Abstract
Canonical Wnt signaling regulates the transcription of T-cell factor (TCF)-responsive genes through the stabilization and nuclear translocation of the transcriptional co-activator, β-catenin. Overexpression of β-catenin features prominently in acute myeloid leukemia (AML) and has previously been associated with poor clinical outcome. Overexpression of γ-catenin mRNA (a close homologue of β-catenin) has also been reported in AML and has been linked to the pathogenesis of this disease, however, the relative roles of these catenins in leukemia remains unclear. Here we report that overexpression and aberrant nuclear localization of γ-catenin is frequent in AML. Significantly, γ-catenin expression was associated with β-catenin stabilization and nuclear localization. Consistent with this, we found that ectopic γ-catenin expression promoted the stabilization and nuclear translocation of β-catenin in leukemia cells. β-Catenin knockdown demonstrated that both γ- and β-catenin contribute to TCF-dependent transcription in leukemia cells. These data indicate that γ-catenin expression is a significant factor in the stabilization of β-catenin in AML. We also show that although normal cells exclude nuclear translocation of both γ- and β-catenin, this level of regulation is lost in the majority of AML patients and cell lines, which allow nuclear accumulation of these catenins and inappropriate TCF-dependent transcription.
Keywords: Wnt signaling, catenin, TCF/LEF, acute myeloid leukemia, nuclear translocation, transcription
Introduction
Wnt signaling is an evolutionary conserved pathway critical for normal developmental processes in both the embryo and adult, including cell growth and differentiation.1 In the absence of an external Wnt ligand, the canonical pathway is maintained in a state of suppression through constitutive degradation of the central mediator, β-catenin. This degradation is mediated through a destruction complex consisting of CK-1, GSK-3β, axin-1 and APC, which promotes its proteosomal degradation in the cytoplasm. The canonical pathway becomes activated upon binding of an exogenous Wnt ligand to the frizzled/lipoprotein receptor-related protein family of receptors. Activation of these receptors leads to the recruitment of disheveled, a failure of destruction complex assembly and consequent stabilization of β-catenin. A second requirement for canonical Wnt signaling is that β-catenin enters the nucleus where it associates with members of the T-cell factor (TCF)/LEF family of DNA-bound transcription factors on TCF-binding elements to mediate activation of Wnt target genes, such as c-myc and cyclin D1.1
Wnt signaling is known to be active in normal hematopoietic development. The expression of multiple Wnt genes has been identified in human hematopoiesis, and Wnt factors can influence the proliferation and differentiation of hematopoietic stem/progenitor cells (HSPCs).2 Despite these findings, it remains controversial whether Wnt signaling is actually required for hematopoiesis because catenin knockout studies have demonstrated functional redundancy.3, 4, 5 Recently, it has been proposed that canonical Wnt signaling is required for hematopoietic stem cell (HSC) self-renewal but the level of signaling required for normal HSC reconstitution is very low. In support of this, conditional overexpression of a stabilized form of β-catenin in murine models leads to a block in multilineage differentiation and a transient expansion of the HSC pool, but also caused exhaustion of long-term HSCs.6
Dysregulation of Wnt signaling has been reported in a number of hematological malignancies, including chronic lymphocytic leukemia,7 chronic myelogenous leukemia8 and acute lymphoblastic leukemia,9 and has been identified as one of the key signaling networks dysregulated in acute myeloid leukemia (AML).10 Specifically, overexpression of the key mediator, β-catenin, has been identified in myeloid cell lines, as well as primary AML blasts,11, 12, 13, 14, 15 where it has been associated with poor survival.16, 17, 18 The role of β-catenin overexpression in the pathogenesis of AML is unclear; however, it has previously been associated with the establishment and maintenance of leukemia-initiating cells.8, 19, 20 The close structural and functional homologue of β-catenin, γ-catenin (aka plakoglobin, JUP), has also been found to be dysregulated in AML.21, 22, 23 Like β-catenin, γ-catenin is regulated by the same destruction complex and has established roles in cell adhesion; however, its role as a transcriptional activator of Wnt signaling is more contentious.24 Ectopic expression of γ-catenin was found to accelerate the cell-cycle progression of murine HSPC and to promote self-renewal in vitro and leukemogenic capacity when transplanted in vivo.22 Despite these studies, there has been no systematic examination of γ-catenin expression (and corresponding analysis of β-catenin) at the protein level in AML, which might indicate whether γ-catenin represents an alternative, or additional, mechanism of transcriptional activation in AML. Here we have undertaken a comprehensive analysis of cytosolic and nuclear expression of γ- and β-catenin expression in AML and show that the expression and translocation of these proteins is linked in this disease. γ-Catenin expression in the context of myeloid leukemia cells promotes the stabilization of β-catenin and its nuclear translocation, leading to increased TCF-dependent transcription. We also show that the ability of γ-catenin to influence β-catenin expression is abnormal, because their expression is independently regulated in HSPC. Further, although nuclear accumulation of both catenins is not observed in normal HSPC, the majority of leukemia cells allow the nuclear accumulation of γ- and β-catenin and the consequent activation of TCF-dependent transcription.
Materials and methods
Normal and AML patient cells
Peripheral blood mononuclear cells (MNCs) or bone marrow MNCs were collected from AML patients enrolled in the UK Medical Research Council/National Cancer Research Institute 10–15 AML clinical trials at point of diagnosis; see Supplementary Table S1. Normal human CD34+ HSPCs were isolated from cord blood MNC using MiniMACS (Miltenyi Biotec, Bisley, UK). All samples were obtained with informed consent and with approval from the South East Wales Research Ethics Committee in accordance with the 1964 Declaration of Helsinki. MNCs used in this study were separated on Ficoll-Hypaque (Sigma-Aldrich, Poole Dorset, UK) as previously described.23
Retroviral transduction and flow cytometric analysis
Normal human CD34+ HSPCs were retrovirally transduced with PINCO-γ-catenin (kindly provided by Martin Ruthardt22) on retronectin-coated tissue culture plates as previously described.23 The leukemic cell lines K562, U937, HEL and THP-1 (European Collection of Cell Cultures, Salisbury, UK) were cultured as recommended and retrovirally transduced with pBabe-puro γ-catenin using a similar protocol as above. Prior to γ-catenin transduction, K562 and U937 cells were lentivirally transduced with the β-catenin-activated reporter (BAR) system (kindly provided by RT Moon, Washington, USA25). pBARVUbR contains a concatamer of 12 TCF response elements upstream of a β-globin minimal promoter linked to Venus (a variant of EYFP) and constitutively expresses DsRed as a selectable marker. As a control for specificity, K562 cells were transduced with the ‘found unresponsive' BAR (pfuBARVUbR) reporter, which contains mutated TCF response elements. K562 cells overexpressing γ-catenin were further transduced with a lentivirus encoding β-catenin small hairpin RNA (a gift from Bob Weinberg; Addgene plasmid 18803 (Cambridge, MA, USA)). Catenin or control-transduced cell lines were assessed for TCF reporter activity by flow cytometric analysis. Cells expressing the BARV reporter system (as identified by coexpression of DsRed marker) were analyzed using the Accuri C6 cytometer (Accuri Cytometers, Ann Arbor, MI, USA). Data analysis was performed using FCS Express version 4 (De Novo Software, Los Angeles, CA, USA).
Assessment of catenin mRNA expression using quantitative real-time PCR
Assessment of catenin protein expression using western blotting
Total protein lysates were prepared as previously described.26 Nuclear and cytosolic protein fractions were prepared using a cell fractionation kit (Biovision, Mountain View, CA, USA) according to the manufacturer's instructions. Sodium dodecyl sulfate polyacrylamide gel electrophoresis and electroblotting was performed as previously described26 using anti-γ-catenin (Clone 15; BD, Oxford, UK) or anti-β-catenin (Clone 14; BD). Equal protein loading and purity of cytosolic/nuclear fractions was assessed using anti-β-actin (mAbcam 8226; Abcam, Cambridge, UK), glyceraldehyde 3-phosphate dehydrogenase (6C5; Santa Cruz Biotechnology, Santa Cruz, CA, USA) and histone H1 (AE-4; AbD Serotec, Kidlington, UK), respectively. Densitometry was performed using Advanced Image Data Analyzer software v4.5 (Raytek Scientific, Sheffield, UK) and relative catenin levels correlated using a Spearman's coefficient (R). Equivalent amounts of cytosolic and nuclear protein were loaded for each patient sample and catenin protein expression values were normalized to a standard loading of K562 lysate. In some experiments, cells were treated for 16 h with 2.5 μℳ of the GSK inhibitor BIO ((2'Z,3'E)-6-Bromoindirubin-3'-oxime; Tocris, Bristol, UK) prior to fractionation.
Assessment of γ-catenin localization using confocal laser scanning microscopy
AML MNCs or normal CD34+ HSPCs (up to 2 × 106) were fixed in 2% paraformaldehyde (Sigma-Aldrich) and permeabilised using 0.1% Triton X-100, prior to 30 min incubation with IgG2a monoclonal anti-γ-catenin antibody (M111; Abcam). Subsequently, cells were incubated with monoclonal rat anti-mouse-IgG2a-FITC (LO-MG2a-9; AbD Serotec) for 30 min at 4 °C. Cells were finally suspended in FACSFlow (BD) containing 165 nℳ TO-PRO-3 iodide (Invitrogen, Paisley, UK) and analyzed using DMIRBE2 light microscope (Leica, Buckinghamshire, UK). Confocal immunofluorescence was analyzed using the resonant scanning head of a TCS SP2 confocal laser microscope (Leica) with a × 63 oil immersion objective NA 1.32 (HCX-PL-APO). The threshold for catenin fluorescence was determined using isotype-matched primary control antibodies. Z-sections, exhibiting representative cytosolic and nuclear areas, were collected for 50 cells per patient and post-acquisition analysis of γ-catenin localization was performed using Leica confocal software ‘Lite' version.
Statistical analyses
Significance of difference using the Student's t-test, and strength of correlation using a Spearman's or Pearson's coefficient (R), were performed using GraphPad Prism v5.01 (GraphPad Software, Inc., San Diego, CA, USA).
Results
γ-Catenin protein is frequently overexpressed in AML
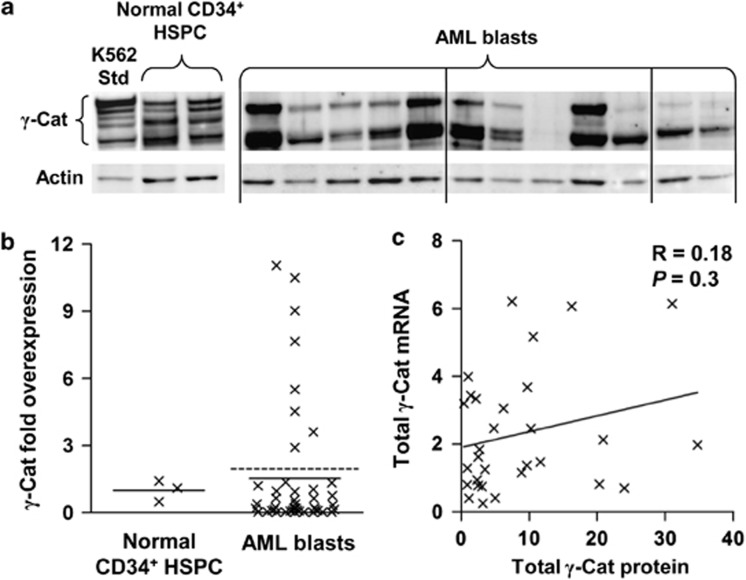
Previously, we have shown γ-catenin mRNA to be frequently overexpressed in AML patients,23 however, γ-catenin expression is also regulated at the protein level via the axin–CK-1–GSK3β–APC complex.24 Therefore, we examined whether there was evidence of γ-catenin overexpression at the protein level and whether this correlated with γ-catenin mRNA expression in AML patients. We characterized γ-catenin expression in 44 AML patients (Cohort 1 of Supplementary Table S1 and S2). γ-Catenin protein expression in AML blasts exhibited a multiple banding pattern (Figure 1a), which has been observed previously for β-catenin13, 16, 27 and is attributed to posttranslational modifications.11, 28 This pattern was found to be identical regardless of antibody used for detection (Supplementary Figure S1) and was also observed when γ-catenin was ectopically expressed (see below). Expression of γ-catenin was found to be highly heterogeneous, being undetectable in 20% (9/44) of patients and overexpressed in 18% (8/44) of patients relative to normal CD34+ HSPC (Figure 1b). This frequency of overexpression was much lower than that observed at the mRNA level from our previous analysis (62% 114/184).23 We observed no correlation with French–American–British type and no preferential overexpression of γ-catenin in the small number of patients assayed (4/44) with core-binding factor abnormalities, which have previously been reported to have high γ-catenin mRNA.21, 23
Figure 1.
γ-Catenin protein is frequently overexpressed in AML blasts. (a) Representative western blot data showing γ-catenin (γ-Cat) protein expression in AML patient blast samples and normal CD34+ HSPC. K562 lysate was used to standardize the analysis. (b) Summary of γ-catenin fold overexpression in AML blasts (n=44) relative to normal CD34+ HSPC (n=3). Bars represent the mean expression from each cohort; the dashed bar represents the threshold of γ-catenin overexpression (mean of normal HSPC+2 s.d.). (c) Correlation between γ-catenin mRNA expression (derived from a validated Affymetrix probeset (Santa Clara, CA, USA); see Supplementary Figure S2) and γ-catenin protein expression (arbitrary densitometric units; see Materials and Methods) in AML blasts (n=30).
To better understand the mechanism of γ-catenin overexpression in AML blasts (that is, transcriptional vs post-transcriptional), we correlated mRNA and protein levels in these patients. Though we found correlation of mRNA and protein levels for some patients, we also observed patients that displayed high protein levels despite of low mRNA expression and, conversely, patients with high mRNA but low levels of protein (Figure 1c). Overall, this resulted in a poor correlation (R=0.18, n=30). These data suggest that although mRNA level may be influential, γ-catenin protein expression is strongly affected by post-transcriptional mechanisms.
γ-Catenin is aberrantly localized in AML
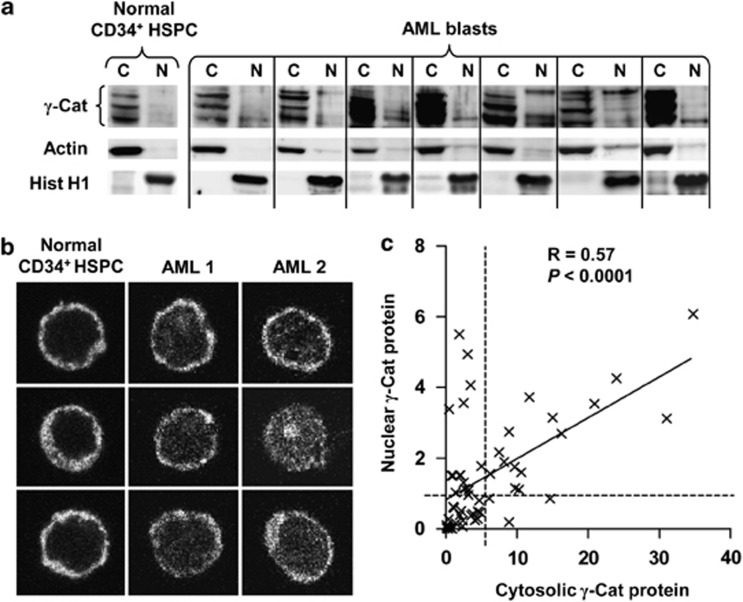
γ-Catenin serves functions in both the cytoskeleton and in transcriptional control in the nucleus, therefore subcellular location can act as an indicator of how γ-catenin functions in AML. We therefore examined the subcellular localization of γ-catenin protein in both normal CD34+ HSPC and primary AML patient blasts. Using nuclear/cytosolic fractionation and western blotting of 59 AML patients (cohort 2 of Supplementary Tables S1 and S3), nuclear-localized γ-catenin was found at very low levels in normal CD34+ HSPC (Figure 2a). In contrast, 35/59 (59%) of AML patients exhibited significantly higher levels of nuclear-translocated γ-catenin, an observation confirmed by confocal analysis (Figure 2b, Supplementary Figure S3 and Supplementary Table S4). Furthermore, nuclear γ-catenin was found to correlate (R=0.57, P<0.0001) with corresponding cytosolic expression level in AML blasts (Figure 2c). High levels of nuclear γ-catenin were also observed in patients with normal cytosolic levels, therefore the frequency of aberrant nuclear localization of γ-catenin was much higher than its frequency of overexpression (59% vs 18%). These data suggest that although nuclear localization of γ-catenin is not observed in normal HSPC, it readily accumulates in the nuclei of AML blasts.
Figure 2.
γ-Catenin subcellular localization is dysregulated in AML blasts. (a) Representative western blot data showing subcellular localization (C, cytosolic and N, nuclear) of γ-catenin protein in normal CD34+ HSPC and AML blasts. Actin and histone H1 (Hist H1) show relative protein loading and fraction purity. (b) Representative examples of confocal laser scanning microscopy Z-sections demonstrating the subcellular localization of γ-catenin immunofluorescence within normal CD34+ HSPC and AML patient blasts (original magnification × 63). Color-merged images showing nuclear localization are shown in Supplementary Figure S3. (c) Summary data showing the correlation between cytosolic γ-catenin and nuclear γ-catenin protein expression within AML blasts (n=59; arbitrary densitometric units). The dashed lines represent the threshold of γ-catenin overexpression for nuclear and cytosolic fractions (mean of normal HSPC+2 s.d.).
γ-Catenin protein expression correlates with that of β-catenin in AML blasts
The data above demonstrate stabilization of γ-catenin protein independent of mRNA expression level, suggesting in many patients that this resulted from post-transcriptional dysregulation. Because both γ-catenin and β-catenin protein stability is regulated by the destruction complex, this implies that patients overexpressing γ-catenin may also overexpress β-catenin. This could arise either because a defect in the degradation complex might mutually stabilize these proteins (as observed in colon cancer29) and/or because high levels of γ-catenin may saturate the destruction complex indirectly stabilizing β-catenin.30 To assess this, we reanalysed the cohort above for nuclear and cytosolic levels of β-catenin. Given the structural homology shared between γ- and β-catenin molecules,31 we first confirmed no cross-reactivity between the respective antibodies before such analyses were performed (data not shown). We determined whether, like γ-catenin, β-catenin protein expression was independent of mRNA level. This proved to be the case (R=0.23, n=30; data not shown), suggesting that β-catenin protein levels are also predominantly post-transcriptionally regulated. We also found β- and γ-catenin mRNA levels correlated poorly (R=0.27, n=30). On the other hand, we found a close correlation between γ- and β-catenin protein expression in AML blasts (R=0.51, P<0.01; Figures 3a and b, Supplementary Table S5). Furthermore, as with γ-catenin (R=0.57, P<0.0001), we also observed a significant correlation between β-catenin expression and its nuclear localization (R=0.56, P<0.001; Figure 3c). These data suggest that, like γ-catenin, β-catenin protein expression in AML cells is heavily influenced by post-transcriptional processes and that γ- and β-catenin are mutually stabilized and translocated to the nucleus in these cells.
Figure 3.
γ-Catenin protein expression correlates with β-catenin protein expression in AML blasts. (a) Representative western blot images demonstrating the localization of γ-catenin and β-catenin (β-Cat) protein within normal CD34+ HSPC and AML patient blasts. Protein loading and fraction purity were assessed by actin and histone H1 detection. (b) Summary data showing the correlation between total γ-catenin protein and total β-catenin protein in AML blasts (n=59; arbitrary densitometric units). (c) Summary data showing the correlation between cytosolic levels of β-catenin and the corresponding level of nuclear β-catenin protein in AML blasts (n=59; arbitrary densitometric units).
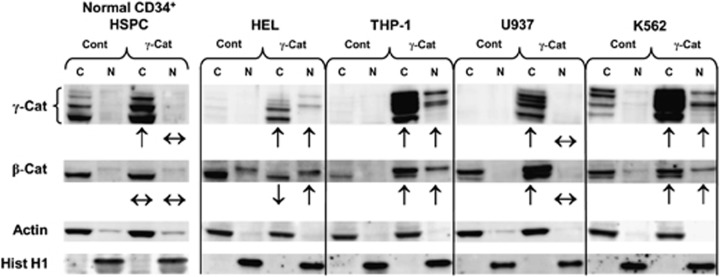
Ectopic γ-catenin expression stabilizes β-catenin in AML blasts but not in normal cells
To establish whether the correlation between γ- and β-catenin protein expression observed in primary AML blasts was functionally driven, we examined the effect of ectopic γ-catenin expression on β-catenin level and the localization in both normal CD34+ HSPC cells and leukemia cell lines. Ectopic expression of γ-catenin in CD34+ HSPC did not promote nuclear localization of this protein, nor did it influence the expression or nuclear translocation of β-catenin, suggesting that in normal human hematopoietic cells, the translocation and expression of these proteins is tightly and independently regulated (Figure 4). We next examined the effect of ectopic γ-catenin overexpression in four myeloid leukemia cell lines. In all the cell lines, except U937 cells, ectopic expression of γ-catenin promoted its nuclear localization (Figure 4). This contrasts with normal cells and suggests that in most leukemia cells the mechanisms preventing nuclear localization of γ-catenin have been disturbed. These data also support our observations in primary AML (above) showing a link between the level of expression and nuclear localization. Examination of the effects on β-catenin expression showed that in each case, except HEL cells, overexpression of γ-catenin also promoted the stabilization of β-catenin. This is again consistent with the observations of AML blasts and highlights a further abnormality of leukemic cells, because in normal HSPC, γ-catenin overexpression did not influence β-catenin expression. As with γ-catenin, stabilization of β-catenin in AML cells tended to promote its nuclear localization (the principal exception being U937 cells where nuclear localization of both catenins was not detected).
Figure 4.
γ-Catenin overexpression promotes β-catenin stabilization and translocation in leukemic blasts, but not in normal hematopoietic cells. Representative western blot data showing the subcellular expression of γ-catenin and β-catenin in normal CD34+ HSPC and myeloid leukemia cell lines transduced with either empty vector (Cont) or γ-catenin. Arrows summarize the effect of γ-catenin overexpression on cytosolic and nuclear levels of γ- and β-catenin. Protein loading and fraction purity are indicated by actin and histone H1.
To investigate whether nuclear localization of γ-catenin might be promoting β-catenin transcription, we examined whether γ-catenin overexpression influenced β-catenin mRNA levels in these cells. Comparison of β-catenin mRNA expression between control and γ-catenin-transduced cell lines showed no change in β-catenin mRNA levels (data not shown). This was in agreement with our analysis of AML patients and suggested that the stabilization of β-catenin was mediated through a post-transcriptional mechanism. In support of this, we found that β-catenin levels were acutely sensitive to the GSK inhibitor, BIO, implying rapid turnover of this protein in leukemia cell lines (Supplementary Figure S4).
Together, these data suggest that AML cells can differ in two respects compared with normal cells. First, that expression of γ-catenin promotes the stabilization of β-catenin and second, that the stabilization of catenins also leads to their nuclear translocation.
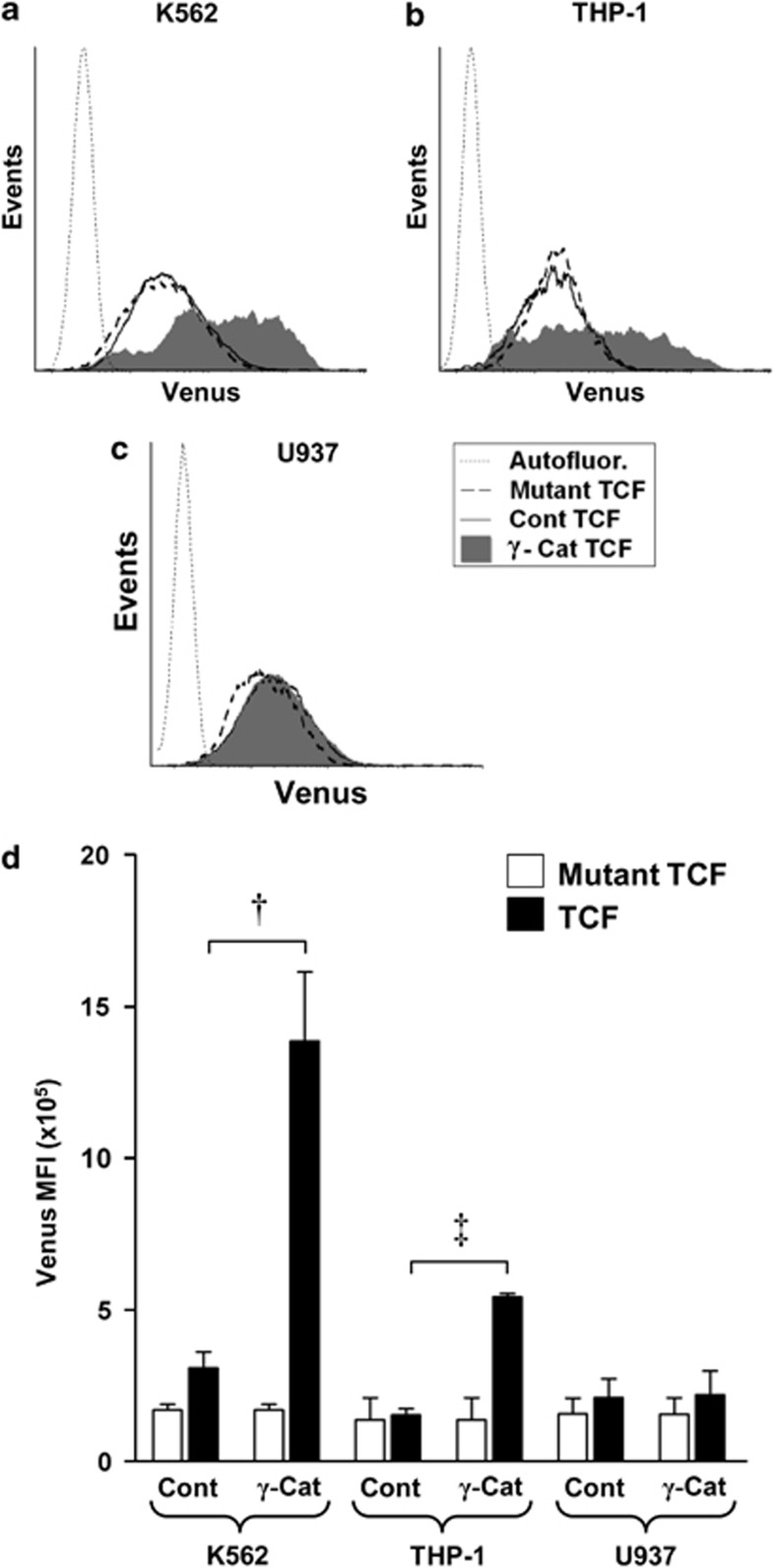
γ-Catenin overexpression promotes TCF transcription both directly, and indirectly through β-catenin stabilization
The data above predict that overexpression of γ-catenin will promote TCF-mediated transcription in leukemia cells, which respond through coordinate stabilization of β-catenin expression and nuclear localization of both catenins. To investigate this, we examined the consequence of catenin dysregulation on TCF-dependent transcription in K562, THP-1 and also U937 cells (which retained the resistance to catenin translocation characteristic of normal cells). To accomplish this, these cells were transduced with the pBARVUbR lentiviral construct, which reports TCF transcription through expression of Venus EYFP. As shown in Figure 5, no cell line examined demonstrated significant endogenous TCF reporter activity compared with control (reporter incorporating mutant TCF sites). However, overexpression of γ-catenin in K562 and THP-1 cells significantly promoted reporter expression 4.5- and 3.9-fold over control cells, respectively (Figures 5a). As expected, overexpression of γ-catenin in U937 cells gave no detectable response relative to controls (Figures 5c and d), reflecting the resistance of these cells to catenin translocation despite their cytosolic stabilization (Figure 4).
Figure 5.
γ-Catenin overexpression promotes TCF/LEF-mediated transcription in leukemic cells. (a, b, c) Representative flow cytometric histograms showing TCF reporter activity (Venus EYFP) in control (Cont TCF) or γ-catenin-transduced (γ-Cat TCF) K562, THP-1 and U937 cells. Autofluorescence levels (Autofluor.) were established using parental cells. Background fluorescence arising from construct integration is indicated by control cells transduced with an identical construct bearing mutated TCF-binding sites (Mutant TCF). (d) Summary data showing the relative Venus EYFP intensity obtained from control or γ-catenin-transduced K562, THP-1 and U937 cell lines. Data represents mean±1 s.d., n=3. Statistical significance is denoted by †P<0.01 and ‡P<0.05, as analyzed by Student's t-test.
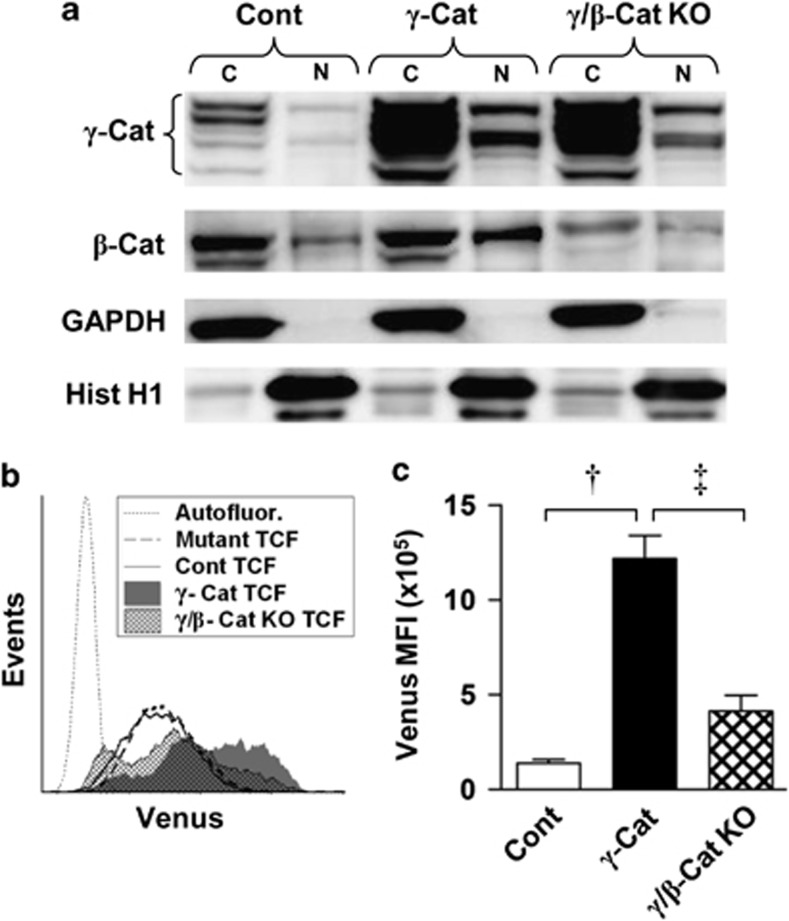
These data showed that, as predicted, nuclear translocation of catenins was able to promote TCF-dependent transcription; however, it remained unclear whether the effect of γ-catenin on TCF activity was direct or indirect through its stabilization of β-catenin. To address this, we used lentiviral small hairpin RNA to inhibit β-catenin expression in K562 cells overexpressing γ-catenin (which displayed the largest shift in reporter activity; Figure 5d). Western blotting confirmed that nuclear β-catenin was reduced below control levels in these cells, whereas γ-catenin expression remained largely unaffected (Figure 6a). We then examined the effect of β-catenin knockdown on reporter gene expression. We found that, TCF reporter activity was significantly reduced (by 66%) but not entirely ablated upon β-catenin knockdown (Figures 6b and c). These data indicate that γ-catenin acts both directly and indirectly (through stabilization of β-catenin) to promote TCF-dependent transcription in hematopoietic cells.
Figure 6.
γ-Catenin promotes TCF/LEF-mediated transcription in leukemic cells both directly and indirectly through stabilization of β-Cat. (a) Representative western blot showing the expression and localization of γ- and β-catenin protein in K562 cells transduced with empty vector (Cont), γ-Catenin or γ-catenin together with β-catenin small hairpin RNA (γ/β-Cat KO). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and histone H1 demonstrate relative protein loading and fraction purity. (b) Representative flow cytometric histograms showing TCF reporter activity in K562 cell lines. (c) Summary data showing the background-subtracted TCF reporter signal (n=3). Data represents mean±1 s.d. Statistical significance is denoted by †P<0.01 and ‡P<0.05, as analyzed by Student's t-test.
Discussion
This study is the first to demonstrate dysregulated γ-catenin protein expression in AML or indeed in any hematological malignancy, with 18% of patient samples exhibiting overexpression. Though overexpression of γ-catenin mRNA has been previously reported,21, 23 this had a minor contribution to protein overexpression in our study, suggesting that post-translational mechanisms are dominant in driving overexpression. Discordance with mRNA has been previously reported in a variety of other contexts for both γ-32, 33, 34 and β-catenin14, 16, 35 and is consistent with the view that their expression is principally regulated through the activity of (and their susceptibility to) the destruction complex.24 Mutations affecting these factors are the principal drivers of β-catenin overexpression in epithelial cancers;1 however, such mutations have not been identified in hematopoietic malignancies, suggesting that alternative mechanisms are responsible. Here we demonstrate that overexpression of γ-catenin is able to fulfill that role in myeloid leukemia. γ-Catenin expression correlated with β-catenin expression in AML blasts, and ectopic expression of γ-catenin was able to stabilize β-catenin expression in a variety of myeloid leukemia cell lines. The mechanism driving overexpression of γ-catenin has not currently been resolved but is likely to be multifactorial. Though mRNA levels correlated poorly with protein level overall, they did appear to be influential in some patients. In other tissues, previous reports have indicated that γ-catenin can be stabilized through increased ser/thr phosphorylation,36 though without suitable antibodies this is difficult to establish in primary material. The costabilization of β-catenin has been reported previously in other contexts and has been proposed to arise through competitive inhibition of the destruction complex, given that γ-catenin is less efficiently degraded than β-catenin.24, 37 This alone may be an insufficient explanation, however, given that this costabilization of β-catenin was not detectable in normal cells. Thus, in addition to γ-catenin overexpression, the costabilization of β-catenin by γ-catenin is also an abnormal feature of AML blasts.
A further abnormality demonstrated by AML cells was the degree of nuclear translocation of both catenins. Overexpression of catenins alone is insufficient to promote their transcriptional activity, which requires their translocation and retention in the nucleus. According to the standard model of canonical Wnt signaling, cytosolic stabilization of catenins is sufficient to promote their translocation and this was indeed seen for both AML blasts and the majority of cell lines examined. It is clear from our data, however, that this represents an aberration of AML cells because normal cells do not show nuclear accumulation of γ-catenin when overexpressed. Similarly, stabilization of β-catenin through inhibition of the destruction complex in HSPC also fails to promote its nuclear localization (Supplementary Figure S4). HSPC do not therefore adhere to the standard model, suggesting that an additional level of regulation exists in hematopoietic cells that restricts the nuclear translocation of catenins. Given the evidence that high levels of canonical Wnt signaling are deleterious to HSC,38 this additional level of regulation may be necessary to protect self-renewal capacity. Evidently, this is not the case for most AML blasts and cells lines, which either tolerate or benefit from high levels of nuclear catenin. Interestingly, one of the cell lines examined (U937) retained the resistance to nuclear accumulation of catenin displayed by normal cells; similarly, Figure 3c shows a subgroup of patients with comparatively high cytosolic β-catenin levels but with undetectable nuclear expression, again indicating that loss of translocational control of catenins is not a universal feature of AML. The movement of large proteins, such as catenins, requires active transport across the nuclear membrane. Both β- and γ-catenin lack nuclear localization/export signals39 and do not depend on importins or karyopherins for nuclear translocation.40 A large number of proteins have been implicated in the nuclear transport and retention of catenins, including LEF-1,37 TCF4,41 APC39 and FoxM1,42 but their involvement is highly context-dependent and the processes mediating nuclear localization of catenins are generally poorly understood. In the hematopoietic context, it has been reported that FLT3-ITD may promote translocation of β-catenin through tyrosine phosphorylation at Y654.43, 44 We were unable to confirm an association between FLT3-ITD and nuclear localization of β-catenin in our patient cohort (9/39 FLT3-ITD patients P=0.75). We also found no association between Y654 phosphorylation and β-catenin nuclear localization in our cell line panel (data not shown). Whatever the mechanisms controlling nuclear localization of catenins, it is likely that these are at least partly shared between γ- and β-catenin because all the leukemic cell lines assayed, and the majority of AML patient samples, demonstrated concurrent translocation of these catenins. The coordinated localization of these proteins to the nucleus has previously been observed in other contexts.37, 45
The coordinate translocation of each catenin raised the issue of what role nuclear localization of γ-catenin might have in TCF-dependent transcriptional activation. Previous studies in epithelial cells have indicated that γ-catenin can activate transcription, though with less efficiency than β-catenin.37, 45, 46, 47, 48, 49, 50 Studies have also shown that γ-catenin is transcriptionally active on β-catenin null backgrounds.45, 46, 51 In this study, selective knockdown of β-catenin in γ-catenin-overexpressing cells showed that γ-catenin contributes both directly and indirectly (through stabilization of β-catenin) to promote TCF-dependent transcription myeloid leukemia cells. Functionally, γ-catenin has been shown to promote self-renewal and leukemia in mouse HSC22 (properties that have also been attributed to β-catenin52). Though the corresponding effects on β-catenin expression were not reported in these studies, our data suggest that the effects of γ-catenin overexpression could be mediated, at least in part, through stabilization of β-catenin.
In summary, this study has identified three key abnormalities associated with canonical Wnt signaling in AML: the overexpression of γ-catenin, the costabilization of β-catenin and the permissive nuclear localization of both catenins. These abnormalities combine to promote TCF-dependent transcription in AML and stand in contrast to catenin regulation in normal cells, which appear to be regulated to strongly resist high levels of Wnt signaling. Further investigations are now required in order to elucidate the mechanisms regulating catenin nuclear transport in normal and leukemic cells, which could be of therapeutic interest.
Acknowledgments
This study was funded by the Leukemia and Lymphoma Research, UK, and the Medical Research Council, UK.
Author contributions
RGM, RLD and AT cowrote the manuscript and analyzed the data. LP, SLD and KL provided technical assistance. AKB provided resources and clinical insight. RGM designed and executed experiments, and RLD and AT designed experiments and provided project direction.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Leukemia website (http://www.nature.com/leu)
Supplementary Material
References
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Staal FJ, Luis TC. Wnt signaling in hematopoiesis: crucial factors for self-renewal, proliferation, and cell fate decisions. J Cell Biochem. 2010;109:844–849. doi: 10.1002/jcb.22467. [DOI] [PubMed] [Google Scholar]
- Cobas M, Wilson A, Ernst B, Mancini SJ, Macdonald HR, Kemler R, et al. Beta-catenin is dispensable for hematopoiesis and lymphopoiesis. J Exp Med. 2004;199:221–229. doi: 10.1084/jem.20031615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch U, Wilson A, Cobas M, Kemler R, Macdonald HR, Radtke F. Simultaneous loss of beta- and gamma-catenin does not perturb hematopoiesis or lymphopoiesis. Blood. 2008;111:160–164. doi: 10.1182/blood-2007-07-099754. [DOI] [PubMed] [Google Scholar]
- Jeannet G, Scheller M, Scarpellino L, Duboux S, Gardiol N, Back J, et al. Long-term, multilineage hematopoiesis occurs in the combined absence of β-catenin and γ-catenin. Blood. 2008;111:142–149. doi: 10.1182/blood-2007-07-102558. [DOI] [PubMed] [Google Scholar]
- Kirstetter P, Anderson K, Porse BT, Jacobsen SE, Nerlov C. Activation of the canonical Wnt pathway leads to loss of hematopoietic stem cell repopulation and multilineage differentiation block. Nat Immunol. 2006;7:1048–1056. doi: 10.1038/ni1381. [DOI] [PubMed] [Google Scholar]
- Lu D, Zhao Y, Tawatao R, Cottam HB, Sen M, Leoni LM, et al. Activation of the Wnt signaling pathway in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2004;101:3118–3123. doi: 10.1073/pnas.0308648100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson CH, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351:657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- Khan NI, Bradstock KF, Bendall LJ. Activation of Wnt/beta-catenin pathway mediates growth and survival in B-cell progenitor acute lymphoblastic leukaemia. Br J Haematol. 2007;138:338–348. doi: 10.1111/j.1365-2141.2007.06667.x. [DOI] [PubMed] [Google Scholar]
- Majeti R, Becker MW, Tian Q, Lee TL, Yan X, Liu R, et al. Dysregulated gene expression networks in human acute myelogenous leukemia stem cells. Proc Natl Acad Sci USA. 2009;106:3396–3401. doi: 10.1073/pnas.0900089106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung EJ, Hwang SG, Nguyen P, Lee S, Kim JS, Kim JW, et al. Regulation of leukemic cell adhesion, proliferation, and survival by beta-catenin. Blood. 2002;100:982–990. doi: 10.1182/blood.v100.3.982. [DOI] [PubMed] [Google Scholar]
- Serinsoz E, Neusch M, Busche G, Wasielewski R, Kreipe H, Bock O. Aberrant expression of beta-catenin discriminates acute myeloid leukaemia from acute lymphoblastic leukaemia. Br J Haematol. 2004;126:313–319. doi: 10.1111/j.1365-2141.2004.05049.x. [DOI] [PubMed] [Google Scholar]
- Simon M, Grandage VL, Linch DC, Khwaja A. Constitutive activation of the Wnt/beta-catenin signalling pathway in acute myeloid leukaemia. Oncogene. 2005;24:2410–2420. doi: 10.1038/sj.onc.1208431. [DOI] [PubMed] [Google Scholar]
- Gandillet A, Park S, Lassailly F, Griessinger E, Vargaftig J, Filby A, et al. Heterogeneous sensitivity of human acute myeloid leukemia to beta-catenin down-modulation. Leukemia. 2011;25:770–780. doi: 10.1038/leu.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siapati EK, Papadaki M, Kozaou Z, Rouka E, Michali E, Savvidou I, et al. Proliferation and bone marrow engraftment of AML blasts is dependent on beta-catenin signalling. Br J Haematol. 2011;152:164–174. doi: 10.1111/j.1365-2141.2010.08471.x. [DOI] [PubMed] [Google Scholar]
- Ysebaert L, Chicanne G, Demur C, De TF, Prade-Houdellier N, Ruidavets JB, et al. Expression of beta-catenin by acute myeloid leukemia cells predicts enhanced clonogenic capacities and poor prognosis. Leukemia. 2006;20:1211–1216. doi: 10.1038/sj.leu.2404239. [DOI] [PubMed] [Google Scholar]
- Xu J, Suzuki M, Niwa Y, Hiraga J, Nagasaka T, Ito M, et al. Clinical significance of nuclear non-phosphorylated beta-catenin in acute myeloid leukaemia and myelodysplastic syndrome. Br J Haematol. 2008;140:394–401. doi: 10.1111/j.1365-2141.2007.06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Gau JP, You JY, Lee KD, Yu YB, Lu CH, et al. Prognostic significance of beta-catenin and topoisomerase IIalpha in de novo acute myeloid leukemia. Am J Hematol. 2009;84:87–92. doi: 10.1002/ajh.21334. [DOI] [PubMed] [Google Scholar]
- Wang Y, Krivtsov AV, Sinha AU, North TE, Goessling W, Feng Z, et al. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327:1650–1653. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung J, Esposito MT, Gandillet A, Zeisig BB, Griessinger E, Bonnet D, et al. beta-Catenin mediates the establishment and drug resistance of MLL leukemic stem cells. Cancer Cell. 2010;18:606–618. doi: 10.1016/j.ccr.2010.10.032. [DOI] [PubMed] [Google Scholar]
- Muller-Tidow C, Steffen B, Cauvet T, Tickenbrock L, Ji P, Diederichs S, et al. Translocation products in acute myeloid leukemia activate the Wnt signaling pathway in hematopoietic cells. Mol Cell Biol. 2004;24:2890–2904. doi: 10.1128/MCB.24.7.2890-2904.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Beissert T, Kukoc-Zivojnov N, Puccetti E, Altschmied J, Strolz C, et al. Gamma-catenin contributes to leukemogenesis induced by AML-associated translocation products by increasing the self-renewal of very primitive progenitor cells. Blood. 2004;103:3535–3543. doi: 10.1182/blood-2003-09-3335. [DOI] [PubMed] [Google Scholar]
- Tonks A, Pearn L, Musson M, Gilkes A, Mills KI, Burnett AK, et al. Transcriptional dysregulation mediated by RUNX1-RUNX1T1 in normal human progenitor cells and in acute myeloid leukaemia. Leukemia. 2007;21:2495–2505. doi: 10.1038/sj.leu.2404961. [DOI] [PubMed] [Google Scholar]
- Zhurinsky J, Shtutman M, Ben-Ze'ev A. Plakoglobin and beta-catenin: protein interactions, regulation and biological roles. J Cell Sci. 2000;113 (Pt 18:3127–3139. doi: 10.1242/jcs.113.18.3127. [DOI] [PubMed] [Google Scholar]
- Biechele TL, Moon RT. Assaying beta-catenin/TCF transcription with beta-catenin/TCF transcription-based reporter constructs. Methods Mol Biol. 2008;468:99–110. doi: 10.1007/978-1-59745-249-6_8. [DOI] [PubMed] [Google Scholar]
- Hole PS, Pearn L, Tonks AJ, James PE, Burnett AK, Darley RL, et al. Ras-induced reactive oxygen species promote growth factor-independent proliferation in human CD34+ hematopoietic progenitor cells. Blood. 2010;115:1238–1246. doi: 10.1182/blood-2009-06-222869. [DOI] [PubMed] [Google Scholar]
- Brown AL, Salerno DG, Sadras T, Engler GA, Kok CH, Wilkinson CR, et al. The GM-CSF receptor utilizes beta-catenin and Tcf4 to specify macrophage lineage differentiation. Differentiation. 2012;83:47–59. doi: 10.1016/j.diff.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SG, Lee HC, Trepel JB, Jeon BH. Anticancer-drug-induced apoptotic cell death in leukemia cells is associated with proteolysis of beta-catenin. Leuk Res. 2002;26:863–871. doi: 10.1016/s0145-2126(02)00018-8. [DOI] [PubMed] [Google Scholar]
- Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- Miller JR, Moon RT. Analysis of the signaling activities of localization mutants of beta-catenin during axis specification in Xenopus. J Cell Biol. 1997;139:229–243. doi: 10.1083/jcb.139.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solanas G, Miravet S, Casagolda D, Castano J, Raurell I, Corrionero A, et al. Beta-catenin and plakoglobin N- and C-tails determine ligand specificity. J Biol Chem. 2004;279:49849–49856. doi: 10.1074/jbc.M408685200. [DOI] [PubMed] [Google Scholar]
- Bradley RS, Cowin P, Brown AM. Expression of Wnt-1 in PC12 cells results in modulation of plakoglobin and E-cadherin and increased cellular adhesion. J Cell Biol. 1993;123:1857–1865. doi: 10.1083/jcb.123.6.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk AP, Palka HL, Luu HH, Nilles LA, Anderson JE, Wheelock MJ, et al. Posttranslational regulation of plakoglobin expression. Influence of the desmosomal cadherins on plakoglobin metabolic stability. J Biol Chem. 1994;269:31214–31223. [PubMed] [Google Scholar]
- Papagerakis S, Shabana AH, Depondt J, Pibouin L, Blin-Wakkach C, Berdal A. Altered plakoglobin expression at mRNA and protein levels correlates with clinical outcome in patients with oropharynx squamous carcinomas. Hum Pathol. 2004;35:75–85. doi: 10.1016/j.humpath.2003.08.018. [DOI] [PubMed] [Google Scholar]
- Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. Beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasdar M, Li Z, Chlumecky V. Plakoglobin: kinetics of synthesis, phosphorylation, stability, and interactions with desmoglein and E-cadherin. Cell Motil Cytoskeleton. 1995;32:258–272. doi: 10.1002/cm.970320403. [DOI] [PubMed] [Google Scholar]
- Simcha I, Shtutman M, Salomon D, Zhurinsky J, Sadot E, Geiger B, et al. Differential nuclear translocation and transactivation potential of beta-catenin and plakoglobin. J Cell Biol. 1998;141:1433–1448. doi: 10.1083/jcb.141.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis TC, Ichii M, Brugman MH, Kincade P, Staal FJ. Wnt signaling strength regulates normal hematopoiesis and its deregulation is involved in leukemia development. Leukemia. 2012;26:414–421. doi: 10.1038/leu.2011.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson BR, Fagotto F. The ins and outs of APC and beta-catenin nuclear transport. EMBO Rep. 2002;3:834–839. doi: 10.1093/embo-reports/kvf181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagotto F, Gluck U, Gumbiner BM. Nuclear localization signal-independent and importin/karyopherin-independent nuclear import of beta-catenin. Curr Biol. 1998;8:181–190. doi: 10.1016/s0960-9822(98)70082-x. [DOI] [PubMed] [Google Scholar]
- Krieghoff E, Behrens J, Mayr B. Nucleo-cytoplasmic distribution of beta-catenin is regulated by retention. J Cell Sci. 2006;119:1453–1463. doi: 10.1242/jcs.02864. [DOI] [PubMed] [Google Scholar]
- Zhang N, Wei P, Gong A, Chiu WT, Lee HT, Colman H, et al. FoxM1 promotes beta-catenin nuclear localization and controls Wnt target-gene expression and glioma tumorigenesis. Cancer Cell. 2011;20:427–442. doi: 10.1016/j.ccr.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiguchi T, Chung EJ, Lee S, Stine A, Kiyoi H, Naoe T, et al. FLT3 regulates beta-catenin tyrosine phosphorylation, nuclear localization, and transcriptional activity in acute myeloid leukemia cells. Leukemia. 2007;21:2476–2484. doi: 10.1038/sj.leu.2404923. [DOI] [PubMed] [Google Scholar]
- Kajiguchi T, Katsumi A, Tanizaki R, Kiyoi H, Naoe T. Y654 of beta-catenin is essential for FLT3/ITD-related tyrosine phosphorylation and nuclear localization of beta-catenin. Eur J Haematol. 2012;88:314–320. doi: 10.1111/j.1600-0609.2011.01738.x. [DOI] [PubMed] [Google Scholar]
- Maeda O, Usami N, Kondo M, Takahashi M, Goto H, Shimokata K, et al. Plakoglobin (gamma-catenin) has TCF/LEF family-dependent transcriptional activity in beta-catenin-deficient cell line. Oncogene. 2004;23:964–972. doi: 10.1038/sj.onc.1207254. [DOI] [PubMed] [Google Scholar]
- Kolligs FT, Kolligs B, Hajra KM, Hu G, Tani M, Cho KR, et al. Gamma-catenin is regulated by the APC tumor suppressor and its oncogenic activity is distinct from that of beta-catenin. Genes Dev. 2000;14:1319–1331. [PMC free article] [PubMed] [Google Scholar]
- Williams BO, Barish GD, Klymkowsky MW, Varmus HE. A comparative evaluation of beta-catenin and plakoglobin signaling activity. Oncogene. 2000;19:5720–5728. doi: 10.1038/sj.onc.1203921. [DOI] [PubMed] [Google Scholar]
- Zhurinsky J, Shtutman M, Ben-Ze'ev A. Differential mechanisms of LEF/TCF family-dependent transcriptional activation by beta-catenin and plakoglobin. Mol Cell Biol. 2000;20:4238–4252. doi: 10.1128/mcb.20.12.4238-4252.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conacci-Sorrell ME, Ben-Yedidia T, Shtutman M, Feinstein E, Einat P, Ben-Ze'ev A. Nr-CAM is a target gene of the beta-catenin/LEF-1 pathway in melanoma and colon cancer and its expression enhances motility and confers tumorigenesis. Genes Dev. 2002;16:2058–2072. doi: 10.1101/gad.227502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga Y, Liu H, Shimizu M, Komiya S, Kawasuji M, Nagafuchi A. Defining the roles of beta-catenin and plakoglobin in cell-cell adhesion: isolation of beta-catenin/plakoglobin-deficient F9 cells. Cell Struct Funct. 2005;30:25–34. doi: 10.1247/csf.30.25. [DOI] [PubMed] [Google Scholar]
- Kim YM, Ma H, Oehler VG, Gang EJ, Nguyen C, Masiello D, et al. The gamma catenin/CBP complex maintains survivin transcription in beta-catenin deficient/depleted cancer cells. Curr Cancer Drug Targets. 2011;11:213–225. doi: 10.2174/156800911794328420. [DOI] [PubMed] [Google Scholar]
- Reya T, Duncan AW, Ailles L, Domen J, Scherer DC, Willert K, et al. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.