This study by Wong et al. investigates tumor progression in mouse skin and uncovers a previously undiscovered pathway leading to the epithelial–mesenchymal transition (EMT). The authors identify two types of epidermal tumors and show that separate pathways—inflammation and Hras signaling—can generate invasive and metastatic cancers in vivo. This study suggests that inhibition of inflammation and Hras pathways may select for invasive tumors that have gone down an alternative pathway leading to EMT and metastasis.
Keywords: EMT, Hras, inflammation, skin carcinogenesis, stem cells
Abstract
Epithelial–mesenchymal transition (EMT) is thought to be an important, possibly essential, component of the process of tumor dissemination and metastasis. About 20%–30% of Hras mutant mouse skin carcinomas induced by chemical initiation/promotion protocols have undergone EMT. Reduced exposure to TPA-induced chronic inflammation causes a dramatic reduction in classical papillomas and squamous cell carcinomas (SCCs), but the mice still develop highly invasive carcinomas with EMT properties, reduced levels of Hras and Egfr signaling, and frequent Ink4/Arf deletions. Deletion of Hras from the mouse germline also leads to a strong reduction in squamous tumor development, but tumors now acquire activating Kras mutations and exhibit more aggressive metastatic properties. We propose that invasive carcinomas can arise by different genetic and biological routes dependent on exposure to chronic inflammation and possibly from different target cell populations within the skin. Our data have implications for the use of inhibitors of inflammation or of Ras/Egfr pathway signaling for prevention or treatment of invasive cancers.
The propensity of tumor cells to undergo widespread dissemination to distant sites is the major cause of cancer mortality. Consequently, this process has been extensively studied, but the exact nature of the events that may contribute to, or are essential for, metastasis remains controversial. Epithelial–mesenchymal transition (EMT) has been variously proposed to be a necessary prerequisite for metastasis or to simply promote local invasion without playing a major causal role in tumor spread to secondary sites (Cardiff 2010; Chaffer and Weinberg 2011). In particular, much of what we have learned about the process of EMT has come from elegant studies of tumor cell lines, while the characterization of the pathways leading to EMT in vivo is much less clear. Cells that have undergone EMT can clearly be seen in vitro, but most human metastases derived from epithelial primary tumors display morphologic and immunophenotypic characteristics that are similar to the original tumor and rarely display mesenchymal morphology. This has been interpreted as evidence that EMT either is not required for metastasis or is reversible and is followed by mesenchymal–epithelial transition (MET) after dissemination (Thiery et al. 2009; Chaffer and Weinberg 2011).
The clinical importance of metastasis has prompted many investigations into the mechanisms of this transition. Studies of cell lines and mouse models of cancer have demonstrated an important role for the TGFβ signaling pathway in promoting EMT, leading to increased tumor invasion (Hosobuchi and Stampfer 1989; Caulin et al. 1995; Cui et al. 1996; Derynck and Akhurst 2007; Padua and Massague 2009; Thiery et al. 2009). Causal roles in promoting EMT have been proposed for Ras (Oft et al. 1996) and p53 mutations (Adorno et al. 2009) in addition to a range of transcription factors—including Smads, Twist, Snail (Snai1), Slug (Snai2), and Zeb1/2—that either activate or repress components of the EMT pathway (Peinado et al. 2007). Parallels have also been drawn between the properties of tissue stem cells and cells that have undergone EMT, due to co-expression of several putative stem cell markers (Polyak and Weinberg 2009; Chaffer and Weinberg 2011). A major unanswered question in cancer biology is whether tumors with some characteristics of stem cells arise from a normal stem cell population (Bonnet and Dick 1997) or whether these properties are acquired due to accumulation of genetic events leading to a process similar to EMT in vivo. The latter scenario has been supported by evidence linking changes in the microenvironment—particularly induction of inflammatory cytokines—with expression of EMT and stem cell markers (Gonzalez-Suarez et al. 2010; Schramek et al. 2010). How these processes are coordinated and whether there is heterogeneity in the cellular and molecular routes to EMT that depend on the presence of prior genetic events have not been clarified.
Mouse models of cancer have been crucial for our understanding of the causal genetic and biological events in the initiation, promotion, and progression of tumors. Studies of skin models demonstrated an important promoting role for inflammatory agents in squamous cell carcinoma (SCC) development (Viaje et al. 1977). Similar conclusions were reached using other models of human cancer, including the colon (Tanaka et al. 2003) and pancreas (Guerra et al. 2011). Collectively, these observations have led to strategies targeting inflammation for cancer prevention or treatment (for review, see Balkwill and Mantovani 2012).
Skin carcinogenesis is presumed to proceed in a linear fashion through distinct stages from inflammation-dependent benign papillomas to malignant carcinomas, some of which undergo EMT and progress to undifferentiated spindle cell carcinomas (Klein-Szanto 1989). Mutation analysis has demonstrated that squamous and spindle carcinomas arising through this classical pathway (henceforth class A carcinomas) are clonally related (Burns et al. 1991) and that this transition can be driven by elevated Ras and Tgfβ signaling (Cui et al. 1996; Oft et al. 2002). We demonstrate here that most malignant carcinomas with EMT-like characteristics (henceforth class B carcinomas) arise by a route that is distinct from the classical pathway, as they show reduced dependence on the duration of exposure to the tumor promoter TPA and on the presence of an intact functional Hras gene. We propose that these tumors may arise from a target cell that does not require such extensive reprogramming by inflammatory and tumor-promoting stimuli.
Results
Gene expression analysis of mouse skin tumors
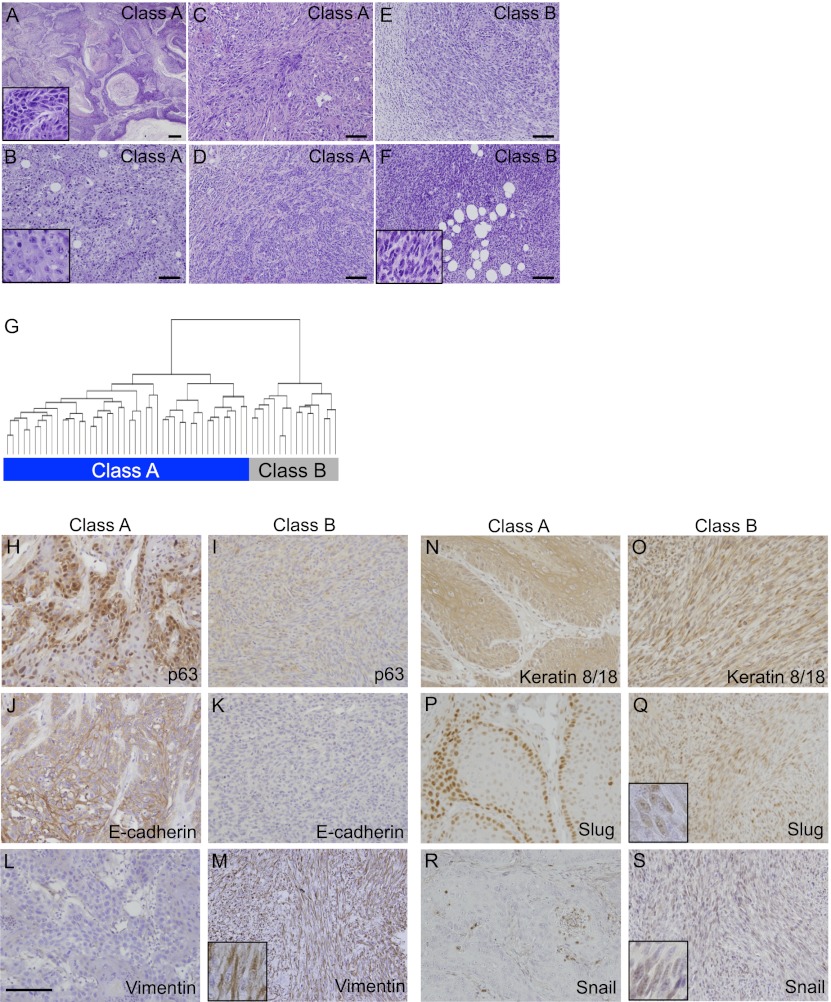
We previously described a strategy involving crosses between different strains and species of mice to recapitulate the genetic heterogeneity that is more typically seen in outbred mouse and human populations (Nagase et al. 1995). Histological analysis of 60 carcinomas from an interspecific [SPRET/Ei × FVB/N] × FVB/N backcross population (FVBBX) demonstrated a range of morphologies from well to poorly differentiated SCCs and spindle cell carcinomas with differing proportions of SCC nests within the tumors (Fig. 1A–F). Unsupervised clustering of the gene expression profiles generated from these carcinomas (Quigley et al. 2011) identified two distinct molecular categories (Fig. 1G). The class A carcinomas included the pure SCCs and those with a smaller but variable spindle component. Class B carcinomas mainly comprised the pure spindle cell carcinomas and those tumors with only small numbers of neoplastic cells with squamous/epithelial morphology.
Figure 1.
FVB/Spret backcross carcinomas can be separated into two different categories. (A) Low-power representative view of well-differentiated SCC demonstrating invasive nests of squamous cells showing heavy keratinization and surrounded by a stromal desmoplastic response. (Insert) High-power view of well-differentiated SCC with ample cytoplasm (low N:C ratio) and well-defined cell borders. Intercellular bridges are evident. (B) Medium-power example of poorly differentiated SCC without spindle morphology. Note the lack of keratinization; however, the tumor cells retain an epithelioid appearance consisting of abundant eosinophilic cytoplasm with defined cellular borders. (Insert) At high power, intercellular bridges are not readily identified. (C,D) Examples of poorly differentiated SCCs with a mixture of epithelioid tumor cells and tumor cells acquiring subtle spindled morphology. (E) Low-power view of poorly differentiated SCCs demonstrating nests of epithelioid squamous cells transitioning into a focus of infiltrating malignant spindle cells. (F) Low-power views of pure malignant spindle cells consistent with spindle cell carcinoma (the insert shows a high-power view of spindle cells). Carcinoma category is shown at the top right corner of the pictures (bars, 100 μm). (G) Unsupervised hierarchical clustering of all microarray probe sets expressed above background separates the carcinomas into two categories. Class A (44 carcinomas) is shown on the left, and class B (16 carcinomas) is shown on the right. IHC on carcinoma sections using antibodies against p63 (H,I), E-cadherin (J,K), Vimentin (L,M), Keratin 8/18 (N,O), Slug (P,Q), and Snail (R,S), with class A carcinomas depicted on the right, and class B carcinomas depicted on the left (bar, 100 μm).
A number of features of EMT have been characterized in multiple tumor types, including loss of E-cadherin, epidermal keratins, and transcription factors such as p63, with gain of Vimentin. We examined expression of these markers in both tumor classes and confirmed by immunohistochemistry (IHC) that epithelial markers p63, E-cadherin (Fig. 1H–K), and Keratin 14 (data not shown) were highly expressed in class A tumors but were very low in class B tumors, while the opposite was seen for the mesenchymal marker Vimentin (Fig. 1L,M). Although negative for the epidermal keratins 5 and 14 (data not shown), the class B spindle tumors stained positively for Krt8/18 (Fig. 1N,O), indicative of their epithelial origin. This observation is compatible with a model where class B tumors have undergone EMT.
Further evidence linking the class B carcinomas to EMT came from gene expression correlation analysis of the complete panel of carcinomas. The Snai1 transcription factor has been positively linked with invasion and EMT, together with a panel of other genes, including Zeb1, Vimentin, and Vcan (Ricciardelli et al. 2009). These genes are positively correlated with Snai1 (ρ > 0.8) in the carcinomas as well as with components of the TGFβ signaling pathway (Tgfbr3 and Tgfb1i1) and are also relatively overexpressed in class B compared with class A tumors (Supplemental Table 1). Interestingly, while Snai1 and Snai2 (Slug) are both commonly thought to function as activators of EMT, only Snai1 is up-regulated in the class B carcinomas, while Snai2 is relatively overexpressed in the class A tumors (Fig. 1P–S) and is positively correlated in expression with epithelial markers, including Krt14 and p63 (Supplemental Table 2). While Snai1 is linked to this major class of tumors that have undergone EMT, Snai2 may play a different role or be involved in transformation of rare cells within the class A carcinoma that adopt a mesenchymal cell fate (Burns et al. 1991).
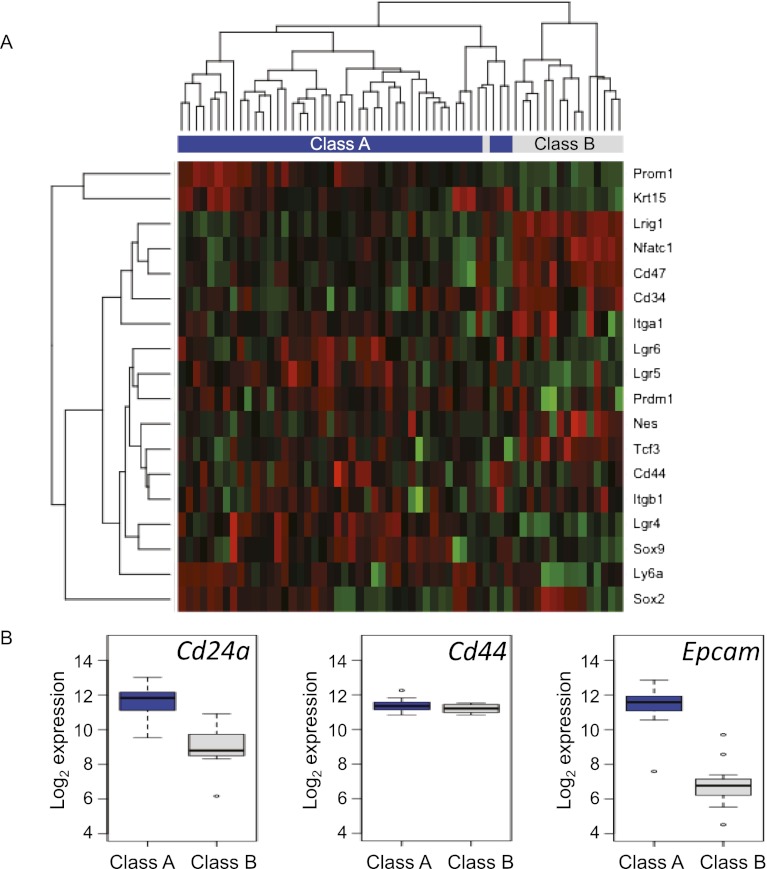
Several studies have proposed that induction of EMT is associated with expression of markers of normal tissue stem cells (Chaffer and Weinberg 2011; Scheel and Weinberg 2011). A range of markers of different stem cell populations has been identified in mouse skin, including the hair follicle bulge markers CD34 (Trempus et al. 2003; Blanpain et al. 2004), NfatC1 (Horsley et al. 2008), Tcf3/4 (Nguyen et al. 2006, 2009), and Krt15 (Liu et al. 2003) as well as markers located mostly outside the bulge, such as Sox9 (Vidal et al. 2005; Nowak et al. 2008), Sox2 (Driskell et al. 2011) Lgr5 (Jaks et al. 2008), Lgr6 (Snippert et al. 2010), and Lrig1 (Jensen et al. 2009). We examined the expression of these and other putative stem cell markers in the carcinomas and found that a subset of stem cell markers found in the hair follicle bulge—e.g., CD34, Tcf3, and Nfatc1 as well as Lrig1, which was proposed to be a marker of epidermal stem cell fate located in the infundibulum—is expressed at significantly higher levels in the class B spindle cell tumors (Fig. 2A; Supplemental Table 3). Expression of CD34 was also verified by immunohistochemical analysis of class A and B tumors (Supplemental Fig. 1). Staining in SCCs was seen mainly in basal cells or small clusters of cells throughout the tumors, as previously demonstrated by others (Malanchi et al. 2008; Lapouge et al. 2012), but was quite extensive in certain cases (e.g., Supplemental Fig. 1, top right panel). The pattern in class B spindle carcinomas was variable between individual tumors, but on average, the CD34 expression was higher and more uniform, with a larger proportion of tumor cells staining positively.
Figure 2.
A subset of skin stem cell markers is expressed at higher levels in class A than in class B carcinomas. (A) Unsupervised hierarchical clustering of skin carcinoma gene expression levels using stem cell marker genes. Green indicates lower expression, and red indicates higher expression. (B) Box plots of gene expression levels of CD24a, CD44, and Epcam in primary mouse skin carcinomas measured by microarray. Class A carcinomas are shown in blue, and class B carcinomas are shown in gray.
In contrast, alternative stem cell markers, including Lgr5, Lgr6, and Krt15, are preferentially expressed in the class A SCCs. Interestingly, class B tumors show the expression pattern of CD44hi/CD24lo/Epcam− (Fig. 2B), previously used for enrichment of cells with tumor-propagating characteristics (Al-Hajj et al. 2003; Al-Hajj and Clarke 2004; Ponti et al. 2005; Fillmore and Kuperwasser 2008), and have a molecular profile very similar to the subset of human breast cancers with a “claudin low signature” (Supplemental Fig. 2; Prat et al. 2010) that have previously been proposed to exhibit stem cell features, including EMT (Visvader 2009). We conclude that the class B carcinomas recapitulate many features of aggressive human cancers that have undergone EMT. Whether these tumors arise from a specific subpopulation of cells within one of the known stem cell compartments is unknown.
Suppression of Egfr autocrine signaling in class B carcinomas
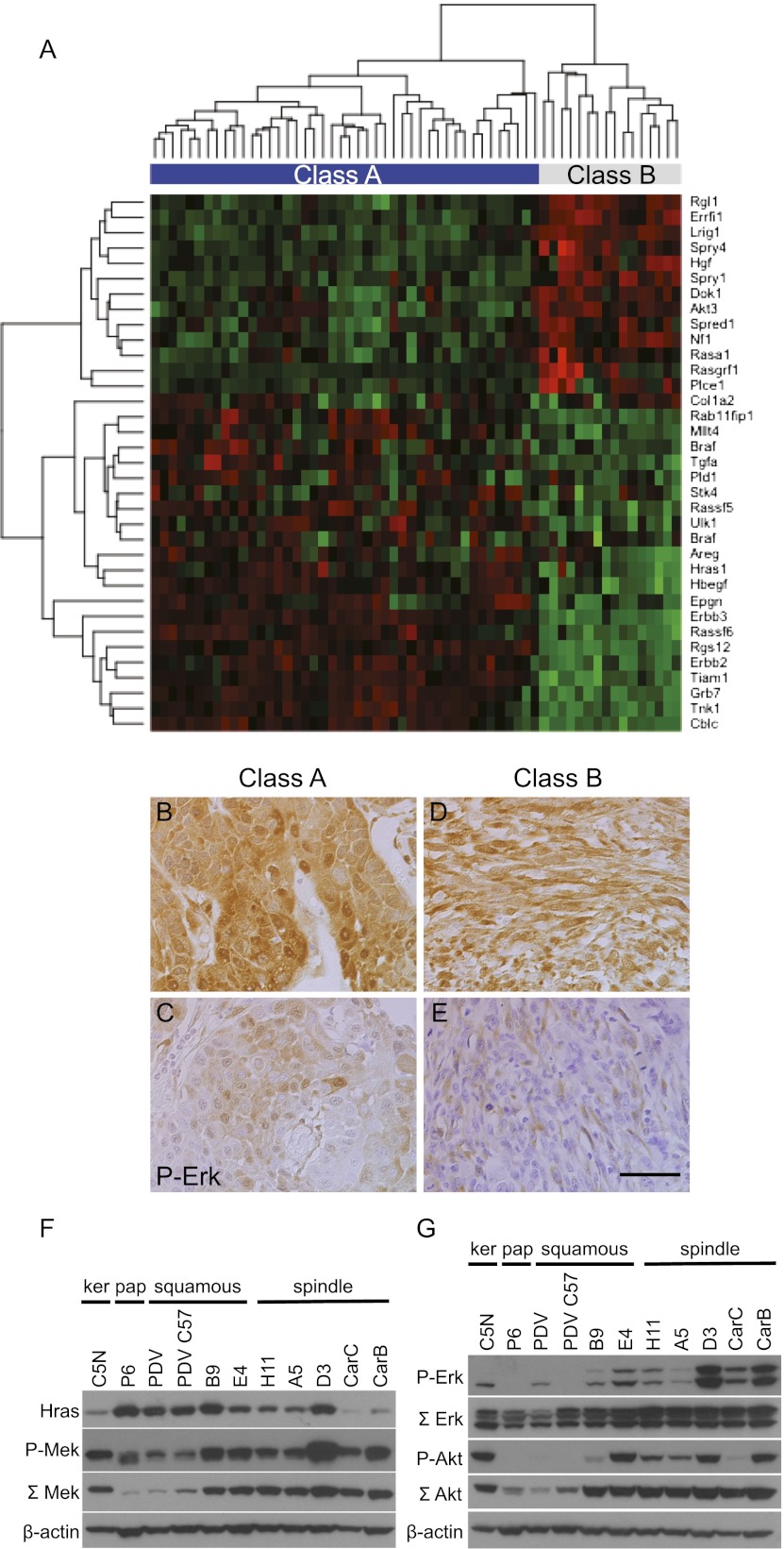
Overexpression of mutant Ras has previously been shown to induce EMT, particularly in the context of elevated Tgfβ/Smad signaling (Oft et al. 1996, 2002). We therefore investigated the expression levels of genes known to be involved in the Ras pathway in this panel of primary mouse skin tumors. Tumor expression profiles were clustered using a panel of genes known to be involved positively or negatively in Ras signaling collected from the literature. This analysis clearly demonstrated a significant difference in the patterns of Ras pathway signaling between class A and class B tumors (Fig. 3A). Paradoxically, Hras1 mRNA levels were lower in the more aggressive class B carcinomas than in the class A SCCs, as were several well-characterized EGF receptor ligands, including Hbegf, Tgfa, Areg, and Epgn (Schneider et al. 2008). The EGF family receptor ErbB2 was also down-regulated in the class B tumors, as were the downstream pathway mediator Grb7 and the Rac exchange factor Tiam1. In contrast, negative regulators of the EGF receptor/Ras pathway such as Lrig1 (Gur et al. 2004; Laederich et al. 2004) Errfi1 (Fiorentino et al. 2000; Hackel et al. 2001; Anastasi et al. 2003; Xu et al. 2005), Sprouty family members 1 and 4, and Spred1 (Guy et al. 2009) as well as the Ras GTPase-activating proteins Nf1 and Rasa1 (RasGap) (Iwashita and Song 2008) were overexpressed relatively in class B tumors. Staining of class A and B tumors with antibodies recognizing ErbB2 and Lrig1 confirmed the patterns predicted from mRNA analysis (Supplemental Fig. 3A–D). This coordinated down-regulation of positive activators of Egfr signaling and up-regulation of negative regulators may be linked to the progressive loss of differentiation capacity of the class B tumors, as the Egfr pathway is associated with commitment to the epidermal lineage and differentiation (Schneider et al. 2008).
Figure 3.
Components of the Ras/EGFR signaling pathway are down-regulated in class B spindle compared with class A SCCs. (A) Unsupervised hierarchical clustering of skin carcinoma gene expression levels using genes involved in the Ras and EGFR pathways. Red indicates higher expression, and green indicates lower expression. (B–E) IHC on class A (B,C) and class B (D,E) carcinoma sections using an antibody against P-Erk1/2 (bar, 100 μm). (F,G) Western blots using a panel of cell lines representing different stages of skin tumor progression: C5N immortalized keratinocytes; P6 papilloma cells; PDV, PDVC57, B9, and E4 SCC cell lines; and H11, A5, D3, CarC, and CarB spindle cell lines. (F) Protein levels of Hras (first lane), P-Mek (second lane), total Mek (third lane), and β-actin as loading control (fourth lane). (G) Protein levels of P-Erk (first lane), total Erk (second lane), P-Akt (third lane), total Akt (fourth lane), and β-actin as loading control (fifth lane).
Although the class B tumors show reduced levels of Egfr signaling components, other classical Ras-activated pathways were strongly stimulated. Immunohistochemical analysis of both class A and B tumors showed that most expressed P-Erk1/2, some at high levels (Fig. 3B–E). Similar conclusions were reached from analysis of Ras signaling in cell lines derived from different stages of skin carcinogenesis (Bremner and Balmain 1990; Quintanilla et al. 1991; Linardopoulos et al. 1995).The cell lines B9 (squamous) and A5 and D3 (spindle) are single-cell clones derived from the same squamous carcinoma with a spindle component (Burns et al. 1991). The CarB and CarC cells are independent spindle cell lines derived from primary spindle carcinomas from NIH/Swiss mice (Linardopoulos et al. 1995). Although the Hras codon 61 mutations in both CarB and CarC cell lines are homozygous, the levels of mutant Hras protein are very low in comparison with other less malignant cell lines (Fig. 3F, first lane), confirming the general picture described for the primary class B spindle cell tumors above.
In spite of the differences in mutant Hras levels, signaling through P-Erk and P-Akt was elevated in almost all of the cell lines that had undergone EMT (Fig. 3G, first and third lanes). In particular, the CarB and CarC cells with the lowest levels of mutant Hras still expressed high levels of P-Erk. The data on total Akt, showing that the protein levels increase with increasing degree of malignancy and spindle cell morphology from left to right in the lower panel (Fig. 3G, fourth lane), exactly parallel the data obtained from the gene expression array analysis, which identified Akt3 expression levels as being significantly up-regulated in class B spindle cell carcinomas (Fig. 3A). Interestingly, levels of total Mek also increased roughly in parallel with malignancy (Fig. 3E, third lane), but neither Akt nor Mek showed a positive relationship to levels of mutant Hras across the panel of cells. A similar lack of correlation between Ras activation and stimulation of P-Erk has been noted previously (Mao et al. 2004; Trejo et al. 2012). We conclude that activation of Ras/Erk is positively, but autocrine Egfr signaling is negatively, correlated with tumor progression in this model.
Ink4/Arf genetic changes distinguish class A from class B tumors
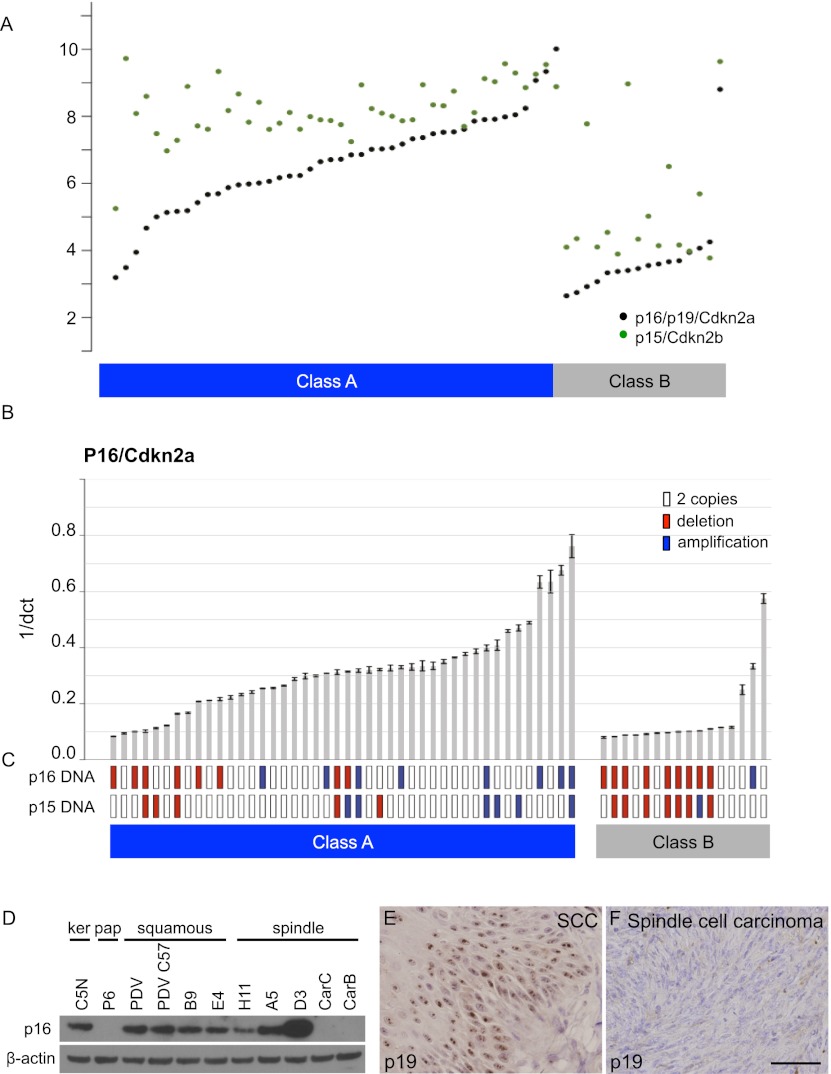
Previously, comparative genomic hybridization (CGH) analysis of genome-wide aberrations in skin carcinomas identified a subset of tumors with deletions or reduced copy number on chromosome 4 (Quigley et al. 2011). The Ink4/Arf locus located within these deleted regions encodes three genes: p16/Cdkn2a, p15/Cdkn2b, and p19/Arf, from now on referred to as p16, p15, and p19, respectively. This locus was a very good candidate driver of these changes, as homozygous deletions have previously been seen in a small subset of invasive mouse skin tumor cell lines (Linardopoulos et al. 1995), and inactivation of Ink4/Arf in the mouse germline causes an increased incidence of skin carcinomas (Krimpenfort et al. 2007). Preliminary inspection of the Affymetrix array data on carcinomas showed that expression of p16/p19 and p15 was indeed very low in most of the class B tumors but elevated in most class A tumors (Fig. 4A). We developed specific TaqMan real-time PCR assays to measure both expression levels and gene copy number of p16, p15, and p19 in all of the mouse skin carcinomas (Fig. 4B,C; Supplemental Fig. 4)
Figure 4.
p16 expression levels as well as p16/p19 and p15 gene copy numbers differ between class A and class B carcinomas. (A) Gene expression of p16 (black points) and p15 (green points) measured by microarrays in class A and class B carcinomas. The Y-axis shows log2 expression level. Carcinomas are sorted according to p16 array expression levels. (B) Quantitative RT–PCR (qRT–PCR) for p16 in class A and class B carcinomas. Error bars represent standard deviation from three replicate measurements. Carcinomas are sorted according to p16 qRT–PCR expression levels. (C) Gene copy number analysis of p16/p19 (top row) and p15 (bottom row). Amplifications of the genes are marked with blue, deletions are marked with red, and two copies are indicated with white boxes. (D) Western blot depicting levels of p16 (lane 1) and β-actin as loading control (lane 2) in a panel of cell lines representing different stages of skin tumor progression. Immunohistochemistry on class A (E) and class B (F) carcinoma sections using an antibody against p19/Arf (bar, 100 μm).
The data shown in Figure 4C demonstrate that deletions of p16/p19 and p15 genes are predominantly seen in class B tumors compared with their class A counterparts (P = 0.007 and P = 0.025, respectively, Fisher exact test). Expression levels of p16 and p15 were significantly elevated in class A tumors (P = 0.0003 and P = 0.009, respectively, t-test). Interestingly, copy number gains of p16/p19 and/or p15 were also seen, particularly in the class A tumors that had high expression of these genes, but the significance of this observation is presently unclear. Separate analyses using specific probes for p19 showed a picture very similar to that seen for p16 (Supplemental Fig. 4B). Although IHC using antibodies against p16 was unsuccessful for technical reasons, an antibody against p19/Arf clearly supported the conclusion from the mRNA data, in that the class A tumors had high expression levels, while the majority of class B tumors were completely negative (Fig. 4E,F). We conclude that expression of several genes in the Ink4 locus is a major distinguishing feature of class A versus class B tumors and may play a causal role in development of these distinct tumor types. Of note, a range of skin tumors arises spontaneously in p15/p16/p19 triple-knockout mice (Krimpenfort et al. 2007). Others have shown that loss of p19/Arf (Kelly-Spratt et al. 2004) or p16/Ink4a (Sharpless et al. 2001, 2002, 2004) promotes skin tumor development and progression, but comparison of these individual studies is difficult because of the various tumor induction protocols and genetic backgrounds used. It is therefore not possible to deduce at present which specific combinations of these genes on chromosome 4 play a causal role in induction of the class B carcinomas.
Class A and B carcinomas differ in dependence on tumor promotion
Squamous papillomas and carcinomas are highly dependent on chronic exposure to inflammatory and tumor-promoting agents. Both the duration and frequency of tumor promoter treatment as well as the use of anti-inflammatory agents profoundly affect the incidence of squamous papillomas in two-stage carcinogenesis experiments (Viaje et al. 1977; Verma and Boutwell 1980). Moreover, in transgenic mouse models of Ras-induced skin tumor formation, papillomas often appear at body sites (such as the ear or base of the tail) that are subject to wounding or chronic irritation due to scratching or fighting (Bailleul et al. 1990; Greenhalgh et al. 1993). We therefore carried out a two-stage carcinogenesis experiment using groups of FVB mice that were initiated with the same dose of DMBA but treated with TPA biweekly for either 5, 10, or 20 wk. Animals were then monitored for a period of >1 yr to document carcinoma incidence as well as the morphology and stage of tumor progression. Papilloma formation was drastically decreased by the reduced duration of TPA treatment: Six of 10 animals in the 5-wk treatment group developed no papillomas after 20 wk, and the remaining four developed only one to two papillomas. In contrast, the 20-wk treatment group demonstrated a significantly increased incidence of papillomas (mean 22.4, SD 9.2; P = 3.5 × 10−5, t-test). These data are similar to the results of previous experiments (Hennings et al. 1985) and demonstrate the critical role of inflammation in benign papilloma development. Whether these observations are due to the induction of inflammation by TPA or to some other pathway is presently unclear. Gene expression analysis of known markers of inflammation shows clear differences in levels of expression between class A and B tumors (Supplemental Fig. 5), but further studies will be required in order to determine whether these immune system components play a causal role in the selection and outgrowth of the different tumor classes.
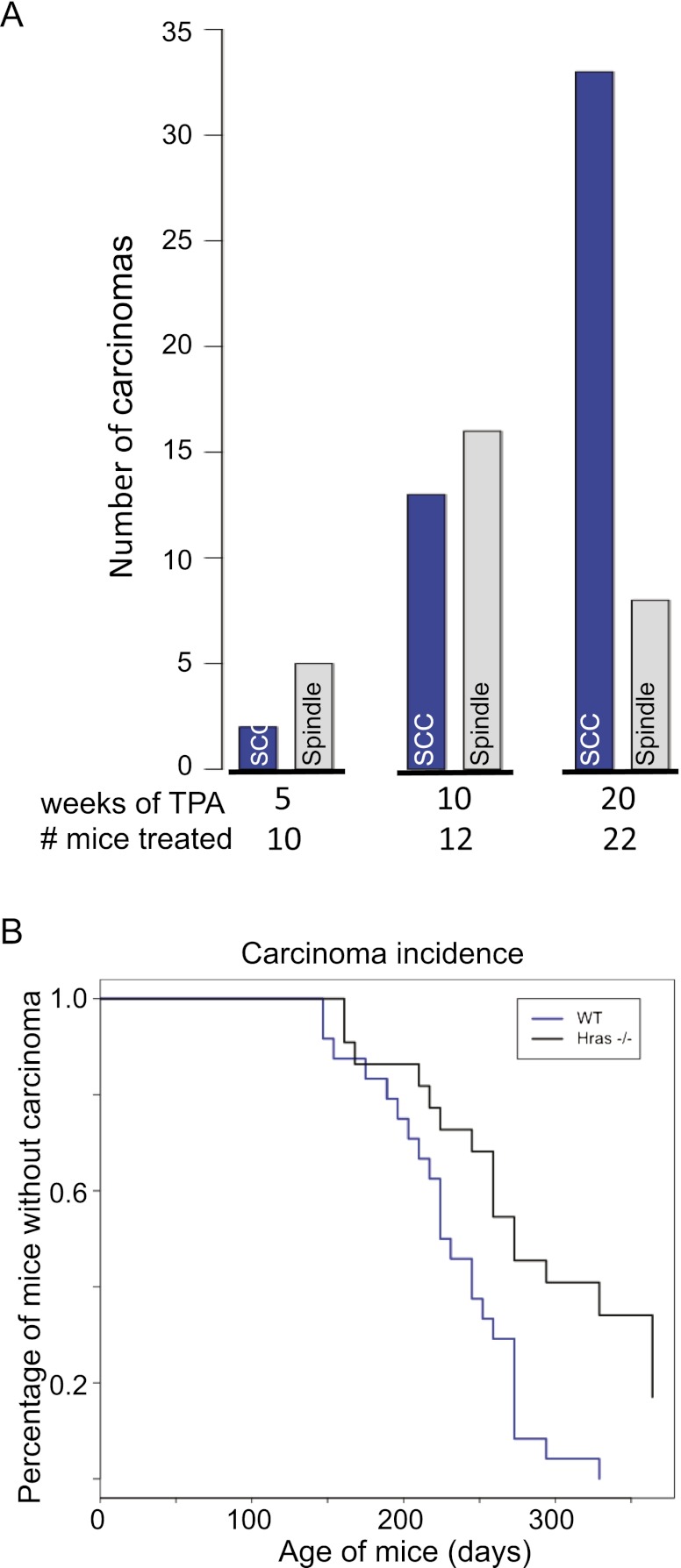
Carcinomas were also strongly dependent on TPA promotion, but a striking difference was seen in the ratio of SCCs to spindle cell tumors. In the 5-wk treatment group, the ratio of SCCs to spindle cell carcinomas was 1:2.5, whereas in the 10-wk group, the ratio was ∼1:1.2, and after 20 wk of TPA treatment, it was 4:1 (Fig. 5A). This 20-wk ratio is very similar to historical controls as well as the data in Figure 1, and the association between tumor ratio and weeks of treatment was statistically highly significant (P = 0.0004, χ2 test for trend in proportions).
Figure 5.
Carcinoma incidence and dependence on inflammation. (A) Numbers of SCCs (blue bars) and spindle cell carcinomas (gray bars) observed in wild-type FVB/N mice after a single dose of DMBA and subsequent biweekly TPA treatments for 5 wk (n = 10 mice), 10 wk (n = 12 mice), or 20 wk (n = 22 mice). All groups of mice were killed when carcinomas reached a minimum size of 1.5 cm in diameter. (B) Kaplan-Meier survival curves showing carcinoma incidence of wild-type mice (blue line; n = 23 mice) and Hras−/− mice (black line; n = 21 mice) treated with TPA for 20 wk. The difference between the curves was statistically significant (P = 0.0006). A very similar result was obtained in a completely independent experiment involving 24 wild-type and 22 Hras−/− mice (P = 0.0024).
In order to verify that the tumor categories obtained using inbred FVB mice reflected what was seen in the original backcross study, we analyzed gene copy number for p16/p19 and p15 in a total of 59 tumors from this experiment. The results demonstrated a statistically significant correlation between the class B tumor morphology, as assessed by two independent investigators, and reduced gene copy number for p16/p19 and p15 (P = 0.003 and P = 0.003, respectively, Fisher exact test) (Supplemental Fig. 6A). The most likely conclusion is that class A and B carcinomas arise independently from one another and that the papillomas and class A SCCs are more dependent on exogenous promotion by exposure to inflammatory agents.
Squamous papillomas and carcinomas are dependent on Hras signaling
The reduced levels of mutant Hras as well as the altered signaling through the Egfr/Ras pathway prompted us to ask whether mice in which the Hras gene has been deleted show an altered tumor spectrum after DMBA/TPA treatment. Hras-null mice are viable and fertile but show a reduced incidence of skin papillomas in a mixed 129/C57BL/6 background (Ise et al. 2000). Previous results on this background showed that the incidence of skin papillomas was reduced by a factor of four to five, but no detailed analysis was carried out of tumor progression or the effects on carcinoma morphology or behavior. We first bred the Hras knockout allele onto a pure FVB background through >20 generations and carried out the standard skin carcinogen treatment. On the FVB background, the number of papillomas after 20 wk of TPA treatment was significantly reduced in Hras-null mice (mean 2.2 vs. mean 22.4 in wild-type mice; P = 4.7 × 10−5, t-test). Surprisingly, however, in spite of their very low papilloma incidence, the majority of the Hras-null mice developed malignant carcinomas, as seen in two independent experiments (Fig. 5B). Histological and IHC assessment of these carcinomas (Supplemental Fig. 7) showed that they were primarily of the spindle type (17 of 24 tumors), classified above as class B carcinomas. Molecular analysis of these tumors showed that, in agreement with a previous study for papillomas (Ise et al. 2000), most of the skin tumors, both papillomas and carcinomas, carried mutations in the Kras gene (Supplemental Table 4). Although skin tumors in wild-type mice rarely exhibit activating Kras mutations, clearly, in the context of inactive signaling through Hras, Kras can adapt and accomplish some but not all of the functions of Hras in skin. Notably, conditional activation of Kras mutations in mouse skin using keratin promoters to drive Cre-recombinase results in the formation of skin papillomas, some of which, in the absence of p53, progress to squamous and, in some cases, spindle carcinomas (White et al. 2011; Lapouge et al. 2012). We propose that Hras mutations in wild-type animals are essential for the development of class A squamous papillomas and carcinomas but dispensable for class B tumors, which can develop through the alternative pathway driven by activation of Kras. A further parallel between the spindle carcinomas in the Hras knockout model and the class B carcinomas in wild-type mice described above was confirmed by gene copy number analysis of Ink4/Arf deletions. These were enriched in carcinomas from Hras knockout compared with their wild-type FVB counterparts and in spindle tumors versus SCCs (Supplemental Fig. 6B).
Both class A and B carcinomas give rise to distant metastases
The mechanisms of metastasis in vivo are complex and controversial (Cardiff 2010; Roussos et al. 2010; Chaffer and Weinberg 2011; Chaffer et al. 2011) In the skin model, analysis of metastasis is frequently not carried out, as the animals have to be killed when primary tumors reach a maximum size that is often attained before overt metastases are detectable. To circumvent this problem, we induced skin carcinomas in wild-type FVB and Hras knockout mice, surgically removed the primary tumors when they reached a size of 1.0–1.5 cm in diameter, and monitored survival of the animals over several months. Most animals eventually succumbed to lymph node and/or distant metastases, primarily to the lung but also to a number of additional sites, including the spleen, liver, kidney, adrenal cortex, soft tissue, and chest wall. A summary of these data is shown in Supplemental Table 5. Histological comparison of the primary and metastatic tumors indicated that both class A and B carcinomas are capable of metastasis, and in some but not all cases, the morphology of the metastases was similar to that of the primary tumor. In cases where the morphology was discordant, it is not possible to exclude the presence of additional small carcinomas that were the true precursors of these metastases, as some mice developed second independent primaries of a different morphology after surgery. From a total of 22 wild-type FVB mice that underwent surgery, 10 metastases (not including Lymph nodes) were detected in the lung or at distant sites. From 13 Hras knockout mice in the same study, 26 metastases were found (mean 0.5 in wild type vs. 1.9 in knockout; P = 0.007, Mann-Whitney test), suggesting that carcinomas from the Hras-deficient mice were more aggressive. Whether this more aggressive behavior is due to the higher frequency of spindle tumors or the presence of alternative genetic events such as Kras mutations is presently unclear.
Alternative models for tumor progression in vivo
Our combined data suggest the existence of a novel pathway leading to malignant progression that is less dependent on inflammation and Hras signaling. In the classical skin carcinogenesis model, papillomas and malignant carcinomas are dependent on chronic inflammation and almost all have Hras mutations. Reduction of inflammation or loss of Hras leads to a strong reduction in overall tumor development but allows emergence of highly malignant class B tumors that exhibit a high frequency of Ink4 genetic alterations. These tumors could arise by deletion or silencing of the Ink4 locus in pre-existing class A carcinomas (Fig. 6A). However, almost all class A SCCs express elevated levels of p16 and p19 (Fig. 4A,B; Supplemental Fig. 4B), and it seems unlikely that the mechanisms leading to this overexpression would be reversed to allow progression through EMT. Class B tumors could also arise from alternative genetic events possibly involving Ink4A at the papilloma stage (Fig. 6B). However p16/p19 levels were elevated in almost all papillomas compared with normal skin, similar to the situation seen with the class A carcinomas (Supplemental Fig. 5), suggesting that the class B tumors might represent a different category that diverges after initiation (Fig. 6C) or comes from a different stem cell type within the skin (Fig. 6D). The latter possibility is analogous to a model proposed for the origin of the “triple-negative” category of human breast cancers, to which the class B spindle cell carcinomas have many similarities (Visvader 2009).
Figure 6.
Schematic models of the possible origins of class B carcinomas. (A) Class B carcinomas could arise from class A squamous tumors. (B) Both class A and class B carcinomas could arise from similar papillomas that undergo separate genetic events. (C) Class A and class B carcinomas could have the same cell of origin but diverge at an early stage, giving rise to different kinds of premalignant lesions. (D) Class A and class B carcinomas could have two completely different cells of origin in the skin.
Discussion
Cancer of the skin is the most common form of human cancer, and in spite of possibilities for early detection and intervention, incidence is still increasing (Alam and Ratner 2001). Squamous carcinomas of the skin, head and neck, and lung have a strong link to carcinogen exposure, exhibit similar morphological and molecular features, and rank among the largest causes of death from cancer worldwide (Stransky et al. 2011; Wang et al. 2011; Hammerman et al. 2012). Tumors of these types have a significant frequency of Hras mutations, including the same codon 61 mutation that is induced by DMBA in mouse skin (Arnault et al. 2012; Oberholzer et al. 2012; Su et al. 2012). Deletion and/or mutation of Cdkn2a has also been specifically linked to poor survival of human skin cancer patients (Kusters-Vandevelde et al. 2010). The mouse skin model used here, which involves carcinogen-induced or spontaneous mutations in many of the same genes, may therefore provide an important approach to the study of the cell lineages and pathways leading to squamous carcinoma development and metastasis.
In the present study, we show that the major category of invasive carcinomas in skin arises through a separate pathway that is independent from the classical route. Evidence comes from several sets of experiments. First, gene expression (Fig. 1) and genomic analysis (p16/p19 copy number) (Fig. 4) identified major differences in the molecular characteristics of class A and B carcinomas arising from skin carcinogenesis studies in vivo. Second, reduced inflammation or elimination of the Hras signaling pathway causes a dramatic reduction in papillomas and class A carcinomas but causes no proportional decrease in class B carcinomas, suggesting that the latter do not arise from pre-existing class A tumors. We conclude that divergence in these two pathways must take place at an early stage of carcinogenesis or even by separate events involving mutations of Ras genes in distinct target cells (Fig. 6D).
Altered Ras/Egfr signaling in spindle cell carcinomas
In spite of prior evidence showing the importance of high levels of mutant Ras in driving the spindle phenotype (Oft et al. 1996, 2002), the class B tumors express lower levels of Hras mRNA (Fig. 3A), and the corresponding cell lines express lower levels of Hras protein (Fig. 3F, first lane) than less aggressive tumor cell types. The class B tumors also have an attenuated pattern of signaling through autocrine pathways involving ligands and receptors of the Egfr family. Expression levels of a number of ligands, receptors, and other Ras pathway effector components are significantly lower in class B spindle cell carcinomas than in squamous tumors, while inhibitors of Egfr/Ras signaling, including Lrig1, Errfi1, Nf1, and Rasa (RasGap), are significantly higher in class B tumors. This suggests that the many upstream components of the Egfr signaling pathway are down-regulated, in conjunction with the lack of any differentiation characteristics in this tumor type. Either this program has been switched off in spindle cell tumors or its absence reflects a feature of the target cell from which these tumors arose. However, the persistence of elevated Erk1/2 and Akt signaling in both primary class B carcinomas and corresponding cell lines suggests that activated Ras is still important in driving these tumors.
These observations have parallels in a number of human tumor types. Studies of lung and pancreatic cancers driven by Kras identified cell lines that showed differential dependence on Ras signaling that correlated with a mesenchymal EMT morphology (Singh et al. 2009). The Kras-independent tumor cell lines expressed several markers of EMT, including high levels of Zeb1 and strongly reduced E-cadherin, features also characteristic of the spindle cell carcinomas in this study. Our data on down-regulation of canonical Egfr signaling in spindle cell tumors are compatible with the previously reported resistance of cells with EMT properties to inhibitors of the EGFR tyrosine kinase (Thomson et al. 2008), leading to clinical resistance of “EMT” tumors to inhibitors of this pathway. It is conceivable that treatment of some epithelial Ras mutant tumors with EGFR inhibitors may in fact select for less differentiated EMT-like cells and thus contribute to the generally poor prognosis noted in clinical trials of Tarceva in patients with Kras mutant lung tumors (Eberhard et al. 2005).
EMT and stem cell markers
It has been shown that induction of EMT in mammary epithelial cells by treatment with Tgfβ or with effectors of other developmental pathways (Scheel and Weinberg 2011) leads to the adoption of some cancer stem cell properties, including ability to grow as mammospheres, self-renewal capacity, and propensity for metastasis. The cells that display such properties can be enriched using a combination of markers (CD44hi/CD24lo/Epcam−), which selects for cells with low adhesion properties and high growth capacity (Al-Hajj et al. 2003; Al-Hajj and Clarke 2004; Ponti et al. 2005; Fillmore and Kuperwasser 2008). This combined expression profile is enriched in the class B spindle cell carcinomas (Fig. 2B), suggesting that this tumor category may have a high proportion of tumor stem/propagating cells. The high expression of CD34 in the class B tumors (Fig. 2A; Supplemental Fig. 1) also supports this interpretation, as Lapouge et al. (2012) have demonstrated that skin carcinomas with high CD34 can be serially transplanted and contain a high proportion of tumor-propagating cells. Direct functional studies of cell lines representing class B carcinomas (CarB and CarC cells) have furthermore shown that they have the highest levels of invasion and chemotaxis (Buchmann et al. 1991) and clonogenic capacity (data not shown). The ease of generation and manipulation of these stem cell-like tumors in vivo may provide new opportunities for development of therapeutic strategies aimed at this aggressive tumor class.
The hair follicle bulge has long been thought to be a reservoir of epidermal stem cells capable of repopulating the follicular or interfollicular epidermis, and several genes expressed in the bulge (e.g., CD34, Nfatc1, and Tcf4) are enriched in gene expression profiles of the spindle cell carcinomas. However another bulge marker, Krt15, is not expressed in these tumor cells, while a putative stem cell marker in the infundibulum, the ErbB pathway inhibitor Lrig1 (Wong et al. 2012), is highly expressed. The segregation of these different stem cell markers across the two categories of malignant tumors is compatible with a model in which these tumors arise from alternative normal stem/progenitor cell populations in the skin epithelium. Detailed linage tracing experiments would, however, be required to test these possibilities. The identification of an in vivo immunocompetent mouse model system for reproducible development of these tumor categories and their metastases offers new opportunities to identify and test combinatorial approaches to therapy targeted against malignant tumors or their component cancer stem cell populations.
Materials and methods
Mouse breeding and induction of carcinogenesis
To obtain backcross mice, we crossed male SPRET/Ei and female FVB/N mice from the Jackson Laboratory. Female F1 hybrids were then mated to male FVB/N mice. Eight-week-old to 12-wk-old mice received one dose of DMBA (25 mg per mouse in acetone) applied to back skin. One week after initiation, tumors were promoted by biweekly topical treatment with TPA (200 μL of a 10−4 M solution in acetone) for 20 wk in the case of backcross mice or 5, 10, and 20 wk in the case of FVB/N wild-type or Hras−/− mice. Primary carcinomas from FVB/N wild-type or Hras−/− mice that reached a size of 1.0–1.5 cm in diameter were surgically removed, and additional carcinomas and metastases were collected when the mice needed to be killed. All mouse experiments were approved by the University of California at San Francisco Laboratory Animal Resource Center.
Microarray preparation and data analysis
Microarrays (Affymetrix M430 2.0, annotation NA31) are available at the Gene Expression Omnibus, accession number GSE21264. Briefly, one piece of each tumor was snap-frozen, and RNA was isolated using TRIzol (Invitrogen) according to the manufacturer's instructions. Residual contaminating genomic DNA was removed by DNase treatment (Ambion). Statistical analysis was performed with R version 2.13 (R Core Development Team., 2010), using the affy package for RMA normalization (Gautier et al. 2004) and samr for differential expression analysis (Tusher et al. 2001). Code and data required to reproduce all statistical results are available on request.
IHC
Five-micrometer tissue sections were deparaffinized and stained (see the Supplemental Material). The following antibodies were used: p63 4A4 (mouse; Millipore), E-cadherin (rabbit; BD Pharmingen), Vimentin (rabbit; Abcam), K8/18 (guinea pig; Progen), Slug (rabbit; Cell Signaling Technology), Snail (guinea pig; generous gift from Dr. Elaine Fuchs [Rockefeller University, New York, NY]), P-p44/42 MAPK (T202/Y204, rabbit; Cell Signaling Technology) p19/Arf (rabbit; Abcam), and biotinylated secondary antibodies (Vector Laboratories).
Gene expression and gene copy number assays
RNA from carcinomas was isolated as described above, and cDNA was synthesized using random primers and the SuperScript III First-Strand kit from Invitrogen. Primers and probes specific for p16 and p19 were designed using Primer Express software (Applied Biosystems), and p15 and the β-actin housekeeping gene as an internal control were assays on demand (Applied Biosystems). Relative expression levels of p15, p16, and p19 were assessed using TaqMan real-time PCR and derived from the difference in cycle time (CT) for each gene compared with the control β-actin using the formula ΔCT = average CTp15, p16, or p19 − average CTβ-actin. All samples were analyzed in triplicate. Genomic DNA was purified by phenol/chloroform extraction following proteinase K digestion. DNA copy number assays for p15 and p16/p19 were performed by TaqMan real-time PCR. The RORI gene (also on chromosome 4 and determined to have two copies across all samples) was used as a control to normalize DNA quantity. Deletion or gain was determined by ΔCT calculation (described above) and analyzed by Macro program provided by Applied Biosystems. Experiments were performed using the ABI 7900 HT System (Applied Biosystems). For probe and primer information, see the Supplemental Material.
Western blotting
Protein samples were separated on Novex Nupage Tris-glycine gels (Invitrogen), followed by electrophoretic transfer to polyvinylidene difluoride membranes (Millipore) and blocking in 5% nonfat milk in PBST. Immunodetection was performed by enhanced chemiluminescence detection (Amersham). The following primary antibodies were used: p16 (M156), Hras (C-20), Erk1 (C-16), rabbit (Santa Cruz Biotechnology), P-p44/42 MAPK (T202/Y204), Akt (pan), P-Akt (S473), MEK1/2 (47E6), and P-MEK1/2 (S217/221, rabbit; Cell Signaling Technology).
Acknowledgments
We thank Karl Banta for assistance with RNA and DNA extraction. This work was supported by Mouse Models of Human Cancers Consortium (MMHCC) UO1 grant CA084244 (to A.B.) and a grant from the California Institute for Regenerative Medicine (CIRM) (grant no. TG2-01153 to C.E.W). C.E.W was also supported in part by the Swiss National Science Foundation (SNF). A.B. acknowledges support from the Barbara Bass Bakar Chair of Cancer Genetics. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of CIRM or any other agency of the state of California.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.210427.112.
References
- Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, Solari A, Bobisse S, Rondina MB, Guzzardo V, et al. 2009. A mutant-p53/Smad complex opposes p63 to empower TGFβ-induced metastasis. Cell 137: 87–98 [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Clarke MF 2004. Self-renewal and solid tumor stem cells. Oncogene 23: 7274–7282 [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF 2003. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci 100: 3983–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam M, Ratner D 2001. Cutaneous squamous-cell carcinoma. N Engl J Med 344: 975–983 [DOI] [PubMed] [Google Scholar]
- Anastasi S, Fiorentino L, Fiorini M, Fraioli R, Sala G, Castellani L, Alema S, Alimandi M, Segatto O 2003. Feedback inhibition by RALT controls signal output by the ErbB network. Oncogene 22: 4221–4234 [DOI] [PubMed] [Google Scholar]
- Arnault JP, Mateus C, Escudier B, Tomasic G, Wechsler J, Hollville E, Soria JC, Malka D, Sarasin A, Larcher M, et al. 2012. Skin tumors induced by sorafenib; Paradoxic RAS–RAF pathway activation and oncogenic mutations of HRAS, TP53, and TGFBR1. Clin Cancer Res 18: 263–272 [DOI] [PubMed] [Google Scholar]
- Bailleul B, Surani MA, White S, Barton SC, Brown K, Blessing M, Jorcano J, Balmain A 1990. Skin hyperkeratosis and papilloma formation in transgenic mice expressing a ras oncogene from a suprabasal keratin promoter. Cell 62: 697–708 [DOI] [PubMed] [Google Scholar]
- Balkwill FR, Mantovani A 2012. Cancer-related inflammation: Common themes and therapeutic opportunities. Semin Cancer Biol 22: 33–40 [DOI] [PubMed] [Google Scholar]
- Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E 2004. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell 118: 635–648 [DOI] [PubMed] [Google Scholar]
- Bonnet D, Dick JE 1997. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 3: 730–737 [DOI] [PubMed] [Google Scholar]
- Bremner R, Balmain A 1990. Genetic changes in skin tumor progression: Correlation between presence of a mutant ras gene and loss of heterozygosity on mouse chromosome 7. Cell 61: 407–417 [DOI] [PubMed] [Google Scholar]
- Buchmann A, Ruggeri B, Klein-Szanto AJ, Balmain A 1991. Progression of squamous carcinoma cells to spindle carcinomas of mouse skin is associated with an imbalance of H-ras alleles on chromosome 7. Cancer Res 51: 4097–4101 [PubMed] [Google Scholar]
- Burns PA, Kemp CJ, Gannon JV, Lane DP, Bremner R, Balmain A 1991. Loss of heterozygosity and mutational alterations of the p53 gene in skin tumours of interspecific hybrid mice. Oncogene 6: 2363–2369 [PubMed] [Google Scholar]
- Cardiff RD 2010. The pathology of EMT in mouse mammary tumorigenesis. J Mammary Gland Biol Neoplasia 15: 225–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caulin C, Scholl FG, Frontelo P, Gamallo C, Quintanilla M 1995. Chronic exposure of cultured transformed mouse epidermal cells to transforming growth factor-β1 induces an epithelial-mesenchymal transdifferentiation and a spindle tumoral phenotype. Cell Growth Differ 6: 1027–1035 [PubMed] [Google Scholar]
- Chaffer CL, Weinberg RA 2011. A perspective on cancer cell metastasis. Science 331: 1559–1564 [DOI] [PubMed] [Google Scholar]
- Chaffer CL, Brueckmann I, Scheel C, Kaestli AJ, Wiggins PA, Rodrigues LO, Brooks M, Reinhardt F, Su Y, Polyak K, et al. 2011. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci 108: 7950–7955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W, Fowlis DJ, Bryson S, Duffie E, Ireland H, Balmain A, Akhurst RJ 1996. TGFβ1 inhibits the formation of benign skin tumors, but enhances progression to invasive spindle carcinomas in transgenic mice. Cell 86: 531–542 [DOI] [PubMed] [Google Scholar]
- Derynck R, Akhurst RJ 2007. Differentiation plasticity regulated by TGF-β family proteins in development and disease. Nat Cell Biol 9: 1000–1004 [DOI] [PubMed] [Google Scholar]
- Driskell RR, Juneja VR, Connelly JT, Kretzschmar K, Tan DW, Watt FM 2011. Clonal growth of dermal papilla cells in hydrogels reveals intrinsic differences between Sox2-positive and -negative cells in vitro and in vivo. J Invest Dermatol 132: 1084–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard DA, Johnson BE, Amler LC, Goddard AD, Heldens SL, Herbst RS, Ince WL, Janne PA, Januario T, Johnson DH, et al. 2005. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol 23: 5900–5909 [DOI] [PubMed] [Google Scholar]
- Fillmore CM, Kuperwasser C 2008. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorentino L, Pertica C, Fiorini M, Talora C, Crescenzi M, Castellani L, Alema S, Benedetti P, Segatto O 2000. Inhibition of ErbB-2 mitogenic and transforming activity by RALT, a mitogen-induced signal transducer which binds to the ErbB-2 kinase domain. Mol Cell Biol 20: 7735–7750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA 2004. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Suarez E, Jacob AP, Jones J, Miller R, Roudier-Meyer MP, Erwert R, Pinkas J, Branstetter D, Dougall WC 2010. RANK ligand mediates progestin-induced mammary epithelial proliferation and carcinogenesis. Nature 468: 103–107 [DOI] [PubMed] [Google Scholar]
- Greenhalgh DA, Rothnagel JA, Quintanilla MI, Orengo CC, Gagne TA, Bundman DS, Longley MA, Roop DR 1993. Induction of epidermal hyperplasia, hyperkeratosis, and papillomas in transgenic mice by a targeted v-Ha-ras oncogene. Mol Carcinog 7: 99–110 [DOI] [PubMed] [Google Scholar]
- Guerra C, Collado M, Navas C, Schuhmacher AJ, Hernandez-Porras I, Canamero M, Rodriguez-Justo M, Serrano M, Barbacid M 2011. Pancreatitis-induced inflammation contributes to pancreatic cancer by inhibiting oncogene-induced senescence. Cancer Cell 19: 728–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur G, Rubin C, Katz M, Amit I, Citri A, Nilsson J, Amariglio N, Henriksson R, Rechavi G, Hedman H, et al. 2004. LRIG1 restricts growth factor signaling by enhancing receptor ubiquitylation and degradation. EMBO J 23: 3270–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy GR, Jackson RA, Yusoff P, Chow SY 2009. Sprouty proteins: Modified modulators, matchmakers or missing links? J Endocrinol 203: 191–202 [DOI] [PubMed] [Google Scholar]
- Hackel PO, Gishizky M, Ullrich A 2001. Mig-6 is a negative regulator of the epidermal growth factor receptor signal. Biol Chem 382: 1649–1662 [DOI] [PubMed] [Google Scholar]
- Hammerman PS, Hayes DN, Wilkerson MD, Schultz N, Bose R, Chu A, Collisson EA, Cope L, Creighton CJ, Getz G, et al. 2012. Comprehensive genomic characterization of squamous cell lung cancers. Nature 489: 519–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennings H, Shores R, Mitchell P, Spangler EF, Yuspa SH 1985. Induction of papillomas with a high probability of conversion to malignancy. Carcinogenesis 6: 1607–1610 [DOI] [PubMed] [Google Scholar]
- Horsley V, Aliprantis AO, Polak L, Glimcher LH, Fuchs E 2008. NFATc1 balances quiescence and proliferation of skin stem cells. Cell 132: 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosobuchi M, Stampfer MR 1989. Effects of transforming growth factor β on growth of human mammary epithelial cells in culture. In Vitro Cell Dev Biol 25: 705–713 [DOI] [PubMed] [Google Scholar]
- Ise K, Nakamura K, Nakao K, Shimizu S, Harada H, Ichise T, Miyoshi J, Gondo Y, Ishikawa T, Aiba A, et al. 2000. Targeted deletion of the H-ras gene decreases tumor formation in mouse skin carcinogenesis. Oncogene 19: 2951–2956 [DOI] [PubMed] [Google Scholar]
- Iwashita S, Song SY 2008. RasGAPs: A crucial regulator of extracellular stimuli for homeostasis of cellular functions. Mol Biosyst 4: 213–222 [DOI] [PubMed] [Google Scholar]
- Jaks V, Barker N, Kasper M, van Es JH, Snippert HJ, Clevers H, Toftgard R 2008. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet 40: 1291–1299 [DOI] [PubMed] [Google Scholar]
- Jensen KB, Collins CA, Nascimento E, Tan DW, Frye M, Itami S, Watt FM 2009. Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell Stem Cell 4: 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly-Spratt KS, Gurley KE, Yasui Y, Kemp CJ 2004. p19Arf suppresses growth, progression, and metastasis of Hras-driven carcinomas through p53-dependent and -independent pathways. PLoS Biol 2: e242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein-Szanto AJ 1989. Pathology of human and experimental skin tumors. Carcinog Compr Surv 11: 19–53 [PubMed] [Google Scholar]
- Krimpenfort P, Ijpenberg A, Song JY, van der Valk M, Nawijn M, Zevenhoven J, Berns A 2007. p15Ink4b is a critical tumour suppressor in the absence of p16Ink4a. Nature 448: 943–946 [DOI] [PubMed] [Google Scholar]
- Kusters-Vandevelde HV, Van Leeuwen A, Verdijk MA, de Koning MN, Quint WG, Melchers WJ, Ligtenberg MJ, Blokx WA 2010. CDKN2A but not TP53 mutations nor HPV presence predict poor outcome in metastatic squamous cell carcinoma of the skin. Int J Cancer 126: 2123–2132 [DOI] [PubMed] [Google Scholar]
- Laederich MB, Funes-Duran M, Yen L, Ingalla E, Wu X, Carraway KL 3rd, Sweeney C 2004. The leucine-rich repeat protein LRIG1 is a negative regulator of ErbB family receptor tyrosine kinases. J Biol Chem 279: 47050–47056 [DOI] [PubMed] [Google Scholar]
- Lapouge G, Beck B, Nassar D, Dubois C, Dekoninck S, Blanpain C 2012. Skin squamous cell carcinoma propagating cells increase with tumour progression and invasiveness. EMBO J 31: 4563–4575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linardopoulos S, Street AJ, Quelle DE, Parry D, Peters G, Sherr CJ, Balmain A 1995. Deletion and altered regulation of p16INK4a and p15INK4b in undifferentiated mouse skin tumors. Cancer Res 55: 5168–5172 [PubMed] [Google Scholar]
- Liu Y, Lyle S, Yang Z, Cotsarelis G 2003. Keratin 15 promoter targets putative epithelial stem cells in the hair follicle bulge. J Invest Dermatol 121: 963–968 [DOI] [PubMed] [Google Scholar]
- Malanchi I, Peinado H, Kassen D, Hussenet T, Metzger D, Chambon P, Huber M, Hohl D, Cano A, Birchmeier W, et al. 2008. Cutaneous cancer stem cell maintenance is dependent on β-catenin signalling. Nature 452: 650–653 [DOI] [PubMed] [Google Scholar]
- Mao JH, To MD, Perez-Losada J, Wu D, Del Rosario R, Balmain A 2004. Mutually exclusive mutations of the Pten and ras pathways in skin tumor progression. Genes Dev 18: 1800–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagase H, Bryson S, Cordell H, Kemp CJ, Fee F, Balmain A 1995. Distinct genetic loci control development of benign and malignant skin tumours in mice. Nat Genet 10: 424–429 [DOI] [PubMed] [Google Scholar]
- Nguyen H, Rendl M, Fuchs E 2006. Tcf3 governs stem cell features and represses cell fate determination in skin. Cell 127: 171–183 [DOI] [PubMed] [Google Scholar]
- Nguyen H, Merrill BJ, Polak L, Nikolova M, Rendl M, Shaver TM, Pasolli HA, Fuchs E 2009. Tcf3 and Tcf4 are essential for long-term homeostasis of skin epithelia. Nat Genet 41: 1068–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak JA, Polak L, Pasolli HA, Fuchs E 2008. Hair follicle stem cells are specified and function in early skin morphogenesis. Cell Stem Cell 3: 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberholzer PA, Kee D, Dziunycz P, Sucker A, Kamsukom N, Jones R, Roden C, Chalk CJ, Ardlie K, Palescandolo E, et al. 2012. RAS mutations are associated with the development of cutaneous squamous cell tumors in patients treated with RAF inhibitors. J Clin Oncol 30: 316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oft M, Peli J, Rudaz C, Schwarz H, Beug H, Reichmann E 1996. TGF-β1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes Dev 10: 2462–2477 [DOI] [PubMed] [Google Scholar]
- Oft M, Akhurst RJ, Balmain A 2002. Metastasis is driven by sequential elevation of H-ras and Smad2 levels. Nat Cell Biol 4: 487–494 [DOI] [PubMed] [Google Scholar]
- Padua D, Massague J 2009. Roles of TGFβ in metastasis. Cell Res 19: 89–102 [DOI] [PubMed] [Google Scholar]
- Peinado H, Olmeda D, Cano A 2007. Snail, Zeb and bHLH factors in tumour progression: An alliance against the epithelial phenotype? Nat Rev Cancer 7: 415–428 [DOI] [PubMed] [Google Scholar]
- Polyak K, Weinberg RA 2009. Transitions between epithelial and mesenchymal states: Acquisition of malignant and stem cell traits. Nat Rev Cancer 9: 265–273 [DOI] [PubMed] [Google Scholar]
- Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, Coradini D, Pilotti S, Pierotti MA, Daidone MG 2005. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res 65: 5506–5511 [DOI] [PubMed] [Google Scholar]
- Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM 2010. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res 12: R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley DA, To MD, Kim IJ, Lin KK, Albertson DG, Sjolund J, Perez-Losada J, Balmain A 2011. Network analysis of skin tumor progression identifies a rewired genetic architecture affecting inflammation and tumor susceptibility. Genome Biol 12: R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintanilla M, Haddow S, Jonas D, Jaffe D, Bowden GT, Balmain A 1991. Comparison of ras activation during epidermal carcinogenesis in vitro and in vivo. Carcinogenesis 12: 1875–1881 [DOI] [PubMed] [Google Scholar]
- Ricciardelli C, Sakko AJ, Ween MP, Russell DL, Horsfall DJ 2009. The biological role and regulation of versican levels in cancer. Cancer Metastasis Rev 28: 233–245 [DOI] [PubMed] [Google Scholar]
- Roussos ET, Keckesova Z, Haley JD, Epstein DM, Weinberg RA, Condeelis JS 2010. AACR special conference on epithelial-mesenchymal transition and cancer progression and treatment. Cancer Res 70: 7360–7364 [DOI] [PubMed] [Google Scholar]
- Scheel C, Weinberg RA 2011. Phenotypic plasticity and epithelial-mesenchymal transitions in cancer and normal stem cells? Int J Cancer 129: 2310–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider MR, Werner S, Paus R, Wolf E 2008. Beyond wavy hairs: The epidermal growth factor receptor and its ligands in skin biology and pathology. Am J Pathol 173: 14–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramek D, Leibbrandt A, Sigl V, Kenner L, Pospisilik JA, Lee HJ, Hanada R, Joshi PA, Aliprantis A, Glimcher L, et al. 2010. Osteoclast differentiation factor RANKL controls development of progestin-driven mammary cancer. Nature 468: 98–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless NE, Bardeesy N, Lee KH, Carrasco D, Castrillon DH, Aguirre AJ, Wu EA, Horner JW, DePinho RA 2001. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature 413: 86–91 [DOI] [PubMed] [Google Scholar]
- Sharpless NE, Alson S, Chan S, Silver DP, Castrillon DH, DePinho RA 2002. p16(INK4a) and p53 deficiency cooperate in tumorigenesis. Cancer Res 62: 2761–2765 [PubMed] [Google Scholar]
- Sharpless NE, Ramsey MR, Balasubramanian P, Castrillon DH, DePinho RA 2004. The differential impact of p16(INK4a) or p19(ARF) deficiency on cell growth and tumorigenesis. Oncogene 23: 379–385 [DOI] [PubMed] [Google Scholar]
- Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, Settleman J 2009. A gene expression signature associated with ‘K-Ras addiction’ reveals regulators of EMT and tumor cell survival. Cancer Cell 15: 489–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert HJ, Haegebarth A, Kasper M, Jaks V, van Es JH, Barker N, van de Wetering M, van den Born M, Begthel H, Vries RG, et al. 2010. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science 327: 1385–1389 [DOI] [PubMed] [Google Scholar]
- Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, et al. 2011. The mutational landscape of head and neck squamous cell carcinoma. Science 333: 1157–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su F, Viros A, Milagre C, Trunzer K, Bollag G, Spleiss O, Reis-Filho JS, Kong X, Koya RC, Flaherty KT, et al. 2012. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N Engl J Med 366: 207–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Kohno H, Suzuki R, Yamada Y, Sugie S, Mori H 2003. A novel inflammation-related mouse colon carcinogenesis model induced by azoxymethane and dextran sodium sulfate. Cancer Sci 94: 965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA 2009. Epithelial-mesenchymal transitions in development and disease. Cell 139: 871–890 [DOI] [PubMed] [Google Scholar]
- Thomson S, Petti F, Sujka-Kwok I, Epstein D, Haley JD 2008. Kinase switching in mesenchymal-like non-small cell lung cancer lines contributes to EGFR inhibitor resistance through pathway redundancy. Clin Exp Metastasis 25: 843–854 [DOI] [PubMed] [Google Scholar]
- Trejo CL, Juan J, Vicent S, Sweet-Cordero A, McMahon M 2012. MEK1/2 inhibition elicits regression of autochthonous lung tumors induced by KRASG12D or BRAFV600E. Cancer Res 72: 3048–3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempus CS, Morris RJ, Bortner CD, Cotsarelis G, Faircloth RS, Reece JM, Tennant RW 2003. Enrichment for living murine keratinocytes from the hair follicle bulge with the cell surface marker CD34. J Invest Dermatol 120: 501–511 [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma AK, Boutwell RK 1980. Effects of dose and duration of treatment with the tumor-promoting agent, 12-O-tetradecanoylphorbol-13-acetate on mouse skin carcinogenesis. Carcinogenesis 1: 271–276 [DOI] [PubMed] [Google Scholar]
- Viaje A, Slaga TJ, Wigler M, Weinstein IB 1977. Effects of antiinflammatory agents on mouse skin tumor promotion, epidermal DNA synthesis, phorbol ester-induced cellular proliferation, and production of plasminogen activator. Cancer Res 37: 1530–1536 [PubMed] [Google Scholar]
- Vidal VP, Chaboissier MC, Lutzkendorf S, Cotsarelis G, Mill P, Hui CC, Ortonne N, Ortonne JP, Schedl A 2005. Sox9 is essential for outer root sheath differentiation and the formation of the hair stem cell compartment. Curr Biol 15: 1340–1351 [DOI] [PubMed] [Google Scholar]
- Visvader JE 2009. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev 23: 2563–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang NJ, Sanborn Z, Arnett KL, Bayston LJ, Liao W, Proby CM, Leigh IM, Collisson EA, Gordon PB, Jakkula L, et al. 2011. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proc Natl Acad Sci 108: 17761–17766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AC, Tran K, Khuu J, Dang C, Cui Y, Binder SW, Lowry WE 2011. Defining the origins of Ras/p53-mediated squamous cell carcinoma. Proc Natl Acad Sci 108: 7425–7430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong VW, Stange DE, Page ME, Buczacki S, Wabik A, Itami S, van de Wetering M, Poulsom R, Wright NA, Trotter MW, et al. 2012. Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling. Nat Cell Biol 14: 401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Makkinje A, Kyriakis JM 2005. Gene 33 is an endogenous inhibitor of epidermal growth factor (EGF) receptor signaling and mediates dexamethasone-induced suppression of EGF function. J Biol Chem 280: 2924–2933 [DOI] [PubMed] [Google Scholar]