A bacterial effector is delivered into plant cells during infection, is phosphorylated, and binds plant 14-3-3 proteins in a phosphorylation-dependent manner.
Abstract
A key virulence strategy of bacterial pathogens is the delivery of multiple pathogen effector proteins into host cells during infection. The Hrp outer protein Q (HopQ1) effector from Pseudomonas syringae pv tomato (Pto) strain DC3000 is conserved across multiple bacterial plant pathogens. Here, we investigated the virulence function and host targets of HopQ1 in tomato (Solanum lycopersicum). Transgenic tomato lines expressing dexamethasone-inducible HopQ1 exhibited enhanced disease susceptibility to virulent Pto DC3000, the Pto ΔhrcC mutant, and decreased expression of a pathogen-associated molecular pattern-triggered marker gene after bacterial inoculation. HopQ1-interacting proteins were coimmunoprecipitated and identified by mass spectrometry. HopQ1 can associate with multiple tomato 14-3-3 proteins, including TFT1 and TFT5. HopQ1 is phosphorylated in tomato, and four phosphorylated peptides were identified by mass spectrometry. HopQ1 possesses a conserved mode I 14-3-3 binding motif whose serine-51 residue is phosphorylated in tomato and regulates its association with TFT1 and TFT5. Confocal microscopy and fractionation reveal that HopQ1 exhibits nucleocytoplasmic localization, while HopQ1 dephosphorylation mimics exhibit more pronounced nuclear localization. HopQ1 delivered from Pto DC3000 was found to promote bacterial virulence in the tomato genotype Rio Grande 76R. However, the HopQ1(S51A) mutant delivered from Pto DC3000 was unable to promote pathogen virulence. Taken together, our data demonstrate that HopQ1 enhances bacterial virulence and associates with tomato 14-3-3 proteins in a phosphorylation-dependent manner that influences HopQ1’s subcellular localization and virulence-promoting activities in planta.
The ability to detect and mount a defense response against pathogenic microbes is vital for plant survival. Plants rely on both passive and active defenses to ward off microbial pathogens. Physical barriers, such as the cell wall and cuticle, as well as chemical barriers provide a first line of defense against microbial colonization. Unlike animals, plants do not possess a circulating immune system and rely on innate immunity for active defenses against microbial pathogens (Spoel and Dong, 2012). Plants use surface-localized receptors to recognize conserved pathogen-associated molecular patterns (PAMPs), such as bacterial flagellin, resulting in pattern-triggered immunity (PTI; Zipfel et al., 2006). Plants also use primarily intracellular nucleotide-binding domain, Leu-rich repeat containing (NLR) immune receptors to recognize pathogen effectors delivered into host cells during infection (Spoel and Dong, 2012). NLR activation results in effector-triggered immunity (ETI). A signature of ETI is the hypersensitive response (HR), a form of programmed cell death occurring at the site of infection.
In order to cause disease and suppress host defense responses, gram-negative bacterial pathogens deliver effector proteins into host cells via the type III secretion system (TTSS). Plant pathogenic bacteria deliver a large number (20–40) of effectors into host cells during infection (Cui et al., 2009). Collectively, effectors are required for bacterial virulence (Lindgren et al., 1986). However, knockouts affecting individual effectors frequently have phenotypes that are subtle, likely due to functional redundancy (Cunnac et al., 2011). Alternatively, individual effectors may play an important role in bacterial survival under conditions that are not typically analyzed in the laboratory or act cooperatively with one another. Progress in understanding individual effectors’ contributions to virulence has been made by generating transgenic plants that express effectors. Multiple effectors have been shown to suppress plant innate immunity and promote bacterial growth when either transiently or stably expressed in plants (Jamir et al., 2004; Guo et al., 2009). Effector expression can also result in avirulent phenotypes when a plant NLR receptor recognizes a cognate effector and mounts an HR. Such an HR phenotype can be used to dissect important effector domains required for plant recognition and enzymatic activity.
Elucidating effector targets and enzymatic activity is necessary in order to understand how they act to subvert plant immune responses and can provide elegant insight into biological processes. Significant progress has been made in elucidating the enzymatic activity of a subset of effectors. Some of the most well-characterized effectors come from Pseudomonas syringae pv tomato (Pto), the causal agent of bacterial speck on tomato (Solanum lycopersicum) and Arabidopsis (Arabidopsis thaliana). Multiple effectors can suppress immune responses by directly targeting PAMP receptors (AvrPto and AvrPtoB) or by interfering with downstream signaling processes (AvrB, AvrPphB, and HopAI1; Cui et al., 2009, 2010). The HopU1 effector interferes with RNA metabolism (Fu et al., 2007), and the HopI1 effector targets heat-shock proteins in the plant chloroplast (Jelenska et al., 2010).
14-3-3s are conserved eukaryotic proteins that bind a diverse set of phosphorylated client proteins, typically at one of three distinct 14-3-3 binding motifs (Bridges and Moorhead, 2005). There are common recognition motifs for 14-3-3 proteins that contain phosphorylated Ser or Thr residues, but binding to nonphosphorylated ligands and to proteins lacking consensus motifs has been reported (Henriksson et al., 2002; Smith et al., 2011). The 14-3-3 mode I consensus motif is RXXpS/pTX and that of mode II is RXXXpS/pTXP, where X can be any amino acid and p indicates the site of phosphorylation (Smith et al., 2011). 14-3-3 proteins can also bind to the extreme C termini of proteins at the RXXpS/pTX-COOH mode III consensus motif (Smith et al., 2011). Interaction with 14-3-3s can regulate protein activity by influencing client subcellular localization, structure, and protein-protein interactions (Bridges and Moorhead, 2005). Recently, the Xanthomonas campestris XopN effector was shown to target tomato 14-3-3 isoforms, which facilitates its interaction with the tomato atypical receptor kinase1 and suppresses PTI (Kim et al., 2009; Taylor et al., 2012). Other 14-3-3s have also been shown to play a role during plant defense responses. The tomato TFT7 14-3-3 interacts with multiple mitogen-activated protein kinases to positively regulate HR induced by ETI (Oh and Martin, 2011). The Arabidopsis 14-3-3 isoform λ interacts with the RPW8.2 powdery mildew receptor and is required for complete RPW8.2-mediated resistance (Yang et al., 2009).
In this study, we investigated the function of the Pto HopQ1 (for Hrp outer protein Q [also known as HopQ1-1]) effector in tomato. HopQ1 is an active effector that is transcribed and translocated via the TTSS (Schechter et al., 2004). HopQ1 induces cell death when expressed in Nicotiana benthamiana and therefore contributes to differences in host range in P. syringae pathovars on Nicotiana spp. (Wei et al., 2007; Ferrante et al., 2009). HopQ1 was also reported to slightly enhance disease symptoms (approximately 0.2 log) and bacterial virulence on bean (Phaseolus vulgaris) when expressed from P. syringae pv tabaci (Ferrante et al., 2009). Here, we generated transgenic tomato plants expressing HopQ1 that exhibited enhanced susceptibility to virulent Pto as well as the Pto ΔhrcC mutant. HopQ1-interacting proteins were identified from tomato using coimmunoprecipitations coupled with mass spectrometry. Multiple 14-3-3 proteins were identified. HopQ1 possesses a 14-3-3 binding motif whose Ser residue is phosphorylated in planta and affects its association with the tomato 14-3-3s TFT1 and TFT5. Mutation of HopQ1’s 14-3-3 binding motif affected its ability to promote bacterial virulence. Taken together, these results indicate that phosphorylation and subsequent interaction with tomato 14-3-3 proteins affect HopQ1’s virulence-promoting activities and subcellular localization.
RESULTS
HopQ1 Is a Conserved Bacterial Effector and Enhances Bacterial Virulence in Transgenic Tomato Plants
HopQ1 is widely conserved across multiple species of plant pathogenic bacteria. Homologs possess high sequence similarity across their effector domains (E = 10−145) and can be identified in strains of Pseudomonas spp., Xanthomonas spp., Ralstonia spp., and Acidovorax spp. as well as certain Rhizobium spp. symbionts (Supplemental Fig. S1). HopQ1’s central region possesses some homology to nucleoside hydrolases (amino acids 92–384), and its C terminus contains no homology to proteins of known function. We have purified HopQ1 from a variety of hosts (Escherichia coli, insect cells, and transgenic plants) but were not able to detect nucleoside hydrolase activity or nucleoside binding using standard substrates (data not shown).
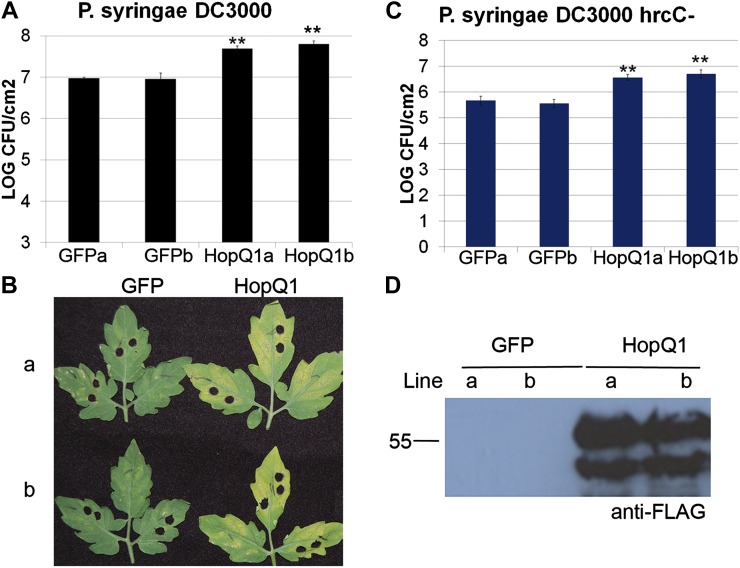
Therefore, we focused on the role of HopQ1’s virulence-promoting activities in tomato. It is often difficult to detect a loss of virulence after deletion of individual type III effectors from virulent P. syringae strains (Collmer et al., 2002). Pto DC3000 hopq1 deletions do not exhibit defects in bacterial virulence on the Arabidopsis ecotype Columbia or the tomato ‘Moneymaker’ cultivar (Wei et al., 2007). In order to identify more robust disease-related phenotypes, we generated transgenic dexamethasone (Dex)-inducible HopQ1 lines with a C-terminal fusion to the 3xFLAG epitope (HopQ1-3xFLAG) in tomato ‘Moneymaker’. Inducing HopQ1 expression by spraying 4-week-old plants with 30 μm Dex did not result in any obvious phenotypic differences in plant growth or health for up to 10 d. When HopQ1 is expressed in plants, both full-length effector and a slightly smaller cleaved version of the effector are detectable by western blot (Fig. 1D). The prevalence and abundance of this smaller cleaved fragment varies depending on plant age, with younger plants (1–3 weeks old) exhibiting more pronounced cleavage (data not shown). In order to test the effect of HopQ1 on bacterial virulence, two homozygous transgenic lines were sprayed with 30 μm Dex to induce HopQ1 expression 24 h before inoculation with Pto DC3000. Transgenic plants expressing inducible GFP were used as the control. Individual transgenic lines expressing HopQ1 exhibited approximately 8- to 10-fold higher Pto DC3000 population sizes than controls (Fig. 1, A and B). These results demonstrate that HopQ1 can act within plant cells to promote bacterial virulence.
Figure 1.
Transgenic tomato plants expressing HopQ1 exhibit enhanced disease susceptibility to Pto. T4 homozygous transgenic tomato plants expressing Dex-inducible HopQ1-3xFLAG or GFP were sprayed with 30 μm Dex 24 h before syringe infiltration with Pto DC3000. A, Growth curve illustrating bacterial population sizes 4 d post inoculation with Pto DC3000 at a concentration of 1 × 105 cfu mL−1. B, Disease symptoms 4 d post inoculation with Pto DC3000. C, Growth curve illustrating bacterial population sizes 4 d post inoculation with Pto DC3000 ΔhrcC at a concentration of 1 × 106 cfu mL−1. For growth curves in A and C, values represent means ± sd (n = 6). The data shown are representative of three independent experiments with similar results. Statistical differences were detected by a two-tailed Student’s t test (α = 0.01). D, Anti-FLAG western blot illustrating HopQ1 protein expression. [See online article for color version of this figure.]
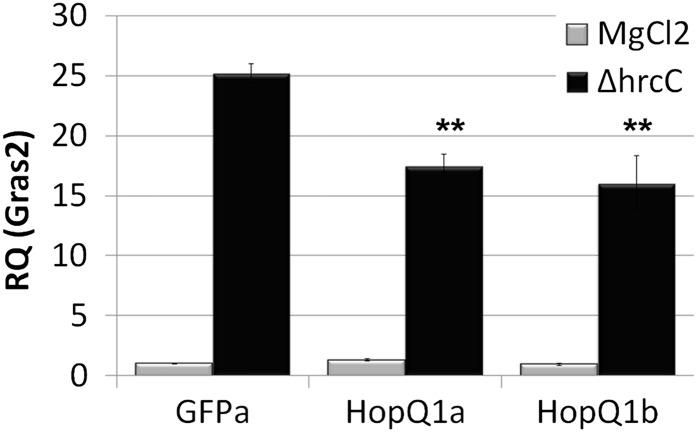
HopQ1-expressing plants were also inoculated with Pto DC3000 ΔhrcC, which is unable to deliver effectors and elicits robust PTI (Collmer et al., 2000). Four days post inoculation, Pto DC3000 ΔhrcC was also able to grow 8- to 10-fold higher on HopQ1-expressing plants compared with controls (Fig. 1C). In order to determine if HopQ1 can suppress PAMP-triggered gene expression in tomato, quantitative real-time PCR was used to analyze the expression of GRAS2. GRAS2 is a transcription factor that has previously been demonstrated to be a marker of PTI in tomato (Kim et al., 2009; Taylor et al., 2012) with links to both biotic and abiotic stress tolerance (Mayrose et al., 2006). In order to monitor changes in gene expression, individual transgenic tomato plants expressing HopQ1 or GFP were vacuum infiltrated with 10 mm MgCl2 or a 2 × 108 colony-forming units (cfu) mL−1 suspension of Pto DC3000 ΔhrcC. Total RNA was isolated from inoculated tissue 6 h post inoculation, and GRAS2 abundance was detected by real-time quantitative reverse transcription (qRT)-PCR (Fig. 2). The expression level of GRAS2 was slightly higher in GFP transgenic plants compared with HopQ1-expressing plants (Fig. 2). Thus, the expression of HopQ1 in planta enhances bacterial virulence.
Figure 2.
HopQ1 suppresses mRNA levels of the GRAS2 marker gene during infection. T4 homozygous transgenic tomato plants expressing Dex-inducible HopQ1-3xFLAG or GFP were sprayed with 30 μm Dex 24 h before syringe infiltration with 2 × 108 cfu mL−1 Pto DC3000 ΔhrcC or 10 mm MgCl2. Total RNA was isolated 6 h post inoculation. qRT-PCR was performed for the GRAS2 PTI marker gene. Actin expression was used to normalize the expression value of each sample. Values represent means ± sd (n = 3). The data shown are representative of three independent experiments with similar results. Statistical differences were detected by a two-tailed Student’s t test (α = 0.01). RQ, Relative quantification.
HopQ1 Interacts with Multiple Tomato 14-3-3 Proteins in a Phosphorylation-Specific Manner
In order to gain insight into HopQ1 function in plants, we investigated components of the HopQ1 protein complex in tomato. Anti-FLAG agarose was used to coimmunoprecipitate HopQ1-3xFLAG-interacting proteins from transgenic cv Moneymaker lines 24 h post Dex application. HopQ1 and associated proteins were eluted with FLAG peptide and subjected to mass spectrometry. Transgenic cv Moneymaker lines expressing Dex-inducible GFP were used as a negative control. Proteins from each sample were analyzed directly using HPLC coupled to tandem mass spectrometry. Proteins were identified using the XTandem algorithm to search the tomato ‘Heinz 1706’ genome (Craig and Beavis, 2004; Tomato Genome Consortium, 2012). Mass spectrometry data identified a large number of spectra matching HopQ1 across three biological replications (Table I; Supplemental Tables S1 and S2). Strikingly, we also identified a number of different tomato 14-3-3 proteins, with spectra specifically matching peptides from the 14-3-3 proteins TFT1 to TFT7, TFT9, and TFT10 (Table I; Supplemental Tables S1 and S2). Of these, unique spectra corresponding to TFT1 and TFT5 were the most abundant.
Table I. HopQ1-interacting proteins identified by mass spectrometry.
T4 homozygous transgenic tomato plants expressing Dex-inducible HopQ1-3xFLAG or GFP were sprayed with 30 μm Dex 24 h before harvesting. One gram of tomato leaf tissue was used for anti-FLAG immunoprecipitations. Values indicate unique spectra for each protein.
| Identified Proteins | Uniprot Identifier | Molecular Mass | HopQ1 (1) | HopQ1 (2) | HopQ1 (3) | GFP (1) | GFP (2) | GFP (3) |
|---|---|---|---|---|---|---|---|---|
| kD | ||||||||
| HopQ1 | Q888Y7 | 49 | 37 | 11 | 20 | 0 | 0 | 0 |
| 14-3-3 protein TFT1 | P93206 | 28 | 7 | 1 | 3 | 0 | 0 | 0 |
| 14-3-3 protein TFT5 | P93210 | 29 | 11 | 2 | 1 | 0 | 0 | 0 |
| 14-3-3 protein TFT4 | P42652 | 29 | 9 | 1 | 1 | 0 | 0 | 0 |
| 14-3-3 protein TFT9 | P93214 | 29 | 6 | 1 | 0 | 0 | 0 | 0 |
| 14-3-3 protein TFT3 | P93209 | 29 | 7 | 0 | 1 | 0 | 0 | 0 |
| 14-3-3 protein TFT6 | P93211 | 29 | 3 | 0 | 0 | 0 | 0 | 0 |
| 14-3-3 protein TFT10 | P93207 | 29 | 4 | 0 | 0 | 0 | 0 | 0 |
| 14-3-3 protein TFT2 | P93208 | 29 | 2 | 0 | 0 | 0 | 0 | 0 |
| 14-3-3 protein TFT7 | P93212 | 29 | 2 | 0 | 0 | 0 | 0 | 0 |
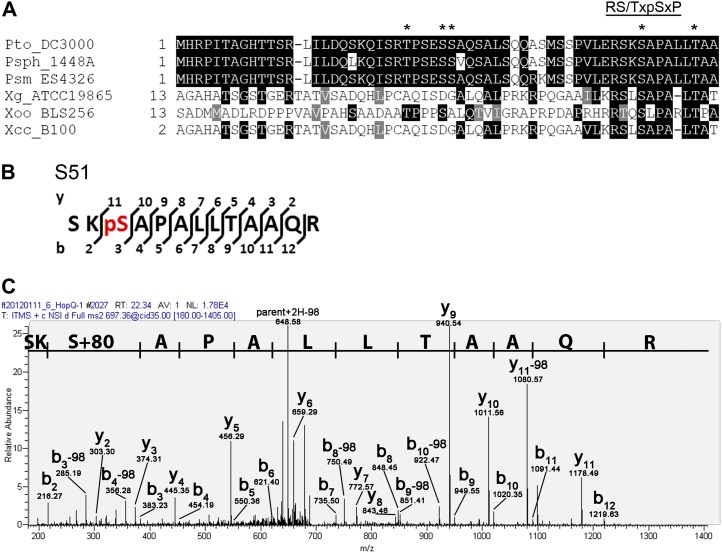
Investigation of HopQ1’s amino acid sequence revealed that it possesses a mode I 14-3-3 binding motif at amino acid residues 48 to 53 (RSKSAP; Fig. 3; Supplemental Fig. S1). Furthermore, HopQ1 homologs present in Pseudomonas spp. and Xanthomonas spp. possess a conserved mode I binding motif at their N termini (Fig. 3). 14-3-3 proteins typically interact with client proteins possessing canonical mode I binding motifs only when these motifs are phosphorylated. In order to determine if HopQ1 is phosphorylated in planta, we performed anti-FLAG pull downs on Dex-inducible HopQ1-3xFLAG transgenic tomato plants. A high-salt wash (300 mm NaCl) was included after binding to anti-FLAG agarose in order to obtain relatively pure protein for mass spectrometry analyses. HopQ1 was eluted from the agarose using competition with excess FLAG peptide, and phosphorylated peptides were enriched using immobilized metal affinity chromatography resin and then subjected to HPLC coupled to tandem mass spectrometry. Raw data were searched with Sequest, and several phosphorylated HopQ1 peptides were identified. Four high-quality tandem mass spectrometry scans unambiguously identified Ser-51 as phosphorylated (Fig. 3, B and C). Additional scans identified other phosphorylated peptides, and Scaffold 3 (Proteome Software) was used to determine the most likely modified amino acid residues within these peptides. Thr-25, Ser-29 or Ser-30, and Thr-57 all appear to be phosphorylated in planta based on this analysis (Supplemental Fig. S2). Examining the conservation of all phosphorylated residues revealed that Ser-51 and Thr-57 are conserved across Pseudomonas spp. and Xanthomonas spp. HopQ1 homologs (Fig. 3A). The Ser-29 residue is also either conserved or substituted with an Asp residue (Fig. 3A). Asp is frequently used as a phosphorylation mimic in mutational analyses because it mimics the negative charge of the phosphate group.
Figure 3.
HopQ1’s 14-3-3 binding motif is widely conserved in homologous effectors and is phosphorylated in tomato. A, Multiple sequence alignment of the N terminus of Pto DC3000 HopQ1 and homologs from Xanthomonas spp. and Pseudomonas spp. Phosphorylated residues of HopQ1 are highlighted with asterisks. The mode I 14-3-3 binding site (RS/TXpSXP) is indicated with a line above the motif. Psph, P. syringae pv phaseolicola 1448A; Psm, P. syringae pv maculicola ES4326; Xg, Xanthomonas gardneri ATCC19865; Xoo, Xanthomonas oryzae pv oryzicola BLS256; Xcc, X. campestris pv campestris B100. B, T4 homozygous transgenic tomato plants expressing Dex-inducible HopQ1-3xFLAG or GFP were sprayed with 30 μm Dex 24 h before harvesting. Two grams of tomato leaf tissue was used for anti-FLAG immunoprecipitations. HopQ1 phosphorylated peptides were identified by mass spectrometry. The y and b ion series are labeled for each spectra, and the phosphorylated amino acids are highlighted in red. C, Manually annotated spectra matching Ser-51 phosphorylation. [See online article for color version of this figure.]
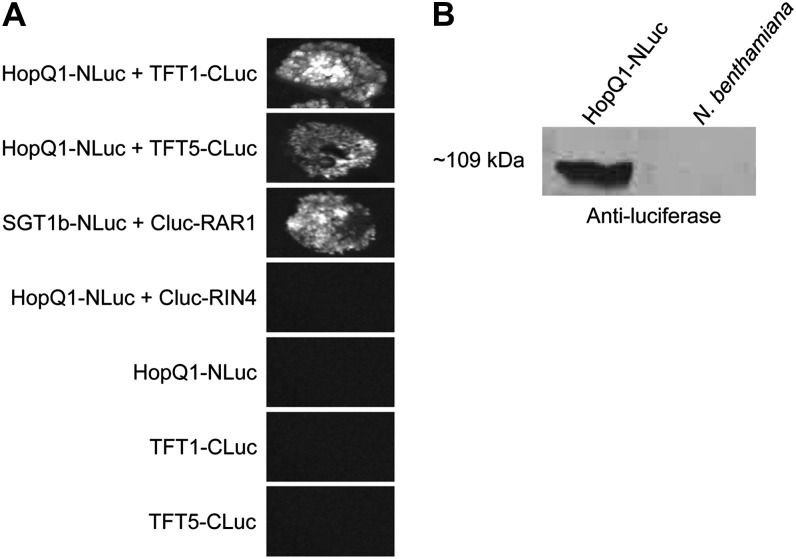
In order to validate that HopQ1 can associate with tomato 14-3-3 proteins, we took advantage of the split-luciferase complementation assay, which enables the detection of bioluminescence if two proteins associate when fused to the N- or C-terminal halves of the firefly luciferase protein (Chen et al., 2008). Using this assay, we could see strong luminescence after coexpressing HopQ1-NLuc with TFT1-CLuc or TFT5-CLuc (Fig. 4). We did not detect luminescence when HopQ1-NLuc was coexpressed with the Arabidopsis RIN4-CLuc protein (Fig. 4). Luminescence was also not detectable when any NLuc- or CLuc-tagged proteins were expressed alone (Fig. 4). These results demonstrate that HopQ1 can associate with both TFT1 and TFT5.
Figure 4.
HopQ1 associates with the tomato 14-3-3 proteins TFT1 and TFT5 using a split-luciferase complementation assay. A, Split-luciferase complementation assay between HopQ1-NLuc, TFT1-CLuc, TFT5-CLuc, and controls. Binary vectors containing split-luciferase constructs were expressed in N. benthamiana using A. tumefaciens-mediated transient expression. SGT1b-Nluc and CLuc-RAR1 as well as HopQ1-NLuc and CLuc-Rin4 were used as positive and negative controls, respectively. Forty hours post inoculation, 1 mm luciferin was infiltrated and the resulting bioluminescence image was captured. B, Western blot demonstrating the expression of HopQ1-NLuc. N. benthamiana leaf tissue was harvested 24 h post inoculation.
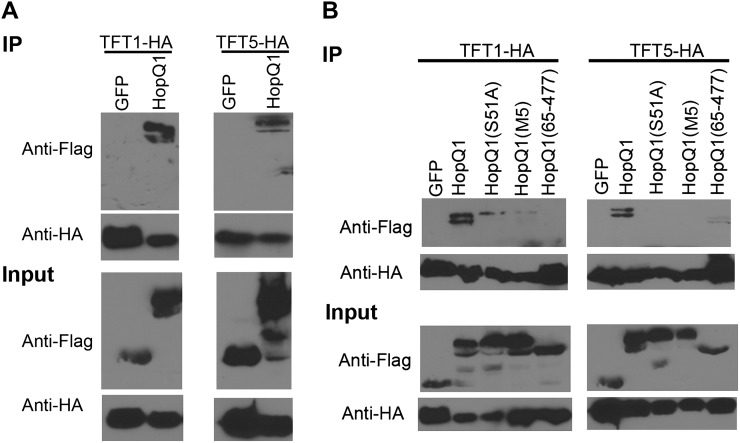
We also verified HopQ1-TFT associations by coimmunoprecipitation in N. benthamiana. We expressed HopQ1-3xFLAG, TFT1-HA (for hemagglutinin), and TFT5-HA in N. benthamiana via Agrobacterium tumefaciens-mediated transient expression. All proteins expressed well in N. benthamiana (Fig. 5). Anti-HA coimmunoprecipitations were used to detect interactions between HopQ1-3xFLAG, TFT1-HA, and TFT5-HA. Both TFT1-HA and TFT5-HA were able to coimmunoprecipitate HopQ1-3xFLAG (Fig. 5A). No interaction was detected between TFT1 or TFT5 and GFP, indicating that this interaction is specific.
Figure 5.
Mutation of HopQ1’s 14-3-3 binding motif affects its association with tomato 14-3-3 proteins. A, HopQ1-3xFLAG and GFP-3xFLAG were transiently expressed with TFT1-HA and TFT5-HA in N. benthamiana using A. tumefaciens-mediated transient expression. Forty hours post inoculation, tissue was harvested and TFT1-HA and TFT5-HA were immunoprecipitated with HA antisera (IP). Associated proteins were detected by immunoblot analyses. B, HopQ1(S51A)-3xFLAG, HopQ1(M5)-3xFLAG, and the HopQ1(65–447) truncation were transiently expressed with TFT1-HA and TFT5-HA in N. benthamiana for immunoprecipitations as described in A.
Ser-51 is located within HopQ1’s 14-3-3 binding motif, and phosphorylation of this Ser residue is predicted to control the association with 14-3-3 proteins. Thus, we mutated Ser-51 to Ala and examined the ability of HopQ1(S51A)-3xFLAG to associate with tomato TFT1-HA and TFT5-HA by coimmunoprecipitation after A. tumefaciens-mediated transient expression in N. benthamiana. Whereas TFT1 and TFT5 were able to strongly coimmunoprecipitate wild-type HopQ1, a very weak interaction was detected with HopQ1(S51A; Fig. 5B). To determine if any of the other phosphorylated residues would impact HopQ1’s association with TFT1 or TFT5, we generated a HopQ1 phosphorylation mutant, where additional residues that were most likely phosphorylated based on mass spectrometry analysis were mutated to Ala (S25A/S29A/S30A/S51A/T57A). This quintuple dephosphorylation mimic is called M5. HopQ1 M5 was unable to associate with TFT5 and had a very weak association with TFT1. An N-terminal truncation of HopQ1 deleting the first 64 amino acids of HopQ1 was unable to associate with TFT1 and had a weak interaction with TFT5 (Fig. 5B). All of the HopQ1 phosphorylation mutants and the HopQ1 N-terminal truncation were expressed in N. benthamiana. Taken together, these data indicate that HopQ1 can interact with 14-3-3 proteins, and the primary determinant of this interaction is through binding HopQ1’s phosphorylated mode I motif.
HopQ1 Phosphorylation Mutants Exhibit Altered Subcellular Localization
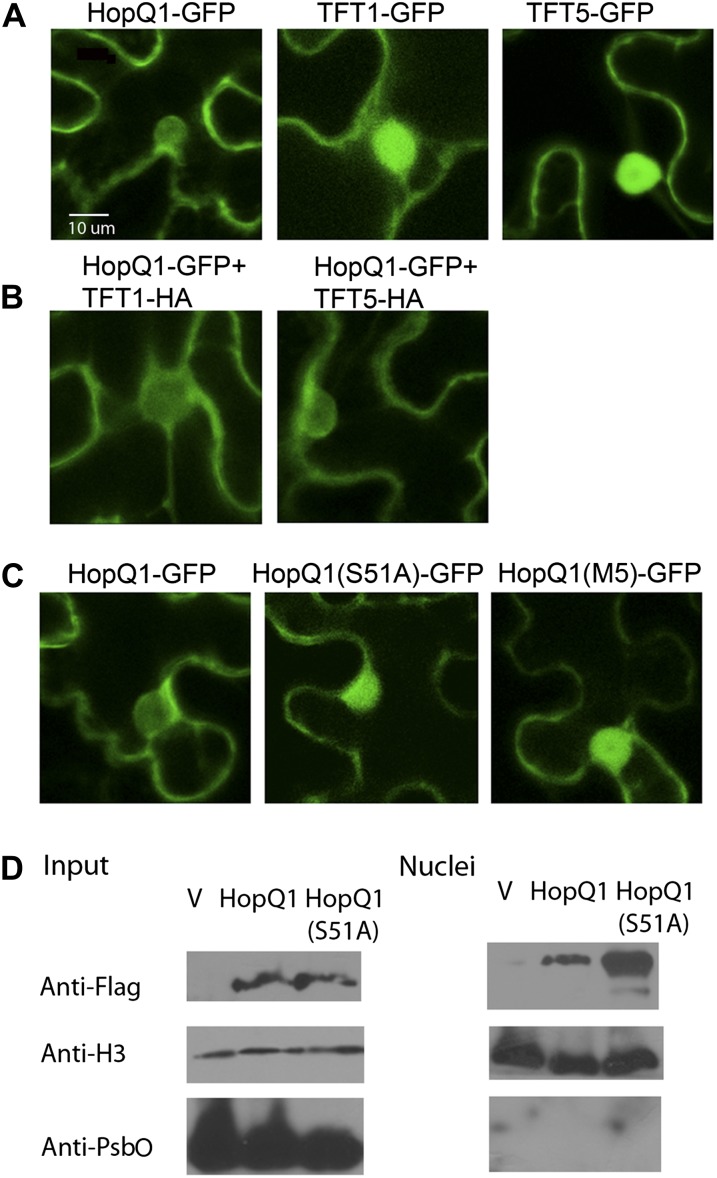
To determine where the interaction between HopQ1 and tomato 14-3-3 proteins occurs, we analyzed their subcellular localization by confocal microscopy. All proteins were transiently expressed in N. benthamiana with a C-terminal GFP fluorescent tag. TFT1 and TFT5 localized to both the nucleus and cytoplasm in epidermal cells (Fig. 6A). This is in agreement with a previous report of yellow fluorescent protein-TFT1 localization to the nucleus and cytoplasm (Kim et al., 2009). HopQ1 primarily exhibited cytoplasmic localization, but a small amount was present in the nucleus as well (Fig. 6A). Examining the amino acid sequence of TFT1, TFT5, and HopQ1 did not reveal any obvious nuclear targeting motif. The predicted molecular mass of the HopQ1-GFP fusion is 76 kD, which is significantly larger than the 40-kD cutoff for passive diffusion through the nuclear pore (Marfori et al., 2011). The molecular mass of TFT1-GFP and TFT5-GFP is 55 and 56 kD, respectively.
Figure 6.
HopQ1’s nucleocytoplasmic localization is influenced by its phosphorylation status. Confocal laser scanning microscopy is shown for N. benthamiana plant leaves transiently expressing HopQ1-GFP and GFP-tagged TFTs. A, HopQ1-GFP, TFT1-GFP, and TFT5-GFP were expressed in N. benthamiana, and confocal images of epidermal cells were taken 48 h post infiltration. B, HopQ1-GFP localization after coexpression with TFT1-HA or TFT5-HA in N. benthamiana. Images were taken as described in A. C, HopQ1(S51A)-GFP and HopQ1(M5)-GFP localization in N. benthamiana epidermal cells. Images were taken as described in A. D, The Pto DC3000 cluster IV deletion transformed with empty pBBR1 vector or pBBR1 expressing HopQ1-3xFLAG or HopQ1(S51A)-3xFLAG was vacuum infiltrated into tomato ‘Moneymaker’ at a concentration of 1 × 108 cfu mL−1. Twelve hours post infiltration, tissue was harvested, nuclei were isolated, and fractions were subjected to anti-FLAG western blotting. Nuclei enrichment was detected by anti-histone H3 western blotting. Nuclei purity was detected by the chloroplast-specific PSII membrane protein PsbO using anti-PsbO western blotting. V, Empty pBBR1 vector.
14-3-3 proteins can affect client protein activity through altering their structure, protein-protein interactions, and subcellular localization. To determine if interaction with TFT1 and TFT5 altered HopQ1’s subcellular localization, we coexpressed TFT1-HA or TFT5-HA with HopQ1-GFP. Coexpression of either TFT1 or TFT5 did not alter the subcellular localization of HopQ1 (Fig. 6B). As 14-3-3 proteins are ubiquitous among eukaryotes, it is likely that HopQ1 is able to interact with endogenous N. benthamiana 14-3-3 proteins, which could mask the effect of coexpression with TFT1 or TFT5. Next, we examined the localization of HopQ1(S51A) and the M5 mutant, which are unable to strongly interact with either TFT1 or TFT5. HopQ1(S51A)-GFP exhibited more pronounced nuclear localization compared with wild-type HopQ1-GFP (Fig. 6C). We also examined the localization of the HopQ1(S51D)-GFP phosphomimetic mutant. The S51D phosphorylation mimic exhibited similar nucleocytoplasmic localization to wild-type HopQ1 (Supplemental Fig. S4). All proteins were expressed in N. benthamiana based on anti-HA and anti-GFP immunoblot analyses (Supplemental Figs. S3 and S4).
It is possible that the altered localization of the S51A mutant may have been influenced by recognition in N. benthamiana. Therefore, we also examined the localization of HopQ1 delivered via the TTSS in the susceptible tomato ‘Moneymaker’ cultivar, which does not recognize any effectors from Pto DC3000. Tomato ‘Moneymaker’ lines were vacuum infiltrated with Pto DC3000 cluster IV polymutant (ΔIV) expressing empty vector, HopQ1-3xFLAG, or HopQ1(S51A)-3xFLAG, and nuclei were purified 12 h post infiltration. HopQ1 and HopQ1(S51A) were expressed at equal levels by anti-FLAG western blotting, but HopQ1(S51A) exhibited enhanced nuclear accumulation in comparison with wild-type HopQ1 (Fig. 6D). These data indicate that HopQ1’s phosphorylation status influences its subcellular localization.
14-3-3 Binding Does Not Affect HopQ1’s Ability to Elicit Cell Death in Nicotiana spp.
HopQ1 is recognized in N. benthamiana, and deletion of HopQ1 from Pto DC3000 enables this bacterium to cause disease on N. benthamiana (Wei et al., 2007). Macroscopic cell death induced by transient expression of HopQ1 in N. benthamiana is variable and is most often detectable by 72 h post inoculation. Thus, we developed A. tumefaciens-mediated transient expression in tobacco (Nicotiana tabacum). HopQ1 elicits a robust cell death 48 h after A. tumefaciens-mediated transient expression in tobacco (Fig. 7A). To examine if HopQ1’s ability to interact with 14-3-3 proteins affects its ability to elicit cell death in tobacco, we examined the phenotype after A. tumefaciens-mediated transient expression of HopQ1(S51A), HopQ1(M5), and HopQ1(65-477). Wild-type HopQ1, HopQ1(S51A), HopQ1(M5), and HopQ1(65-477) all were expressed in tobacco by western-blot analyses (Fig. 7B). Both mutants were able to still elicit robust cell death by 48 h post inoculation (Fig. 7A). HopQ1 and corresponding mutants that were able to elicit cell death in tobacco were also able to elicit cell death in N. benthamiana. Thus, 14-3-3 binding does not affect HopQ1’s ability to elicit cell death in Nicotiana spp.
Figure 7.
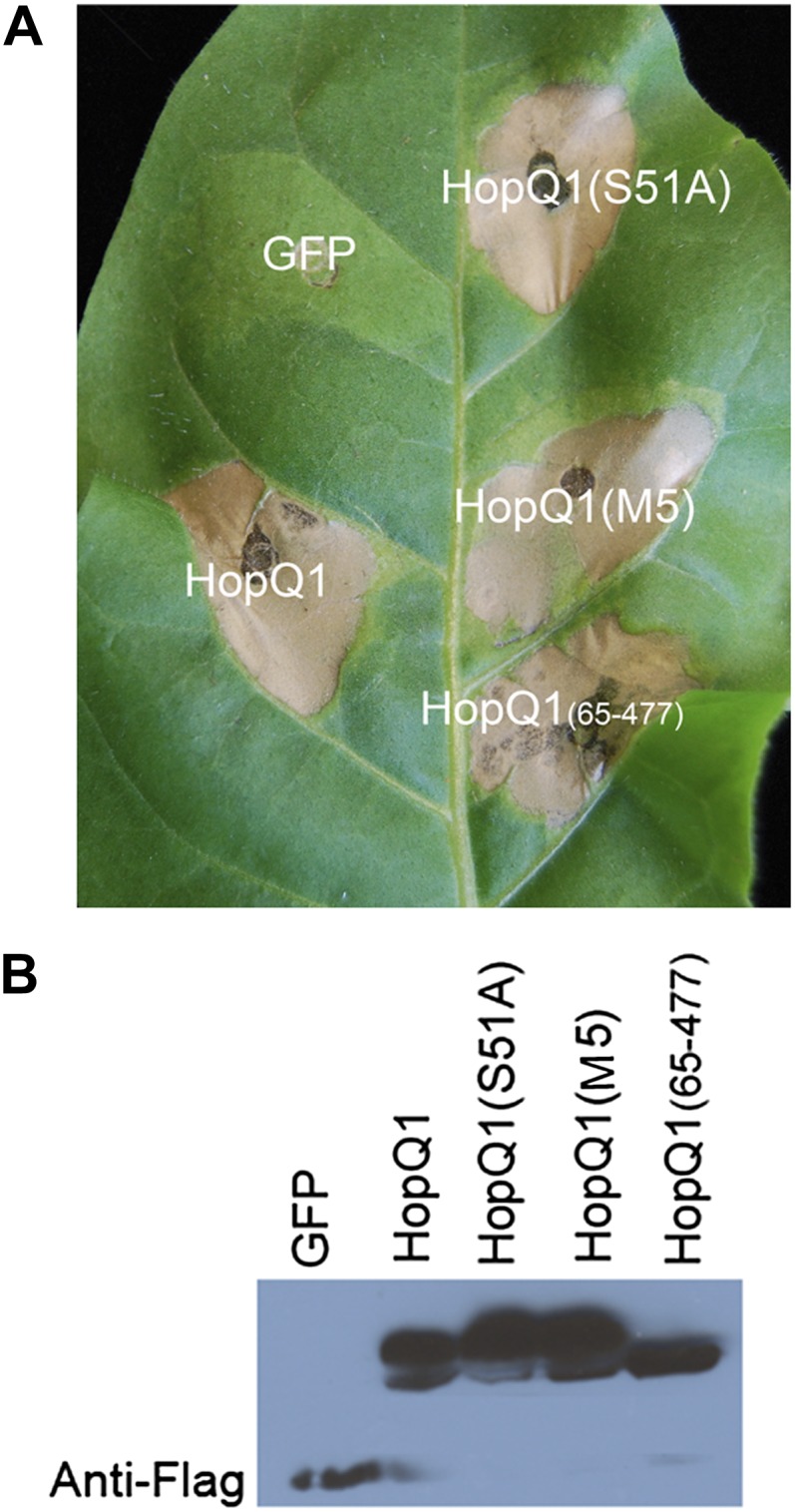
HopQ1’s phosphorylation status does not affect its ability to elicit an HR in tobacco. A, HopQ1 induces an HR in tobacco. Dex-inducible HopQ1-3xFLAG or GFP was expressed in tobacco using A. tumefaciens-mediated transient expression. Thirty micromolars of Dex was applied 24 h post infiltration, and photographs were taken 72 h post infiltration. B, Western blots probed with anti-FLAG showing expression levels of all constructs 40 h post infiltration. [See online article for color version of this figure.]
14-3-3 Binding Affects HopQ1’s Ability to Enhance Bacterial Virulence
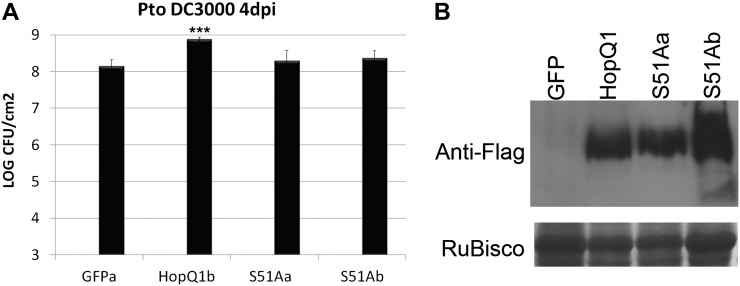
Transgenic plants expressing Dex-inducible HopQ1 exhibit enhanced disease susceptibility to P. syringae (Fig. 1). In order to determine if HopQ1’s interaction with host 14-3-3 proteins affects its ability to promote bacterial virulence, transgenic Dex-inducible HopQ1(S51A) lines in cultivated tomato ‘Moneymaker’ were generated. The growth of Pto DC3000 was compared between transgenic lines expressing GFP, wild-type HopQ1, and HopQ1(S51A). Transgenic lines expressing HopQ1 exhibited approximately 8-fold higher Pto DC3000 population sizes than controls (Fig. 8). Two independent transgenic lines expressing HopQ1(S51A) did not exhibit statistically significant differences in bacterial population sizes compared with the empty-vector control (Fig. 8). Western-blot analyses demonstrated that HopQ1 and HopQ1(S51A) were expressed to similar levels in transgenic tomato (Fig. 8B). Both HopQ1 and HopQ1(S51A) exhibit some cleavage on their N termini in tomato seedlings. However, in 4-week-old plants, only full-length HopQ1 is detectable (Fig. 8B). Thus, HopQ1’s phosphorylation status plays an important role in its virulence-promoting activities.
Figure 8.
The HopQ1(S51A) dephosphorylation mimic cannot promote bacterial virulence in transgenic tomato plants. A, Transgenic tomato ‘Moneymaker’ plants expressing Dex-inducible HopQ1-3xFLAG, HopQ1(S51A)-3xFLAG, or GFP were sprayed with 30 μm Dex 16 h before syringe infiltration with Pto DC3000. The growth curve illustrates bacterial population sizes 4 d post inoculation with Pto DC3000. Values represent means ± sd (n = 6). The data shown are representative of three independent experiments with similar results. Statistical differences were detected by a two-tailed Student’s t test. B, Anti-FLAG western blot illustrating HopQ1 protein expression in all transgenic lines.
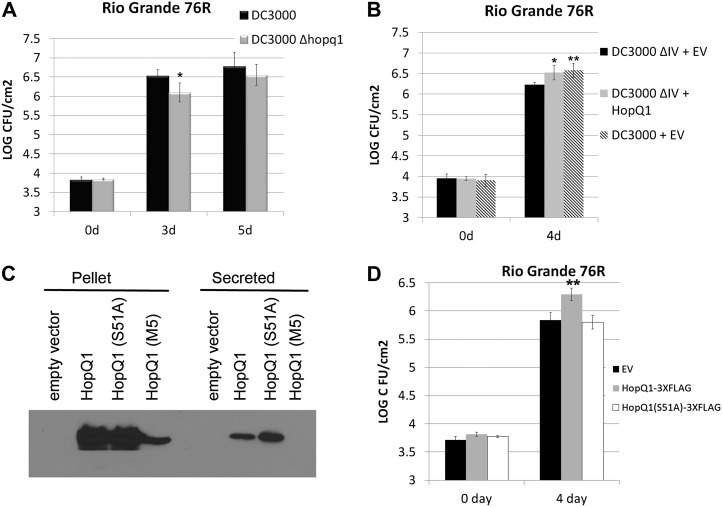
Previously, HopQ1 was reported to have no effect on bacterial virulence after inoculating Pto DC3000 Δhopq1 on tomato ‘Moneymaker’ or Arabidopsis ecotype Columbia with Pto DC3000 (Wei et al., 2007). Therefore, we tested Pto DC3000 Δhopq1 for alterations in bacterial virulence in other tomato genotypes. Tomato ‘Rio Grande 76R’ recognizes the Pto DC3000 effectors AvrPto and AvrPtoB through the protein kinase PTO and the NLR PRF (Salmeron et al., 1996). Pto DC3000 Δhopq1 displayed slightly reduced bacterial growth after inoculation on 76R compared with wild-type Pto DC3000, indicating that HopQ1 can promote bacterial virulence in this tomato line (Fig. 9A). Consistent with these findings, Pto DC3000 ΔIV, lacking the HopQ1, HopD1, and HopR1 effectors (Wei et al., 2007), also exhibited a slight reduction in bacterial growth after inoculation on 76R compared with wild-type Pto DC3000 (Fig. 9B). Furthermore, this decrease in virulence could be complemented by expressing HopQ1 in the Pto DC3000 ΔIV using the pBBR1-MCS5 broad-host-range vector (Fig. 9B). Although the virulence decrease in Pto DC3000 ΔIV is subtle (0.2–0.4 log), we were able to detect a reproducible decrease across multiple replications that could be complemented by expressing HopQ1 (Fig. 9; Supplemental Fig. S5).
Figure 9.
Pto DC3000-delivered HopQ1, but not HopQ1(S51A), can promote bacterial virulence in tomato ‘Rio Grande 76R’. A, The Pto DC3000 Δhopq1 deletion exhibits reduced bacterial virulence during ETI. Tomato ‘Rio Grande 76R’ plants were syringe infiltrated with 1 × 105 cfu mL−1 Pto DC3000 or Pto DC3000 Δhopq1. Growth curves illustrating bacterial population sizes are shown 3 and 5 d post inoculation. B, Expression of HopQ1from the broad-host-range vector pBBR1 can complement the Pto DC3000 cluster IV deletion lacking the HopQ1, HopD1, and HopR1 effectors. Tomato ‘Rio Grande 76R’ plants were syringe infiltrated with 1 × 105 cfu mL−1 bacteria, and growth curves were determined 4 d post inoculation. C, The Pto DC3000 cluster IV deletion transformed with empty pBBR1 vector, or pBBR1 expressing HopQ1-3xFLAG, HopQ1(S51A)-3xFLAG, or HopQ1(M5)-3xFLAG, were grown in hrp-inducing minimal medium for 16 h at 18°C. The resulting bacterial pellet and precipitated secreted proteins were subjected to an anti-FLAG western blot to detect protein expression. D, Expression of HopQ1(S51A) from the broad-host-range vector pBBR1-MCS5 cannot complement the Pto DC3000 cluster IV deletion. Growth curves were conducted as described in B. For all graphs, values represent means ± sd (n = 6). The data shown are representative of three independent experiments with similar results. Statistical differences were detected by a two-tailed Student’s t test. EV, Empty vector.
In order to determine if the genetic background of cv Rio Grande 76R was responsible for the ability to detect a virulence decrease in Pto DC3000 ΔIV, we also determined growth curves on the cv Rio Grande 76S pto mutant (pto11/pto11 Prf/Prf; Salmeron et al., 1994). We were unable to detect a virulence decrease on cv Rio Grande 76S or on the susceptible Moneymaker cultivar (Supplemental Fig. S6). It is possible that HopQ1 may specifically inhibit ETI. Alternatively, the slight virulence decrease of Pto DC3000 ΔIV may be more pronounced when pathogen virulence is decreased. The ability to detect subtle virulence effects using less virulent pathogens has been widely used in Pto DC3000-Arabidopsis research.
Next, we examined if HopQ1(S51A) or M5 could complement Pto DC3000 ΔIV. Wild-type HopQ1, HopQ1(S51A), and HopQ1(M5) were analyzed for their expression and secretion from Pto after induction with hypersensitive response and pathogenicity (hrp)-inducing minimal medium. While wild-type HopQ1 and the S51A mutant were expressed at equal levels and secreted into minimal medium, the M5 mutant was expressed at a lower level (Fig. 9C). Furthermore, we were unable to detect M5 secretion into hrp-inducing minimal medium (Fig. 9C). Therefore, we focused on comparing the virulence contribution of wild-type HopQ1 with the S51A mutant. HopQ1 delivered from Pto DC3000 ΔIV was able to promote bacterial virulence on 76R, while HopQ1(S51A) was not (Fig. 9D; Supplemental Fig. S5). Collectively, these results demonstrate that HopQ1 can promote bacterial virulence and that the phosphorylated Ser-51 residue is crucial for virulence promotion.
DISCUSSION
In this paper, we report the identification of multiple tomato 14-3-3 proteins that can associate with the Pto effector HopQ1. HopQ1 is phosphorylated in planta, and phosphorylation of Ser-51 within its mode I binding motif regulates its ability to promote bacterial virulence and interact with the tomato 14-3-3 proteins TFT1 and TFT5. The data in this paper provide models for the role of HopQ1 during infection.
14-3-3 proteins are highly conserved regulatory eukaryotic protein adapters whose interaction with client proteins can regulate client activity. There are common recognition motifs for 14-3-3 proteins that contain phosphorylated Ser or Thr residues, but binding to nonphosphorylated ligands and to proteins lacking consensus motifs has been reported (Henriksson et al., 2002; Smith et al., 2011). The 14-3-3 mode I consensus motif is RSXpS/pTX and that of mode II is RXF/YXpS/pTXP, where X can be any amino acid and p indicates the site of phosphorylation (Smith et al., 2011). 14-3-3 proteins can also bind to the extreme C termini of proteins at the RXXpS/pTX-COOH mode III consensus motif (Smith et al., 2011).
HopQ1 possesses a conserved mode I motif that is phosphorylated by plant kinase(s) during infection. Mutation of Ser-51 within this mode I motif strongly eliminated or abolished the ability of HopQ1 to interact with TFT1 and TFT5, respectively (Fig. 5). Furthermore, the Ser-51 residue is conserved in HopQ1 homologs in Pseudomonas spp. and Xanthomonas spp., suggesting that phosphorylation and 14-3-3 binding are likely conserved. We were also able to detect two variations potentially matching additional 14-3-3 binding motifs in HopQ1 (residues 24–29 and 73–78; Obenauer et al., 2003). However, we were unable to detect any phosphorylation of Ser-27 or Ser-77 based on our mass spectrometry data (Fig. 3). Furthermore, these Ser residues are not conserved in HopQ1 homologs in Xanthomonas spp. as well as several Pseudomonas spp. homologs (Supplemental Fig. S1). These additional sites may be involved in cooperative binding of 14-3-3 dimers (Bridges and Moorhead, 2005) but are most likely not the primary determinants of the HopQ1-14-3-3 interaction based on our coimmunoprecipitation results.
Although unique spectra matching TFT1 and TFT5 were the most abundant, we were able to detect a number of different 14-3-3 proteins that could associate with HopQ1 by mass spectrometry (Table I). In tomato, there are 12 genes predicted to encode 14-3-3 proteins, which are named in the sequence TFT1 to TFT12 (Xu and Shi, 2006). The tomato 14-3-3 family can be divided into two major groups, the non-ε group (TFT1–TFT6, TFT10, and TFT11) and an ε-like group (TFT7–TFT9 and TFT12; Xu et al., 2012). As TFT1 and TFT5 are both members of the non-ε group, it is possible that isoform specificity exists for the HopQ1 interaction, potentially influenced by TFT1’s and TFT5’s subcellular localization and expression pattern in leaves.
14-3-3 proteins were first implicated in plant-microbe interactions due to the fungal metabolite fusicoccin (Elmore and Coaker, 2011). Fusicoccin is produced by the fungal pathogen Fusicoccum (Phomopsis) amygdale (Ballio et al., 1964). Fusicoccin functions to lock the interaction between 14-3-3 proteins and the C-terminal regulatory domain of the plasma membrane H+-ATPase, leading to constitutive activation of this hydrogen pump and stomatal opening (Jahn et al., 1997; Baunsgaard et al., 1998). More recently, the role of 14-3-3 proteins as effector targets and during plant NLR signaling has been found. The effectors XopN (Kim et al., 2009; Taylor et al., 2012), HopM1 (Nomura et al., 2006), and AvrRxv (Whalen et al., 2008) can associate with host 14-3-3 proteins. Of these effector-14-3-3 associations, XopN was recently demonstrated to bind TFT1 in a phosphorylation-independent manner and target TFT1 to promote pathogen virulence in Xanthomonas spp. (Taylor et al., 2012). Analysis of current Pseudomonas spp. effectors and Xanthomonas spp. effector repertoires using Scansite indicates that a high percentage of effectors possess 14-3-3 binding motifs (data not shown). Thus, it is likely that the targeting of host 14-3-3 proteins and phosphorylation by host kinases are conserved mechanisms employed by multiple effectors to modulate their activity and subcellular localization. Since 14-3-3s bind phosphorylated client proteins, it will be important to determine which host kinase(s) are responsible for effector phosphorylation and elucidate kinase specificity, if any. We were unable to identify any kinases by mass spectrometry, likely because of the transient nature of this interaction coupled with an absence of cross linking. We have clearly demonstrated that HopQ1 is important for bacterial virulence. Moreover, the phosphorylation of HopQ1 is required for promoting bacterial virulence as well as 14-3-3 binding, as the HopQ1(S51A) dephosphorylation mimic is unable to promote bacterial virulence in transgenic tomato plants or after delivery via the TTSS (Figs. 8 and 9; Supplemental Fig. S5).
Transgenic tomato plants expressing HopQ1 exhibited enhanced disease susceptibility to virulent Pto DC3000 as well as the Pto ΔhrcC mutant (Fig. 1). However, we were unable to identify a reproducible virulence phenotype for this effector in Pto Δhopq1 after inoculation of susceptible tomato ‘Moneymaker’ or ‘Rio Grande 76S’ (Supplemental Fig. S6). This result is not surprising, as Pto DC3000 delivers many effectors that can promote bacterial virulence and compromise PTI, commonly resulting in no difference in virulence upon deletion of a single effector (Cunnac et al., 2011). We were able to detect a significant contribution of type III-delivered HopQ1 in cv Rio Grande 76R. Moreover, the virulence contribution of HopQ1 on cv Rio Grande 76R was consistent with the results seen in transgenic plants; HopQ1, but not HopQ1(S51A), can promote bacterial virulence (Figs. 8 and 9; Supplemental Fig. S5). Previous studies have highlighted the role of 14-3-3 proteins during ETI (Konagaya et al., 2004; Yang et al., 2009; Oh et al., 2010; Oh and Martin, 2011). It is possible that HopQ1’s virulence-promoting effect during ETI may be due to interfering with 14-3-3 proteins necessary for robust ETI signaling. Alternatively, subtle virulence effects may be more pronounced after inoculation on cv Rio Grande 76R, as bacterial virulence is decreased in this cultivar. These results highlight the importance of testing for effector virulence promotion on a variety of plant genotypes.
One way in which 14-3-3s can influence signaling is by altering client protein subcellular localization. Using confocal microscopy, we observed that HopQ1-GFP exhibits primarily a cytoplasmic localization pattern, while HopQ1(S51A)-GFP exhibits more pronounced nuclear localization (Fig. 6). Both TFT1 and TFT5 exhibit a nucleocytoplasmic localization pattern. These results indicate that HopQ1 phosphorylation and subsequent 14-3-3 associations regulate this effector’s subcellular localization. HopQ1’s central domain possesses homology to nucleoside hydrolases. However, we have been unable to detect nucleoside hydrolase activity or nucleoside binding using standard substrates with HopQ1 recombinant protein purified from E. coli, insect cells, or after coimmunoprecipitation from plants (data not shown). Thus, it is likely that HopQ1 alters host metabolism to promote pathogen virulence by targeting novel metabolites. Two different models of HopQ1 function are possible. In one model, HopQ1 is delivered into the host cytosol via the TTSS, where it is phosphorylated, and association with host 14-3-3 proteins sequesters it in the cytoplasm, where it acts to promote pathogen virulence by altering plant metabolism. Alternatively, HopQ1 may initially target host nuclei, where it is phosphorylated and then exported into the cytoplasm, possibly in a 14-3-3-dependent manner. Future studies investigating the role of host-induced posttranslational modification and 14-3-3 binding in regulating effector enzymatic activity will significantly advance our understanding of how effectors manipulate their hosts.
MATERIALS AND METHODS
Plasmids and Constructs
HopQ1 site-directed mutants HopQ1(S51A), HopQ1(S51D), and HopQ1(M5) were generated by PCR mutagenesis. The National Center for Biotechnology Information gene identifier for the DC3000 allele of HopQ1 is 1182506. The internal deletion construct HopQ1(65-477) was directly amplified using single-step PCR. All primers used in this study are listed in Supplemental Table S3. All HopQ1 PCR products were cloned into pENTR/D-TOPO (Invitrogen) and then subcloned into respective Gateway vectors.
For inducible expression in tomato (Solanum lycopersicum), HopQ1 and related clones were introduced into the pTA7001 binary vector under the control of a Dex-inducible promoter (Aoyama and Chua, 1997). pTA7001 was modified to be Gateway compatible with a C-terminal 3xFLAG tag. The 3xFLAG amino acid sequence (DYKDHDGDYKDHDIDYKDDDDK) was codon optimized for expression in plants and cloned into the pCR2.1 shuttle vector as a SalI/NotI fragment. The coding sequence for the Gateway recombination cassette (containing the ccdB gene, the CAT chloramphenicol resistance gene, and attR recombination sites) was amplified and cloned in front of the pCR2.1 3xFLAG as an XhoI/SalI fragment. The Gateway cassette:3xFLAG fusion was then cut out of pCR2.1 and ligated into pTA7001 as an XhoI/SpeI fragment to generate pTA7001/des/3xFLAG. For expression in Pseudomonas syringae DC3000, HopQ1 and related clones were introduced into the modified broad-host-range vector pBBR1-MCS5 (Kovach et al., 1995) with a C-terminal 3xFLAG tag under the control of the AvrB promoter. pBRR1-MCS5 was modified to be Gateway compatible with a C-terminal 3xFLAG tag. The Gateway cassette:3xFLAG fusion was then cut out of pCR2.1 and ligated into pBRR1-MCS5 as an XhoI/SpeI fragment to generate pBRR1-MCS5/des/3xFLAG. An area of 256 bp upstream of the translational start codon of avrB was cloned into pENTR/D-TOPO containing HopQ1, and then the insert was transferred to pBRR1-MCS5/des/3xFLAG. For detecting HopQ1 localization in planta, HopQ1 and related mutants were cloned into the binary vector pEarly Gate103, which contains the 35S promoter and a C-terminal fusion to enhanced GFP (Earley et al., 2006).
For split-luciferase complementation experiments, HopQ1, TFT1, TFT5, RIN4 (AT3G25070), SGT1B (AT4G11260), and RAR1 (AT5G51700) were cloned into pDONR (Invitrogen) without stop codons. The resulting clones were then moved into pCAMBIA NLuc and CLuc vectors using Gateway technology (Chen et al., 2008).
TFT1 and TFT5 were amplified from tomato ‘Moneymaker’ complementary DNA (cDNA) and cloned into pENTR/D-TOPO. TFT1 and TFT5 were then subcloned into the binary vectors pEarly Gate103 (Earley et al., 2006) and pMD1 (Tai et al., 1999). The pMD1 destination vector contains the 35S promoter for gene expression and a built-in C-terminal HA tag.
Plant Materials and Growth Conditions
GFP, HopQ1, HopQ1(S51A), HopQ1(M5), and HopQ1(65-477) in the binary vectors pTA7001 and pEarly Gate103 were electroporated into Agrobacterium tumefaciens strain GV3101. Transgenic tomato ‘Moneymaker’ lines expressing Dex-inducible HopQ1-3xFLAG, HopQ1(S51A)-3xFLAG, and GFP were generated at the University of California, Davis, transformation center as described previously (Fillatti et al., 1987). Tomato plants were grown in a greenhouse. Greenhouse growth conditions were 14-h days supplemented with high-intensity sodium lamps, 25°C day temperature, and 60% relative humidity. All experiments were conducted on 5-week-old plants. For transgenic plants, all experiments were conducted on homozygous T3 or T4 lines, with the exception of HopQ1(S51A). Growth curve experiments with HopQ1(S51A) transgenic tomato lines were conducted on clones derived from cuttings from two independent T0 lines. For these experiments, transgenic GFP- and HopQ1-expressing lines were also propagated from cuttings. All experiments were repeated at least three times, with a minimum of three biological replicates per time point. Growth curve experiments had a minimum of six biological replicates per time point.
Bacterial Inoculations and Growth Curve Analyses
Inoculations and bacterial growth curves in tomato were conducted by syringe infiltration. For bacterial growth curves, 5-week-old tomato plants were syringe infiltrated with 1 × 105 cfu mL−1 Pto in 10 mm MgCl2. Growth curves were determined as described previously (Liu et al., 2009). Transgenic tomato plants expressing Dex-inducible HopQ1-3xFLAG or GFP were sprayed with 30 μm Dex containing 0.02% Silwett L-77 using a Preval spray gun 24 h before syringe infiltration with 1 × 105 cfu mL−1 Pto or 1 × 106 cfu mL−1 Pto ΔhrcC in 10 mm MgCl2.
To analyze PAMP-triggered gene expression, transgenic tomato plants expressing Dex-inducible HopQ1-3xFLAG or GFP were sprayed with 30 μm Dex containing 0.02% Silwett L-77 24 h before vacuum infiltration with 2 × 108 cfu mL−1 Pto DC3000 ΔhrcC. Samples for quantitative PCR were collected 6 h post inoculation. To analyze effector expression from Pto DC3000, bacteria were grown overnight in hrp-inducing minimal medium, and proteins were harvested as described previously (Kunkeaw et al., 2010).
RNA Isolation and qRT-PCR
Two leaf discs (9 mm in diameter) were collected to extract total RNA using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. One microgram of total RNA was used to make cDNA (in a total volume of 20 µL) with Moloney murine leukemia virus reverse transcriptase (Promega). Three microliters of the six times diluted cDNA was used for qRT-PCR. qRT-PCR was performed using a CFX96 touch real-time PCR detection system with the SsoFast EvaGreen Supermix kit (Bio-Rad). qRT-PCR conditions consisted of an initial incubation step at 95°C for 30 s, followed by 40 cycles of 10 s of denaturation at 95°C and 15 s of annealing at 60°C. The primers used to amplify GRAS2 were described previously (Kim et al., 2009). Tomato actin was used as an internal control. The resulting quantitative PCR data were analyzed as described previously (Schmittgen and Livak, 2008).
HopQ1 Complex Purification
Dex-inducible HopQ1 and GFP transgenic plants were sprayed with 30 μm Dex containing 0.02% Silwett L-77. Leaf tissue was harvested for protein complex purification 24 h post Dex application. All steps were carried out on ice or at 4°C. Two grams of leaf sample was ground in liquid nitrogen and resuspended in 2 mL of immunoprecipitation (IP) buffer (50 mm HEPES, 50 mm NaCl, 10 mm EDTA, and 0.2% Triton X-100, pH 7.5). The supernatant was incubated with 30 µL of anti-FLAG M2 affinity agarose (Sigma) for 3 h. Immunocomplexes were washed three times with IP buffer and eluted in 3 × 50 µL of 3xFLAG peptide at a concentration of 500 µg mL−1 (Sigma) in IP buffer. The eluted proteins were concentrated to a final volume of 30 µL with StrataClean resin (Stratagene) and loaded onto a single lane on a 10% SDS-PAGE gel. Proteins were run 5 mm into the separating gel and stained with colloidal Coomassie blue (Novex). Trypsin digestions and mass spectrometry were conducted as described previously (Liu et al., 2011).
For the identification of HopQ1 phosphorylation sites, HopQ1 expression was induced by Dex application as described above. Five grams of leaf tissue was ground in 4 mL of IP buffer and incubated with 30 µL of anti-FLAG M2 affinity agarose (Sigma) for 3 h. Immunocomplexes were washed with high salt (50 mm HEPES, 300 mm NaCl, 10 mm EDTA, and 0.2% Triton X-100, pH 7.5), eluted with 3xFLAG peptide, and concentrated with StrataClean resin as described above. Samples were run on a 10% SDS-PAGE gel, stained with colloidal Coomassie blue (Novex), and the HopQ1 band was excised from the gel, and phosphopeptide mapping using mass spectrometry was conducted as described previously (Liu et al., 2011).
A. tumefaciens-Mediated Transient Expression
A. tumefaciens-mediated transient expression in Nicotiana benthamiana and tobacco (Nicotiana tabacum) was performed as described previously (Leister et al., 2005). Agrobacteria were infiltrated into tobacco leaves at an optical density at 600 nm (OD600) = 0.4. Cell-death phenotypes in tobacco were recorded 72 h post inoculation. For subcellular localization experiments, HopQ1 and HopQ1-related clones TFT1 and TFT5 were expressed from the binary vector pEarly Gate103 in N. benthamiana via A. tumefaciens-mediated transient expression.
Coimmunoprecipitation
HopQ1-3xFLAG and its derivatives were expressed from the pTA7001 binary vector. TFT1-HA and TFT5-HA were expressed from the pMD1 binary vector. Genes were expressed in N. benthamiana by A. tumefaciens-mediated transient expression. GFP-FLAG expressed from pTA7001 was used as a negative control. Twenty-four hours post infiltration, 30 µm Dex was applied to induce HopQ1-3xFLAG expression. Samples were collected 16 h post Dex application. Two grams of leaf tissue was collected for protein extraction. Samples were ground in 1 mL of extraction buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.1% Triton X-100, 0.2% Nonidet P-40, and 1× plant protein protease inhibitor [Roche]). Protein extract was incubated at 4°C for 3 h with 10 µL of anti-HA agarose beads (Sigma). After washing with IP buffer three times, beads were resuspended in 100 μL of Laemmli buffer and boiled to release bound proteins.
Split-Luciferase Complementation Assay
All genes were cloned into the pCAMBIA NLuc and CLuc vectors, creating fusions to the amino and carboxy halves of firefly luciferase using Gateway cloning (Chen et al., 2008). The split-luciferase complementation assay was performed as described previously (Chen et al., 2008). pCAMBIA vectors were transformed into A. tumefaciens strain C58C1. A. tumefaciens suspensions containing the respective constructs (OD600 = 0.4) were coinfiltrated into N. benthamiana leaves. Forty hours post infiltration, 1 mm luciferin was infiltrated into each leaf, and the bioluminescence image was captured on a Kodak Image Station 4000R PRO (Carestream Molecular Imaging).
Confocal Microscopy
HopQ1 and HopQ1 mutants TFT1 and TFT5 with C-terminal fusions to enhanced GFP were syringed into N. benthamiana via A. tumefaciens-mediated transient expression at OD600 = 0.4. Forty to 48 h post infiltration, samples were observed with a Leica TCS SP2/MP confocal laser-scanning microscope with a 63× 0.7 numerical aperture. GFP was excited at 500 nm, and fluorescent emissions were measured at 525 nm.
Nuclear Isolation
Tomato ‘Moneymaker’ plants were vacuum infiltrated with 1 × 108 cfu mL−1 Pto ΔIV carrying empty pBRR1-MCS5 vector, pBBR1 expressing HopQ1-3xFLAG, or pBBR1 expressing HopQ1(S51A)-3xFLAG. Twenty grams of tomato leaves was collected for each sample 12 h post inoculation. Nuclei were immediately isolated as described previously (Craig and Beavis, 2004).
Western Blotting
SDS-PAGE and subsequent immunoblotting were performed as described previously (Liu et al., 2011). FLAG immunoblots were performed with monoclonal anti-FLAG conjugated to horseradish peroxidase (HRP; Sigma) at a concentration of 1:1,000. HA immunoblots were performed with anti-HA-HRP (Roche) at a concentration of 1:1,000. GFP immunoblots were performed with anti-GFP-HRP (Miltenyi Biotec) at a concentration of 1:2,000. Anti-histone H3 immunoblots were performed with anti-histone H3 antibody conjugated to HRP (Abcam) at a concentration of 1:1,000. Immunoblots detecting the chloroplast PSII subunit PsbO were performed with rabbit polyclonal anti-PsbO (Abcam) at a concentration of 1:1,000. Goat anti-rabbit IgG-HRP conjugate (Bio-Rad) was used at a concentration of 1:3,000 for the detection of PsbO via enhanced chemiluminescence. Luciferase western blotting was performed using anti-luciferase (Sigma) at a concentration of 1:5,000.
Sequence data from this article can be found in the GenBank data libraries under accession numbers. 1182506 (HopQ1), 543565 (TFT1), X95903.1 (TFT5), 100301918 (GRAS2), 822098 (RIN4), 826728 (SGT1b), and AF192262.1 (RAR1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. ClustalW alignment of HopQ1 and homologs from phytopathogenic bacteria.
Supplemental Figure S2. Spectra of phosphorylated HopQ1 peptides.
Supplemental Figure S3. Western blot of GFP- and HA-tagged proteins used in confocal microscopy.
Supplemental Figure S4. HopQ1(S551D)-GFP and HopQ1-GFP both exhibit nuclear-cytoplasmic localization.
Supplemental Figure S5. HopQ1 can reproducibly complement the Pto DC3000 ΔIV on tomato ‘Rio Grande 76R’.
Supplemental Figure S6. No virulence effect for HopQ1 or HopQ1(S51A) is detected after delivery from Pto DC3000 ΔIV on the susceptible cultivars Moneymaker or Rio Grande 76S.
Supplemental Table S1. Complete list of proteins identified by mass spectrometry with number of unique spectra.
Supplemental Table S2. Complete list of proteins identified by mass spectrometry with number of total spectra.
Supplemental Table S3. Primers used for cloning.
Acknowledgments
All mass spectrometry was performed at the University of California, Davis, Genome Center Proteomics Core Facility. We thank Brett Phinney for use of the mass spectrometry equipment. We thank DongHyuk Lee for cloning HopQ1 into pBBR1-MCS5. We thank Kentaro Inoue for the kind gift of PsbO antiserum.
Glossary
- PAMP
pathogen-associated molecular pattern
- PTI
PAMP-triggered immunity
- NLR
nucleotide-binding domain, leucine-rich repeat containing
- ETI
effector-triggered immunity
- HR
hypersensitive response
- TTSS
type III secretion system
- Pto
Pseudomonas syringae pv tomato
- Dex
dexamethasone
- cfu
colony-forming units
- qRT
quantitative reverse transcription
- HA
hemagglutinin
- ΔIV
cluster IV polymutant
- OD600
optical density at 600 nm
- cDNA
complementary DNA
- IP
immunoprecipitation
- PTI
pattern-triggered immunity
Footnotes
This work was supported by the U.S. Department of Agriculture (grant no. 2010–65108–20527 to G.C.) and by the National Science Foundation (Integrative Graduate Education and Research Traineeship Program graduate research training grant no. DGE–0653984 to J.M.E.).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Aoyama T, Chua NH. (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Ballio A, Chain EB, De Leo P, Erlanger BF, Mauri M, Tonolo A. (1964) Fusicoccin: a new wilting toxin produced by Fusicoccum amygdali Del. Nature 203: 297 [Google Scholar]
- Baunsgaard L, Fuglsang AT, Jahn T, Korthout HA, de Boer AH, Palmgren MG. (1998) The 14-3-3 proteins associate with the plant plasma membrane H(+)-ATPase to generate a fusicoccin binding complex and a fusicoccin responsive system. Plant J 13: 661–671 [DOI] [PubMed] [Google Scholar]
- Bridges D, Moorhead GB. (2005) 14-3-3 proteins: a number of functions for a numbered protein. Sci STKE 296: re10. [DOI] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM. (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collmer A, Badel JL, Charkowski AO, Deng WL, Fouts DE, Ramos AR, Rehm AH, Anderson DM, Schneewind O, van Dijk K, et al (2000) Pseudomonas syringae Hrp type III secretion system and effector proteins. Proc Natl Acad Sci USA 97: 8770–8777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collmer A, Lindeberg M, Petnicki-Ocwieja T, Schneider DJ, Alfano JR. (2002) Genomic mining type III secretion system effectors in Pseudomonas syringae yields new picks for all TTSS prospectors. Trends Microbiol 10: 462–469 [DOI] [PubMed] [Google Scholar]
- Craig R, Beavis RC. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20: 1466–1467 [DOI] [PubMed] [Google Scholar]
- Cui H, Wang Y, Xue L, Chu J, Yan C, Fu J, Chen M, Innes RW, Zhou JM. (2010) Pseudomonas syringae effector protein AvrB perturbs Arabidopsis hormone signaling by activating MAP kinase 4. Cell Host Microbe 7: 164–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Xiang T, Zhou JM. (2009) Plant immunity: a lesson from pathogenic bacterial effector proteins. Cell Microbiol 11: 1453–1461 [DOI] [PubMed] [Google Scholar]
- Cunnac S, Chakravarthy S, Kvitko BH, Russell AB, Martin GB, Collmer A. (2011) Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae. Proc Natl Acad Sci USA 108: 2975–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Elmore JM, Coaker G. (2011) The role of the plasma membrane H+-ATPase in plant-microbe interactions. Mol Plant 4: 416–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante P, Clarke CR, Cavanaugh KA, Michelmore RW, Buonaurio R, Vinatzer BA. (2009) Contributions of the effector gene hopQ1-1 to differences in host range between Pseudomonas syringae pv. phaseolicola and P. syringae pv. tabaci. Mol Plant Pathol 10: 837–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillatti JJ, Kiser J, Rose R, Comai L. (1987) Efficient transfer of a glyphosate tolerance gene into tomato using a binary Agrobacterium tumefaciens vector. Nat Biotechnol 5: 726–730 [Google Scholar]
- Fu ZQ, Guo M, Jeong BR, Tian F, Elthon TE, Cerny RL, Staiger D, Alfano JR. (2007) A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature 447: 284–288 [DOI] [PubMed] [Google Scholar]
- Guo M, Tian F, Wamboldt Y, Alfano JR. (2009) The majority of the type III effector inventory of Pseudomonas syringae pv. tomato DC3000 can suppress plant immunity. Mol Plant Microbe Interact 22: 1069–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson ML, Francis MS, Peden A, Aili M, Stefansson K, Palmer R, Aitken A, Hallberg B. (2002) A nonphosphorylated 14-3-3 binding motif on exoenzyme S that is functional in vivo. Eur J Biochem 269: 4921–4929 [DOI] [PubMed] [Google Scholar]
- Jahn T, Fuglsang AT, Olsson A, Brüntrup IM, Collinge DB, Volkmann D, Sommarin M, Palmgren MG, Larsson C. (1997) The 14-3-3 protein interacts directly with the C-terminal region of the plant plasma membrane H+-ATPase. Plant Cell 9: 1805–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamir Y, Guo M, Oh HS, Petnicki-Ocwieja T, Chen S, Tang X, Dickman MB, Collmer A, Alfano JR. (2004) Identification of Pseudomonas syringae type III effectors that can suppress programmed cell death in plants and yeast. Plant J 37: 554–565 [DOI] [PubMed] [Google Scholar]
- Jelenska J, van Hal JA, Greenberg JT. (2010) Pseudomonas syringae hijacks plant stress chaperone machinery for virulence. Proc Natl Acad Sci USA 107: 13177–13182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-G, Li X, Roden JA, Taylor KW, Aakre CD, Su B, Lalonde S, Kirik A, Chen Y, Baranage G, et al. (2009) Xanthomonas T3S effector XopN suppresses PAMP-triggered immunity and interacts with a tomato atypical receptor-like kinase and TFT1. Plant Cell 21: 1305–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konagaya K-i, Matsushita Y, Kasahara M, Nyunoya H. (2004) Members of 14-3-3 protein isoforms interacting with the resistance gene product N and the elicitor of Tobacco mosaic virus. J Gen Plant Pathol 70: 221–231 [Google Scholar]
- Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, II, Peterson KM. (1995) Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166: 175–176 [DOI] [PubMed] [Google Scholar]
- Kunkeaw S, Tan S, Coaker G. (2010) Molecular and evolutionary analyses of Pseudomonas syringae pv. tomato race 1. Mol Plant Microbe Interact 23: 415–424 [DOI] [PubMed] [Google Scholar]
- Leister RT, Dahlbeck D, Day B, Li Y, Chesnokova O, Staskawicz BJ. (2005) Molecular genetic evidence for the role of SGT1 in the intramolecular complementation of Bs2 protein activity in Nicotiana benthamiana. Plant Cell 17: 1268–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren PB, Peet RC, Panopoulos NJ. (1986) Gene cluster of Pseudomonas syringae pv. “phaseolicola” controls pathogenicity of bean plants and hypersensitivity of nonhost plants. J Bacteriol 168: 512–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Fuglsang AT, Palmgren MG, Staskawicz BJ, Coaker G. (2009) RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol 7: e1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Lin ZJ, Coaker G. (2011) A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 9: 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfori M, Mynott A, Ellis JJ, Mehdi AM, Saunders NF, Curmi PM, Forwood JK, Bodén M, Kobe B. (2011) Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim Biophys Acta 1813: 1562–1577 [DOI] [PubMed] [Google Scholar]
- Mayrose M, Ekengren SK, Melech-Bonfil S, Martin GB, Sessa G. (2006) A novel link between tomato GRAS genes, plant disease resistance and mechanical stress response. Mol Plant Pathol 7: 593–604 [DOI] [PubMed] [Google Scholar]
- Nomura K, Debroy S, Lee YH, Pumplin N, Jones J, He SY. (2006) A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313: 220–223 [DOI] [PubMed] [Google Scholar]
- Obenauer JC, Cantley LC, Yaffe MB. (2003) Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res 31: 3635–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh CS, Martin GB. (2011) Tomato 14-3-3 protein TFT7 interacts with a MAP kinase kinase to regulate immunity-associated programmed cell death mediated by diverse disease resistance proteins. J Biol Chem 286: 14129–14136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh CS, Pedley KF, Martin GB. (2010) Tomato 14-3-3 protein 7 positively regulates immunity-associated programmed cell death by enhancing protein abundance and signaling ability of MAPKKK α. Plant Cell 22: 260–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron JM, Barker SJ, Carland FM, Mehta AY, Staskawicz BJ. (1994) Tomato mutants altered in bacterial disease resistance provide evidence for a new locus controlling pathogen recognition. Plant Cell 6: 511–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron JM, Oldroyd GE, Rommens CM, Scofield SR, Kim HS, Lavelle DT, Dahlbeck D, Staskawicz BJ. (1996) Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell 86: 123–133 [DOI] [PubMed] [Google Scholar]
- Schechter LM, Roberts KA, Jamir Y, Alfano JR, Collmer A. (2004) Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J Bacteriol 186: 543–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Smith AJ, Daut J, Schwappach B. (2011) Membrane proteins as 14-3-3 clients in functional regulation and intracellular transport. Physiology (Bethesda) 26: 181–191 [DOI] [PubMed] [Google Scholar]
- Spoel SH, Dong X. (2012) How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol 12: 89–100 [DOI] [PubMed] [Google Scholar]
- Tai TH, Dahlbeck D, Clark ET, Gajiwala P, Pasion R, Whalen MC, Stall RE, Staskawicz BJ. (1999) Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc Natl Acad Sci USA 96: 14153–14158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KW, Kim JG, Su XB, Aakre CD, Roden JA, Adams CM, Mudgett MB. (2012) Tomato TFT1 is required for PAMP-triggered immunity and mutations that prevent T3S effector XopN from binding to TFT1 attenuate Xanthomonas virulence. PLoS Pathog 8: e1002768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CF, Kvitko BH, Shimizu R, Crabill E, Alfano JR, Lin NC, Martin GB, Huang HC, Collmer A. (2007) A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana. Plant J 51: 32–46 [DOI] [PubMed] [Google Scholar]
- Whalen MC, Richter T, Zakhareyvich K, Yoshikawa M, Al-Azzeh D, Adefioye A, Spicer G, Mendoza LL, Morales CQ, Klassen V, et al. (2008) Identification of a host 14-3-3 protein that interacts with Xanthomonas effector AvrRxv. Physiol Mol Plant Pathol 72: 46–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Shi W, Jia L, Liang J, Zhang J. (2012) TFT6 and TFT7, two different members of tomato 14-3-3 gene family, play distinct roles in plant adaption to low phosphorus stress. Plant Cell Environ 35: 1393–1406 [DOI] [PubMed] [Google Scholar]
- Xu WF, Shi WM. (2006) Expression profiling of the 14-3-3 gene family in response to salt stress and potassium and iron deficiencies in young tomato (Solanum lycopersicum) roots: analysis by real-time RT-PCR. Ann Bot (Lond) 98: 965–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Wang W, Coleman M, Orgil U, Feng J, Ma X, Ferl R, Turner JG, Xiao S. (2009) Arabidopsis 14-3-3 lambda is a positive regulator of RPW8-mediated disease resistance. Plant J 60: 539–550 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]