAH | hceeecCCCc | |
| MAX2 | 662–666 | 0.10 | 657–670 | QLReDYYPAPEnDm | cccccCCCCCCCCC | 669–670 1 bp in grass |
| MAX3 | 109 | 0.03* | 104–113 | DDHGSTVHPL | CCCCCcccec | |
| MAX3 | 284–286 | 0.03* | 279–290 | vsCNAEDMLLPr | EEEccCCCCCCC | 282–285 4 bp relative to outgroup |
| MAX4 | 334–336 | 0.53 | 329–340 | LrAEPTPLYKFe | hcCCCCceeeee |
To identify the degree of intraspecific variation, nonsynonymous single-nucleotide polymorphisms (SNPs) among the Arabidopsis ecotypes were identified in each of the MAX genes (Supplemental Data Set S1). The number of nonsynonymous SNPs per gene in this set of ecotypes varied from three in D14 to 22 in MAX2.
DISCUSSION
SL Pathway Evolution
From the results of this study, the MAX genes can be broadly categorized into two groups based on the taxonomic distribution of their wider gene families. MAX1, MAX3, and MAX4 were each found to be most similar to nonplant genes, while MAX2, D14, and D27 are most similar to other plant-specific genes.
D14 and D27
Of the plant-specific genes, D14 and D27 show similar evolutionary patterns. Canonical D14 and D27 are absent from the genomes of the two most basal species (P. patens and S. moellendorffii), and both have closely related outgroup sequences that are so similar as to have been dubbed D14-like and D27-like (Arite et al., 2009; Lin et al., 2009). This similarity is likely why no significant synapomorphies were detected for D14 and D27 in our sliding-window analysis (Fig. 7). Proteins related to the wider D27-like family have been identified in basal plants and throughout the green algae (Delaux et al., 2012; Waters et al., 2012a), although the biological function of these clades is unknown and they have no known link to SL. By contrast, D14-like is required for responses to karrikins (Waters et al., 2012b). Karrikins are smoke-generated molecules structurally related to SLs that promote germination and photomorphogenesis in species that colonize ground cleared by forest fires. Arabidopsis is sensitive to karrikins despite not being a fire-following species, suggesting that there are other endogenous SL-like molecules that regulate germination and early seedling establishment in a D14-like-dependent manner (Nelson et al., 2010; Waters et al., 2012b). Karrikins require MAX2 for their signal transduction (Nelson et al., 2011), with the D14 and D14-like paralogs apparently providing specificity for SL and karrikin MAX2-mediated responses, respectively (Waters et al., 2012b; Kagiyama et al., 2013). However, as D14-like retains some ability to mediate SL responses, it may be that SL signaling represents the ancestral role for the whole D14 family in plants, including those D14-like members in moss and S. moellendorffii identified as outgroup sequences here and those reported from charophyte algae (Delaux et al., 2012). Consistent with this idea, the moss P. patens, the liverwort M. polymorpha, and the charophyte C. corallina are all responsive to the synthetic strigolactone GR24 (Proust et al., 2011; Delaux et al., 2012).
MAX3 and MAX4
Although three of the six core SL genes are present in a range of taxonomic groups, including Metazoa and Bacteria, closely related algal sequences were detected only for MAX3, although our analysis includes relatively few algal species. Delaux et al. (2012) identified putative MAX4 and MAX3 orthologs from several algal species, but noted that several residues likely to be involved in substrate recognition are not conserved in these orthologs, even those from the Charales, although they did detect SLs in charophytes. As these charophyte orthologs were recovered from transcriptomic, not genomic, data, it is possible that canonical MAX3 and MAX4 orthologs may yet be present in these species. However, they report that MAX4 orthologs are undetectable in the draft M. polymorpha genome (Delaux et al., 2012).
Four residues have been identified as essential for substrate specificity of CCD enzymes, such as MAX3 and MAX4, through structural analysis (Messing et al., 2010). These residues correspond to L-113, I-397, F-489, and F-613 of AtMAX3 and F-128, F-371, M-456, and L-562 of AtMAX4, which, in our analyses, were conserved across the ingroup sequences, with the exception of F-489 of MAX3, which, as noted by Delaux et al. (2012), is not conserved within the embryophytes. The residues that are conserved in the embryophytes are also typically conserved or at least partially conserved in the outgroup sequences, with only one exception in each gene, M-456 in MAX4 and particularly L-113 in MAX3. This latter site coincides with a region in which the average density of synapomorphic sites is significantly elevated and is therefore a candidate site for SL-related specificity.
Thus, although several basal species synthesize SLs, canonical MAX3 and MAX4 do not emerge until the embryophytes, suggesting some promiscuity in the biosynthetic pathway. In this context, it is interesting that P. patens synthesizes a variety of SLs, including strigol and orobanchol (Proust et al., 2011), both of which are also synthesized in higher plants, and strigol, at least, can reduce branching in rice (Yoneyama et al., 2007; Umehara et al., 2008; Kohlen et al., 2011). P patens MAX4/CCD8 knockout mutants show morphological phenotypes that can be rescued by GR24 addition. In these mutants, orobanchol can no longer be detected, but strigol levels are the wild type (Proust et al., 2011). These results suggest that there is MAX4/CCD8-independent strigol synthesis and that wild-type strigol levels are not sufficient to trigger moss SL responses. Orobanchol synthesis has also been shown to be largely MAX4-dependent in Arabidopsis, but here its synthesis is also MAX1 dependent, a function apparently lacking in moss (Kohlen et al., 2011). These factors suggest that discussions of a distinct SL pathway are inappropriate, both in terms of biosynthesis and response. Instead, there may be a diversity of SL-like compounds, the response to which requires a D14/D14-like protein (Flematti et al., 2013). This ancestral system could have been refined into increasingly distinct ligand-specific pathways over evolutionary time.
MAX2
Possibly the most clearly defined component of the modern SL signaling system is MAX2. Like the canonical D27 and D14 clades, it likely arose after the last common ancestor of algae and land plants. MAX2 is present in all surveyed embryophyte genomes and is more diverged from its nearest sister clade than any other member of the SL pathway, inferring two substitutions per site since its divergence from its closest sister gene. This degree of divergence reduces confidence in the alignment of MAX2 to its wider gene family, rendering formal tests of selection impossible; however, it is indicative of strong selection pressure acting on the gene.
Despite the degree of divergence from non-MAX2 sequences, the sequences within the clade are all highly similar, suggesting that there has been little selection pressure for a change to its role in signaling among the different plant species, despite the absence of elements of the SL biosynthetic pathway from some basal species. This is compatible with MAX2 not being the signal receptor, in contrast to other F-box proteins involved in hormone signaling (Dharmasiri et al., 2005; Kepinski and Leyser, 2005; Ueguchi-Tanaka et al., 2005; Katsir et al., 2008), a point supported by the apparent absence of MAX2 orthologs in GR24-responsive charophyte algae (Delaux et al., 2012). Interestingly, the rice d3 mutant (affecting the MAX2 ortholog) has recently been shown to have defective AM colonization, a phenotype not found in the biosynthetic SL mutants or the d14 mutant (Yoshida et al., 2012). These results suggest the interesting hypothesis that the ancestral role for MAX2 is in mycorrhization, and it was subsequently recruited to the SL pathway. Its tight subsequent conservation suggests the increasing number of downstream effects of SLs and SL-related compounds may be mediated by a small number of specific MAX2 targets.
Consistent with this idea, the C-terminal 15% of the MAX2 protein, which is likely to be involved in substrate recognition, is highly conserved and has the highest level of synapomorphy. The original point mutation that defined the locus, max2-1, maps close to this C-terminal domain. The max2-1 mutation is predicted to cause a substitution of D581 for Asn (Stirnberg et al., 2002). This site is part of a longer area of conserved sequence that is dissimilar to that of the wider gene family, and no nonsynonymous SNPs were identified among the Arabidopsis ecotypes within approximately 10 residues of this mutation. The grass MAX2 sequences have a single residue insertion relative to the remaining MAX2 sequences within one of the C-terminal synapomorphic motif regions between residues 669 and 670. Because synapomorphic insertions may be more likely to have affected the protein function than substitutions, these regions provide potentially promising avenues for further research.
MAX1
The contrasting evolutionary history of MAX2 and the upstream genes suggests that MAX2 evolution may be constrained by downstream target recognition, but its activity could be linked to diverse upstream SL and SL-like signals, which have changed during land plant evolution. The MAX1 gene is a particularly strong candidate to mediate diversification in upstream inputs into MAX2 signaling. MAX1 acts late in the SL biosynthetic pathway (Booker et al., 2005; Kohlen et al., 2011), it is absent from the P. patens and algal genomes, it is polyphyletic in monocots (Nelson et al., 2008), and there are currently no mutant phenotypes associated with max1 orthologs from other well-studied systems. There is only limited evidence for conserved MAX1 activity beyond Arabidopsis—a petunia MAX1 ortholog can rescue the Arabidopsis max1 mutant (Drummond et al., 2012). Our observations that the SmMAX1 ortholog can fully rescue the Arabidopsis max1-1 mutant shoot phenotypes (Figs. 4A and 5, A–C) demonstrates that, at least, the catalytic function(s) used in the Arabidopsis SL pathway is/are present in more basal land plants. Likewise, PgMAX1, a potential ortholog identified from the gymnosperm white spruce, which does have axillary branching, is also capable of partially rescuing Arabidopsis max1-1, supporting the wide taxonomic conservation of MAX1 function throughout tracheophytes.
MAX1 Diversification in the Angiosperms
Earlier phylogenetic analyses of MAX1 revealed that the five rice MAX1 genes occur on three distinct branches, each of which has orthologs in maize (Nelson et al., 2008), and our results show that these three separate clades are represented in all surveyed grass genomes. Only two clades are present in the remaining monocotyledon, P. dactylifera, consistent with genome duplications that are likely to have given rise to these clades. The presence of the three separate clades in all four grass species suggests that MAX1 was duplicated between the last common ancestor of the angiosperms (125–145 million years ago) and the last common ancestor of the monocotyledons (112–125 million years ago; De Bodt et al., 2005). MAX1 paralogs were also identified in five dicotyledon species (Mimulus guttatus, soybean, M. truncatula, P. trichocarpa, and strawberry [Fragaria vesca]); however, the extent of duplication and degree of conservation appears to be unique to MAX1 in grasses.
Functional analysis supports the hypothesis that these distinct clades are at least partly redundant at the protein level. Os02g0221900 and Os06g0565100, each representing a different monocot MAX1 clade, have similar functional capability to AtMAX1, although Os02g0221900 cannot fully rescue all of the max1-1 leaf phenotypes. The tested Os01g0701500 allele is incapable of rescuing any of the tested max1 phenotypes, likely due to a premature stop codon, suggesting that this gene may have lost all function and may be degenerating to become a pseudogene, consistent with redundancy within and/or between clades. Functional redundancy among the OsMAX1 orthologs could also explain the lack of a recorded max1 mutant phenotype for rice to date, despite all other SL pathway genes being identified in screens for rice branching mutants (McSteen, 2009). There may be functional diversity in the other members of the Os01g0701500 clade, and there is some variation in the expression of MAX1 orthologs, with some being phosphate starvation inducible and others not (Umehara et al., 2010). This may reflect diversification of the function of SLs and SL-related compounds to roles beyond rhizosphere signaling and developmental responses to nutrient limitation.
There is stronger evidence for diversification at the protein level in M. truncatula. One of its two MAX1 orthologs (Medtr3g104560) was able to rescue fully the Arabidopsis max1-1 phenotypes, but the other showed only weak activity at best (Medtr1g015860). Medtr1g015860 may be degenerating to become a pseudogene, or it may have been co-opted to perform a different reaction. Interestingly, Medtr1g015860, but not Medtr3g104560, is up-regulated in response to nodulation stress (Ruffel et al., 2008), and in pea, SLs have been found to be important for nodulation (Foo and Davies, 2011), suggesting that Medtr1g015860 divergence may be linked to a specific role in SL/SL-like control of nodule development.
There is some evidence for systematic variation in the degree of rescue of the different phenotypes tested. Several orthologs that show strong rescue of PC2 and branching do not rescue leaf size as strongly. However, because the S. moellendorffii ortholog is capable of partial rescue of leaf size, the less robust rescue by the rice orthologs compared with Medtr3g104560 is unlikely to reflect the later evolutionary origin of this function. Instead, it is possible that the leaf size SL phenotype has a higher threshold for successful rescue than branching and PC2.
Molecular Basis for MAX1 Diversity
The max1-1 mutant phenotype is caused by a single point mutation predicted to substitute P117 for Leu (Booker et al., 2005). P117 and its surrounding sequence is widely conserved among the MAX1s; however, the vine sequence and one maize sequence have an Ala at this position, while one rice sequence and the S. moellendorffii sequence have a Thr and a Tyr, respectively. Despite this variation, the catalytic capabilities of the MAX1 orthologs appear to be conserved throughout the tracheophytes, including S. moellendorffii and the rice paralogs, whatever the function of these orthologs (and the SLs they may produce) in planta. This suggests that in grasses and particularly in S. moellendorffii, the surrounding sequence has diverged so much that residues sensitive to mutation in Arabidopsis (such as P117) are replaced with residues with different physicochemical properties.
Likewise, the genetic basis of the divergence in catalytic ability in the M. truncatula paralogs is difficult to infer, as few of the residues (only three) that show high conservation in angiosperms are altered in Medtr1g015860. Of these, two (V43 to Leu and V514 to Ile) are changes between amino acids with very similar properties, and the remaining substitution (D133 Asp to Asn) is at a position in which several of the grass sequences (including Os01g0700900 and Os01g0701400) and S. moellendorffii have hydrophobic residues, suggesting that this residue is unlikely to be the sole source of the change in Medtr1g015860 function. This raises the question of why some of these residues are highly conserved within the dicotyledons. Possible explanations include: (1) These residues are not concerned with the catalytic properties of the MAX1 enzyme, but impact its regulation (a regulation that is overridden in the complementation analysis, possibly by use of a strong promoter), and (2) although the protein conformation required for functionality is maintained between S. moellendorffii and gymnosperm and angiosperm orthologs, the differences in the sequences that create this conformation also mean that these sequences have different regions of sensitivity to mutation, leading to different patterns of selection. Given the wide range of reactions performed by members of the cytochrome p450 gene family (Hannemann et al., 2007), it perhaps reflects an inherent ability of these enzymes to perform their reactions with a wide variety of conformations. In the context of the apparent diversity in SLs and in SL biosynthesis pathways, a related idea is that the phenocritical substrates for MAX1 differ in different species, with concomitant differences in selection pressures.
CONCLUSION
Phylogenetic analysis of the known SL pathway genes has revealed their diverse evolutionary histories (summarized in Table IV). The CCD enzymes, MAX3 and MAX4, are present in all land plant groups and are members of gene families represented across the major domains of life. A crucial innovation appears to have been the recruitment of an F-box protein, MAX2, to a signaling role in the pathway. This protein is the common downstream effector of at least two SL-like signals in Arabidopsis, SLs and karrikins. This, and the recent evidence of SL signaling in the moss P. patens, which has no canonical orthologs of D14, D27, or MAX1, suggests considerable promiscuity in the events upstream of MAX2 in early land plants, with respect both to signaling and synthesis of bioactive compounds. Our analysis of MAX1 suggests that it may have played a significant role in this upstream SL signal diversity and its later refinement.
Table IV. Comparison of the requirements and roles of the six known SL pathway genes in angiosperms, where they were first genetically defined, and early land plants.
The data summarized here are referenced in main text or are from this study.
| Gene | Biochemical Function | Role in Angiosperms | Role in Basal Lineages |
|---|---|---|---|
| D27 | β-carotene isomerase | Required for SL synthesis | Absent from P. patens and basal lineages, despite SL synthesis; D27-like present |
| MAX4 | CCD | Required for SL synthesis | Present in P. patens and required for some SLs, but not required for strigol synthesis |
| MAX3 | CCD | Required for SL synthesis | Present in P. patens; no functional data |
| MAX1 | Cytochrome P450 | Required for SL synthesis in Arabidopsis; evidence for in-paralog redundancy and divergence in rice and M. truncatula | Absent from P. patens and basal lineages, despite SL synthesis; activity present in S. moellendorffii |
| D14 | α/β hydrolase | Required for perception and/or cleavage | Absent from P. patens and basal lineages despite SL responses; D14-like present |
| MAX2 | F-box protein | Required for signaling | Present in P. patens, but absent from more basal lineages; no functional data |
MATERIALS AND METHODS
Data Sources
Published or prerelease genomic sequence data were obtained for 24 plant species (Table V). These included five monocotyledons, 17 dicotyledons, the basal tracheophyte Selaginella moellendorffii, and the bryophyte Physcomitrella patens. For each species, complete sets of predicted coding sequences and corresponding protein sequences were saved locally for use in subsequent analyses. For Arabidopsis (Arabidopsis thaliana), data on intraspecific polymorphisms were obtained by directly querying a local copy of the Ensembl Plants database version 58 (Kersey et al., 2010).
Table V. Taxonomic distribution and sources of plant genomes.
| Taxonomy | Species | National Center for Biotechnology Information Taxon Identification | Genome Version | No. of Predicted Protein-Coding Loci | Genome Size | URL |
|---|---|---|---|---|---|---|
| Mbp | ||||||
| Embryophyta | P. patens | 3218 | 1.1 | 39727 | 480r | http://www.phytozome.net/physcomitrella |
| Trachaeophyta | S. moellendorffii | 88036 | 1 | 22273 | 88–127s | http://genome.jgi-psf.org/Selmo1/Selmo1.home.html |
| Magnoliophyta | ||||||
| Poaceae | ||||||
| BEP clade | B. distachyon | 15368 | 1 | 25532 | 352l | http://www.brachypodium.org/ |
| Rice | 4530 | 6 | 40838 | 420q | http://rice.plantbiology.msu.edu/ | |
| PACCAD clade | S. bicolor | 4558 | 1 | 34496 | 730o,p | http://www.phytozome.net/sorghum |
| Maize | 4577 | 4a.53 | 32540 | 2300g,n | http://www.maizesequence.org/index.html | |
| Arecaceae | P. dactylifera | 42345 | 3 | 28889 | 650b | http://qatar-weill.cornell.edu/research/datepalmGenome |
| Stem eudicotyledons | A. coerulea | 218851 | 1.1 | 24832 | 302b | http://www.phytozome.net/aquilegia |
| Core eudicotyledons | ||||||
| Asterids | M. guttatus | 4155 | 1 | 25530 | 430m | http://www.phytozome.net/mimulus |
| S. lycopersicum | 4081 | 2.3 | 34727 | 738b | http://solgenomics.net/organism/Solanum_lycopersicum/genome | |
| Rosids | V. vinifera | 29760 | 30434 | 483g | http://www.genoscope.cns.fr/spip/Vitis-vinifera-e.html | |
| Brassicales | C. papaya | 3649 | 27332 | 372g | http://asgpb.mhpcc.hawaii.edu/papaya/ | |
| Arabidopsis | 3702 | 9 | 37343 | 157k,l | http://www.arabidopsis.org/ | |
| A. lyrata | 59689 | 1 | 32670a | 206.7b | http://genome.jgi-psf.org/Araly1/Araly1.home.html | |
| Malvids | C. sinensis | 2711 | 1 | 25376 | 319b | http://www.phytozome.net/citrus |
| E. grandis | 71139 | 1.1 | 36376 | 691b | http://www.phytozome.net/eucalyptus | |
| Fabids | C. sativus | 3659 | 21491 | 367g | http://www.phytozome.net/cucumber | |
| Rosaceae | Strawberry | 57918 | 1.0 | 34809 | 240t | http://www.rosaceae.org/projects/strawberry_genome |
| Papilionoideae | Soybean | 3847 | 66153 | 1115g | http://www.phytozome.net/soybean | |
| M. truncatula | 3880 | 3 | 50962 | 454–526e,g | http://www.medicagohapmap.org/?genome | |
| L. japonicus | 34305 | 43051 | 469h,i | http://www.kazusa.or.jp/lotus/ | ||
| Malpighiales | P. trichocarpa | 3694 | 2 | 41377 | 485f | http://www.phytozome.net/poplar |
| Euphorbiaceae | M. esculenta | 3983 | 1 | 47164 | 772c,d | http://www.phytozome.net/cassava |
| R. communis | 3988 | 0.1 | 31221a | 224.9e | http://castorbean.jcvi.org/index.php |
Number of protein-coding transcripts. bData from genome project Web site. cAwoleye et al. (1994). dBennett and Leitch (1997). eBennett and Leitch (1995). fTuskan et al. (2006). gArumuganathan and Earle (1991). hBennett and Smith (1976). iCheng and Grant (1973). kBennett et al. (2003). lBennett and Leitch (2005). mWu et al. (2008). nSchnable et al. (2009). oPaterson et al. (2009). pLaurie and Bennett (1985). qGoff et al. (2002). rReski (1999). sWang et al. (2005). tShulaev et al.(2011).
A more diverse set of taxa was represented by the complete set of protein sequences from the KEGG database (Kanehisa and Goto, 2000). The set of sequences in the KEGG database provided a nonredundant set of protein sequences from a diverse range of species with relatively even taxonomic coverage, within the limitations of the taxonomic distribution of completed genome sequencing projects. While diverse and extensive, the set of included species is not exhaustive, so the resulting data sets were of a manageable size.
Ortholog Prediction
Orthologs, in-paralogs, and paralogs were predicted for each of the six target genes (MAX1, MAX2, MAX3, MAX4, D14, and D27) in 18 of the 24 plant genomes (indicated in Table I) from protein sequence data using the stand-alone InParanoid 4 software (Berglund et al., 2008). Putative ortholog and in-paralog clusters were identified for all possible pairs of these 18 genomes, and the resulting clusters were then combined by grouping all clusters with matching sequences to produce putative ortholog sets encompassing all species. Singleton sequences that were not assigned to any ortholog cluster were added to the ortholog set containing the sequence with which they had the highest BLAST (Altschul et al., 1990) score. Run independently, InParanoid has been shown to exhibit false positive and false negative rates of approximately 0.2 (Chen et al., 2007); combining clusters was expected to reduce the false negative rate at the expense of elevating the false positive rate. For each target gene, the phylogenetic distribution of sequences (see “Phylogenetic Reconstruction”) was considered to identify false positives, which were removed from the final ortholog set. Orthologous sequences in the remaining six genome sequences were identified by direct BLAST searches to identify sequences with greater homology to the putative ortholog clusters than to nonorthologous sequences. Use of BLAST in favor of the more complex InParanoid algorithm reduced the expected false negative rate to 0.04 at the expense of a false positive rate of approximately 0.5 (Chen et al., 2007), so false positives were again filtered during phylogenetic reconstruction.
Gene Families
A more taxonomically diverse set of species was considered to establish the relationships between the genes of the MAX pathway and their wider gene families. Target gene protein sequences from Arabidopsis were used in a BLAST search against the full set of protein sequences in the KEGG database (Kanehisa and Goto, 2000). Protein sequences were obtained for up to 500 hits per query, and the resulting sequences were filtered to remove incomplete proteins (length of <50% of query sequence or missing an initial Met). These sets were further refined during sequence alignment and phylogenetic reconstruction, removing those sequences that could not be aligned without the inference of very long indels. Sequences that were inferred to belong to poorly resolved clades in the phylogeny were also removed, but only if all remaining sequences formed a robustly supported monophyletic clade.
Sequence Alignment
All protein sequences were aligned using MAFFT (Katoh et al., 2005) with the “-auto” flag. This approach allowed the software to determine the exact alignment strategy used according to the characteristics of the input sequences and, in practice, resulted in the most accurate alignment strategies being applied to the ortholog alignments and more efficient strategies being used for the larger gene family alignments.
Phylogenetic Reconstruction
Maximum-likelihood phylogenies were reconstructed using RAxML 7.0.4 (Stamatakis, 2006). For each gene, the protein alignment was used to reconstruct an ortholog set phylogeny under the general time-reversible model (Tavaré, 1986) with γ-distributed rate heterogeneity (GTRGAMMA). The gene family alignment was used to reconstruct a gene family phylogeny using the JTT (Jones et al., 1992) substitution matrix with discrete rate categories, and the final topology was re-evaluated using γ-distributed rate heterogeneity (PROTMIXJTT). Support for the maximum-likelihood phylogenies was estimated from 100 rapid bootstrap resamplings (Stamatakis et al., 2008).
Production of Transgenic Plants
MAX1 homologs from Medicago truncatula accession Jemalong A17 (Medtr1g015860 and Medtr3g104560), rice (Oryza sativa ssp. japonica) ‘Nipponbare’ (Os01g0701500, Os02g0221900, and Os06g0565100), and S. moellendorffii (e_gw1.19.137.1 jgi|Selmo1|97512) were cloned by PCR from complementary DNA (cDNA) on the basis of their predicted sequences using primers listed in Supplemental Table S1. RNA was extracted from Arabidopsis, rice, M. truncatula, and S. moellendorffii using the RNeasy Plant Mini Kit (Qiagen) with all optional steps included according to manufacturer’s instructions. For extraction from white spruce (Picea glauca) and rice shoot, the method described by Azevedo et al. (2003) was used. cDNA was synthesized from 500 ng RNA using SuperScript II Reverse Transcriptase (Invitrogen) according to manufacturer’s instructions, using oligo(dT) as the nonspecific primer, except for construction of RACE libraries and cloning Os06g0565100 from rice shoot. For these, the manufacturer’s instructions for the incubation step were changed to 40 min at 42°C, 10 min at 70°C, readdition of the enzyme, and 20 min of further incubation at 50°C. For Os01g0701500, an early stop codon was confirmed by 3′ RACE as described by Scotto-Lavino et al. (2006). For PgMAX1, an EST (GenBank accession no. BT103061) was identified as a gymnosperm ortholog on the basis of reciprocal BLAST hits from white spruce, and 5′ RACE (as described by Sambrook and Russell [2001]) was used to identify the missing 5′ sequence. Genes were transferred by restriction digestion (see Supplemental Table S2) into the binary vector pART7 (Gleave, 1992) under control of the near-constitutive strong Cauliflower mosaic virus 35S promoter. Constructs were transformed into the Arabidopsis max1-1 mutant in the Columbia background (Stirnberg et al., 2002) via the Agrobacterium tumefaciens floral dip method (Clough and Bent, 1998). For each construct, eight to 11 independent transformant lines were produced, brought to homozygosity, checked for the presence of the max1-1 mutation and for transgene expression (Supplemental Fig. S1), and then phenotyped in the T3 generation.
Quantitative PCR
cDNA was produced from 10 plant pools of 10-d-old Arabidopsis seedlings for each transgenic line carrying 35S:Medtr1g015860, and expression levels of the transgene were quantified using primers listed in Supplemental Table S1. Expression was normalized to the geometric mean of expression of At2g28390 and At1g13320, and data shown is the mean of two biological repeats, each technically repeated three times.
Plant Growth and Materials
Seed were stratified for 2 d at 4°C and germinated, and plants were grown on F2 soil treated with Intercept (Levington Horticulture) in either short-photoperiod conditions in a growth cabinet (8-h light/16-h dark at approximately 80 μmol m–2 s–1 with temperatures of 20°C during the day and 18°C at night) or a long photoperiod (16-h light/8-h dark) between spring and autumn in a greenhouse supplemented with artificial light at approximately 150 μmol m–2 s–1 and temperatures between 15°C and 24°C.
Branching Assay
Branching was assessed using a method adapted from Greb et al. (2003). Seed were germinated and plants grown in P40 4-cm pot trays (Levington Horticulture) in short-day conditions for 4 weeks and then moved to long days to trigger flowering. Bolting stems were decapitated close to the rosette when they reached 10 to 15 cm, and rosette branches longer than 0.5 cm were counted 10 d later. Twenty plants were measured for each of eight to 11 independent transformant lines per construct, except for the three control lines, for which 40 plants each were used. These controls were wild-type Columbia, max1-1 negative control, and a single line of 35S::AtMAX1 previously reported to rescue max1-1 (S. Ward, personal communication).
Leaf Shape Analysis
Seeds were grown in P24 trays (Levington Horticulture) in the greenhouse for 5 weeks or, for the MtMAX1 construct lines (Fig. 4), for 6 weeks. Six to 10 plants of two representative lines were analyzed per construct, except for the MtMAX1 constructs, for which all transformant lines were tested. Leaves were processed as described in Kieffer et al. (2011) and analyzed as described in Danisman et al. (2012); adult rosette leaves were removed in phyllotaxic sequence and scanned using a Scanjet 4370 scanner (Hewlett-Packard) at 300 dpi resolution. Images were saved in TIFF format, and landmarks were assigned and analyzed with LeafAnalyser (Weight et al., 2008). Adult leaves at node 4 and above were used for analysis. Coordinates produced by LeafAnalyser were Procrustes fitted using MorphoJ (Klingenberg, 2011), which also generated the centroid size. LeafAnalyser was then used to produce a Principal Component Analysis eigenvector matrix from a library of 1,500 leaves from 10 Arabidopsis accessions, which had also been Procrustes fitted (Danisman et al., 2012). The eigenvector matrix was used to calculate leaf point models scaled to the sds of the natural accession database.
Synapomorphies
A sliding-window analysis was used to quantify the distribution of uniquely conserved (synapomorphic) sites for each gene across the 24 selected genomes. A synapomorphic site was considered to be a residue in the alignment that was conserved in the majority of the sequences in the ortholog set (mismatches in the alignment were tolerated in a maximum of 10% of sequences) and for which a different amino acid was found in the majority of nonortholog sequences (matching amino acids were tolerated in up to 20% of nonorthologs). A window size of 10 residues was used for all genes, and a 95% statistical significance was calculated against the binomial distribution using the total number of synapomorphic residues and the sequence length.
Structure Prediction
Protein secondary structure features were predicted using the Phyre (Kelley and Sternberg, 2009) Web server. Only one (Arabidopsis) sequence was used to represent each ortholog set, as the method is based on similarity to proteins of known structure, so the effect of differences between species on structure could not be predicted. Results for the consensus of three different structure prediction methods have been presented to indicate regions predicted with high (>70%) and low confidence to form an α-helix, β-sheet, or coil.
Intraspecific Variation
SNP data from 99 Arabidopsis ecotypes in the Ensembl Plants database (19 lines resequenced by Kover et al. [2009] and a further 80 lines resequenced as part of the 1,001 genomes project [Cao et al., 2011]) for each target gene were collated to identify the positions of nonsynonymous substitutions and the ecotypes affected.
Sequences of mRNA used for MAX1 complementation have been deposited at GenBank with the following accession numbers: PgMAX1: JX566695, SmMAX1: JX566702, Os01g0701500: JX566699, Os02g0221900: JX566700, Os06g0565100: JX566698, Medtr1g01586: JX566697, and Medtr3g104560: JX566696.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Semiquantitative PCR of transgene expression in the various MAX1-expressing transgenic lines analyzed compared with a TUBULIN9 control.
Supplemental Figure S2. Phylogenetic trees depicting the relationship between putative orthologs of five of the MAX genes across 24 plant species.
Supplemental Figure S3. Degree of branching rescue compared with transgene expression for individual transgenic lines of 35S::Medtr1g015860 max1-1.
Supplemental Table S1. PCR primers for cloning MAX1 and quantitative PCR.
Supplemental Table S2. Cloning strategies for MAX1 transgenic constructs.
Supplemental Table S3. Summary of phenotypic rescue of max1-1 Arabidopsis carrying putative MAX1 orthologs.
Supplemental Data Set S1. Conserved sites in the MAX1, MAX2, MAX3, MAX4, D14, and D27 proteins.
Acknowledgments
We thank Sally Ward (University of Cambridge) for the 35S::AtMAX1 max1-1 transgenic seeds and Vera Matser and Joe Vaughan (University of York) for assistance with the leaf shape analysis. Thanks for plant material are due to J. A. Banks (Purdue University; for S. moellendorffii), Joan Cottrell (Forest Research; for white spruce), and Michael Schultze (University of York; for M. truncatula). We also thank Stephen Day for critical reading of the manuscript.
Glossary
- SL
strigolactone
- AM
arbuscular mycorrhizal
- SNP
single-nucleotide polymorphism
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- cDNA
complementary DNA
- PC2
principal component 2
- PC3
principal component 3
- CCD
carotenoid cleavage dioxygenase
- RAxML
randomized axelerated maximum likelihood
References
- Akiyama K, Matsuzaki K, Hayashi H. (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827 [DOI] [PubMed] [Google Scholar]
- Alder A, Jamil M, Marzorati M, Bruno M, Vermathen M, Bigler P, Ghisla S, Bouwmeester HJ, Beyer P, Al-Babili S. (2012) The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335: 1348–1351 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J. (2007) DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J 51: 1019–1029 [DOI] [PubMed] [Google Scholar]
- Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J. (2009) d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol 50: 1416–1424 [DOI] [PubMed] [Google Scholar]
- Arumuganathan K, Earle ED. (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Report 9: 208–218 [Google Scholar]
- Awoleye F, van Duren M, Dolezel J, Novak FJ. (1994) Nuclear DNA content and in vitro induced somatic polyploidization cassava (Manihot esculenta Crantz) breeding. Euphytica 76: 195–202 [Google Scholar]
- Azevedo H, Lino-Neto T, Tavares RM. (2003) An improved method for high-quality RNA isolation from needles of adult maritime pine trees. Plant Mol Biol Report 21: 333–338 [Google Scholar]
- Bennett MD, Leitch IJ. (1995) Nuclear DNA amounts in angiosperms. Ann Bot (Lond) 76: 113–176 [Google Scholar]
- Bennett MD, Leitch IJ. (1997) Nuclear DNA amounts in angiosperms—583 new estimates. Ann Bot (Lond) 80: 169–196 [Google Scholar]
- Bennett MD, Leitch IJ. (2005) Nuclear DNA amounts in angiosperms: progress, problems and prospects. Ann Bot (Lond) 95: 45–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD, Leitch IJ, Price HJ, Johnston JS. (2003) Comparisons with Caenorhabditis (approximately 100 Mb) and Drosophila (approximately 175 Mb) using flow cytometry show genome size in Arabidopsis to be approximately 157 Mb and thus approximately 25% larger than the Arabidopsis genome initiative estimate of approximately 125 Mb. Ann Bot (Lond) 91: 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD, Smith JB. (1976) Nuclear DNA amounts in angiosperms. Philos Trans R Soc Lond B Biol Sci 274: 227–274 [DOI] [PubMed] [Google Scholar]
- Berglund A-C, Sjölund E, Östlund G, Sonnhammer ELL. (2008) InParanoid 6: eukaryotic ortholog clusters with inparalogs. Nucleic Acids Res 36: D263–D266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker J, Auldridge M, Wills S, McCarty D, Klee H, Leyser O. (2004) MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr Biol 14: 1232–1238 [DOI] [PubMed] [Google Scholar]
- Booker J, Sieberer T, Wright W, Williamson L, Willett B, Stirnberg P, Turnbull C, Srinivasan M, Goddard P, Leyser O. (2005) MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev Cell 8: 443–449 [DOI] [PubMed] [Google Scholar]
- Cao J, Schneeberger K, Ossowski S, Günther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C, et al. (2011) Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet 43: 956–963 [DOI] [PubMed] [Google Scholar]
- Chen F, Mackey AJ, Vermunt JK, Roos DS. (2007) Assessing performance of orthology detection strategies applied to eukaryotic genomes. PLoS ONE 2: e383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng RI-J, Grant WF. (1973) Species relationships in the Lotus corniculatus group as determined by karyotype and cytophotometric analyses. Can J Genet Cytol 15: 101–115 [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cook CE, Whichard LP, Turner B, Wall ME, Egley GH. (1966) Germination of witchweed (Striga lutea Lour.): isolation and properties of a potent stimulant. Science 154: 1189–1190 [DOI] [PubMed] [Google Scholar]
- Danisman S, van der Wal F, Dhondt S, Waites R, de Folter S, Bimbo A, van Dijk AD, Muino JM, Cutri L, Dornelas MC, et al. (2012) Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol 159: 1511–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S, Maere S, Van de Peer Y. (2005) Genome duplication and the origin of angiosperms. Trends Ecol Evol 20: 591–597 [DOI] [PubMed] [Google Scholar]
- Delaux P-M, Xie X, Timme RE, Puech-Pages V, Dunand C, Lecompte E, Delwiche CF, Yoneyama K, Bécard G, Séjalon-Delmas N. (2012) Origin of strigolactones in the green lineage. New Phytol 195: 857–871 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. (2005) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Drummond RS, Sheehan H, Simons JL, Martínez-Sánchez NM, Turner RM, Putterill J, Snowden KC. (2012) The expression of petunia strigolactone pathway genes is altered as part of the endogenous developmental program. Front Plant Sci 2: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flematti GR, Waters MT, Scaffidi A, Merritt DJ, Ghisalberti EL, Dixon KW, Smith SM. (2013) Karrikin and cyanohydrin smoke signals provide clues to new endogenous plant signaling compounds. Mol Plant 6: 29–37 [DOI] [PubMed] [Google Scholar]
- Foo E, Davies NW. (2011) Strigolactones promote nodulation in pea. Planta 234: 1073–1081 [DOI] [PubMed] [Google Scholar]
- Gleave AP. (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan T-H, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot J-P, Letisse F, Matusova R, Danoun S, Portais J-C, et al. (2008) Strigolactone inhibition of shoot branching. Nature 455: 189–194 [DOI] [PubMed] [Google Scholar]
- Greb T, Clarenz O, Schafer E, Muller D, Herrero R, Schmitz G, Theres K. (2003) Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev 17: 1175–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamiaux C, Drummond RSM, Janssen BJ, Ledger SE, Cooney JM, Newcomb RD, Snowden KC. (2012) DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr Biol 22: 2032–2036 [DOI] [PubMed] [Google Scholar]
- Hannemann F, Bichet A, Ewen KM, Bernhardt R. (2007) Cytochrome P450 systems—biological variations of electron transport chains. Biochim Biophys Acta 1770: 330–344 [DOI] [PubMed] [Google Scholar]
- Humphrey AJ, Beale MH. (2006) Strigol: biogenesis and physiological activity. Phytochemistry 67: 636–640 [DOI] [PubMed] [Google Scholar]
- Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J. (2005) Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol 46: 79–86 [DOI] [PubMed] [Google Scholar]
- Johnson X, Brcich T, Dun EA, Goussot M, Haurogné K, Beveridge CA, Rameau C. (2006) Branching genes are conserved across species. Genes controlling a novel signal in pea are coregulated by other long-distance signals. Plant Physiol 142: 1014–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8: 275–282 [DOI] [PubMed] [Google Scholar]
- Kagiyama M, Hirano Y, Mori T, Kim S-Y, Kyozuka J, Seto Y, Yamaguchi S, Hakoshima T. (2013) Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes Cells 18: 147–160 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28: 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33: 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. (2008) COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci USA 105: 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJE. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4: 363–371 [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Kersey PJ, Lawson D, Birney E, Derwent PS, Haimel M, Herrero J, Keenan S, Kerhornou A, Koscielny G, Kähäri A, et al. (2010) Ensembl Genomes: extending Ensembl across the taxonomic space. Nucleic Acids Res 38: D563–D569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer M, Master V, Waites R, Davies B. (2011) TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J 68: 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg CP. (2011) MorphoJ: an integrated software package for geometric morphometrics. Mol Ecol Resour 11: 353–357 [DOI] [PubMed] [Google Scholar]
- Kohlen W, Charnikhova T, Liu Q, Bours R, Domagalska MA, Beguerie S, Verstappen F, Leyser O, Bouwmeester HJ, Ruyter-Spira C. (2011) Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis. Plant Physiol 155: 974–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kover PX, Valdar W, Trakalo J, Scarcelli N, Ehrenreich IM, Purugganan MD, Durrant C, Mott R. (2009) A Multiparent Advanced Generation Inter-Cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet 5: e1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie DA, Bennett MD. (1985) Nuclear DNA content in the genera Zea and Sorghum. Intergeneric, interspecific and intraspecific variation. Heredity 55: 307–313 [Google Scholar]
- Lin H, Wang R, Qian Q, Yan M, Meng X, Fu Z, Yan C, Jiang B, Su Z, Li J, et al. (2009) DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21: 1512–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen P. (2009) Hormonal regulation of branching in grasses. Plant Physiol 149: 46–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing SAJ, Gabelli SB, Echeverria I, Vogel JT, Guan JC, Tan BC, Klee HJ, McCarty DR, Amzel LM. (2010) Structural insights into maize viviparous14, a key enzyme in the biosynthesis of the phytohormone abscisic acid. Plant Cell 22: 2970–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DC, Flematti GR, Riseborough J-A, Ghisalberti EL, Dixon KW, Smith SM. (2010) Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 7095–7100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DC, Scaffidi A, Dun EA, Waters MT, Flematti GR, Dixon KW, Beveridge CA, Ghisalberti EL, Smith SM. (2011) F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc Natl Acad Sci USA 108: 8897–8902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DR, Ming R, Alam M, Schuler MA. (2008) Comparison of cytochrome P450 genes from six plant genomes. Trop Plant Biol 1: 216–235 [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al. (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Proust H, Hoffmann B, Xie X, Yoneyama K, Schaefer DG, Yoneyama K, Nogué F, Rameau C. (2011) Strigolactones regulate protonema branching and act as a quorum sensing-like signal in the moss Physcomitrella patens. Development 138: 1531–1539 [DOI] [PubMed] [Google Scholar]
- Reski R. (1999) Molecular genetics of Physcomitrella. Planta 208: 301–309 [Google Scholar]
- Rice P, Longden I, Bleasby A. (2000) EMBOSS: the European molecular biology open software suite. Trends Genet 16: 276–277 [DOI] [PubMed] [Google Scholar]
- Ruffel S, Freixes S, Balzergue S, Tillard P, Jeudy C, Martin-Magniette ML, van der Merwe MJ, Kakar K, Gouzy J, Fernie AR, et al. (2008) Systemic signaling of the plant nitrogen status triggers specific transcriptome responses depending on the nitrogen source in Medicago truncatula. Plant Physiol 146: 2020–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyter-Spira C, Kohlen W, Charnikhova T, van Zeijl A, van Bezouwen L, de Ruijter N, Cardoso C, Lopez-Raez JA, Matusova R, Bours R, et al. (2011) Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiol 155: 721–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Schüssler A, Schwarzott D, Walker C. (2001) A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol Res 105: 1413–1421 [Google Scholar]
- Scotto-Lavino E, Du G, Frohman MA. (2006) 3′ end cDNA amplification using classic RACE. Nat Protoc 1: 2742–2745 [DOI] [PubMed] [Google Scholar]
- Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, Delcher AL, Jaiswal P, Mockaitis K, Liston A, Mane SP, et al. (2011) The genome of woodland strawberry (Fragaria vesca). Nat Genet 43: 109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon L, Bousquet J, Lévesque RC, Lalonde M. (1993) Origin and diversification of endomycorrhizal fungi and coincidence with vascular land plants. Nature 363: 67–69 [Google Scholar]
- Snowden KC, Simkin AJ, Janssen BJ, Templeton KR, Loucas HM, Simons JL, Karunairetnam S, Gleave AP, Clark DG, Klee HJ. (2005) The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell 17: 746–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorefan K, Booker J, Haurogné K, Goussot M, Bainbridge K, Foo E, Chatfield S, Ward S, Beveridge CA, Rameau C, et al. (2003) MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev 17: 1469–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690 [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. (2008) A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol 57: 758–771 [DOI] [PubMed] [Google Scholar]
- Stirnberg P, Furner IJ, Leyser HMO. (2007) MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J 50: 80–94 [DOI] [PubMed] [Google Scholar]
- Stirnberg P, van De Sande K, Leyser HMO. (2002) MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129: 1131–1141 [DOI] [PubMed] [Google Scholar]
- Tavaré S. (1986) Some probabilistic and statistical problems in the analysis of DNA sequences. Lect Math Life Sci 17: 57–86 [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, et al. (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Magome H, Takeda-Kamiya N, Yamaguchi S. (2010) Contribution of strigolactones to the inhibition of tiller bud outgrowth under phosphate deficiency in rice. Plant Cell Physiol 51: 1118–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, et al. (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200 [DOI] [PubMed] [Google Scholar]
- Wang W, Tanurdzic M, Luo M, Sisneros N, Kim HR, Weng J-K, Kudrna D, Mueller C, Arumuganathan K, Carlson J, et al. (2005) Construction of a bacterial artificial chromosome library from the spikemoss Selaginella moellendorffii: a new resource for plant comparative genomics. BMC Plant Biol 5: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Brewer PB, Bussell JD, Smith SM, Beveridge CA. (2012a) The Arabidopsis ortholog of rice DWARF27 acts upstream of MAX1 in the control of plant development by strigolactones. Plant Physiol 159: 1073–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Nelson DC, Scaffidi A, Flematti GR, Sun YK, Dixon KW, Smith SM. (2012b) Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139: 1285–1295 [DOI] [PubMed] [Google Scholar]
- Weight C, Parnham D, Waites R. (2008) LeafAnalyser: a computational method for rapid and large-scale analyses of leaf shape variation. Plant J 53: 578–586 [DOI] [PubMed] [Google Scholar]
- Wu CA, Lowry DB, Cooley AM, Wright KM, Lee YW, Willis JH. (2008) Mimulus is an emerging model system for the integration of ecological and genomic studies. Heredity (Edinb) 100: 220–230 [DOI] [PubMed] [Google Scholar]
- Yoneyama K, Xie X, Kim HI, Kisugi T, Nomura T, Sekimoto H, Yokota T, Yoneyama K. (2012) How do nitrogen and phosphorus deficiencies affect strigolactone production and exudation? Planta 235: 1197–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama K, Yoneyama K, Takeuchi Y, Sekimoto H. (2007) Phosphorus deficiency in red clover promotes exudation of orobanchol, the signal for mycorrhizal symbionts and germination stimulant for root parasites. Planta 225: 1031–1038 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Kameoka H, Tempo M, Akiyama K, Umehara M, Yamaguchi S, Hayashi H, Kyozuka J, Shirasu K. (2012) The D3 F-box protein is a key component in host strigolactone responses essential for arbuscular mycorrhizal symbiosis. New Phytol 196: 1208–1216 [DOI] [PubMed] [Google Scholar]
- Zou JH, Zhang SY, Zhang WP, Li G, Chen ZX, Zhai WX, Zhao XF, Pan XB, Xie Q, Zhu LH. (2006) The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J 48: 687–698 [DOI] [PubMed] [Google Scholar]