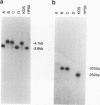
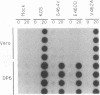
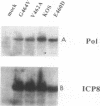
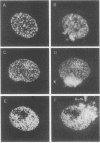
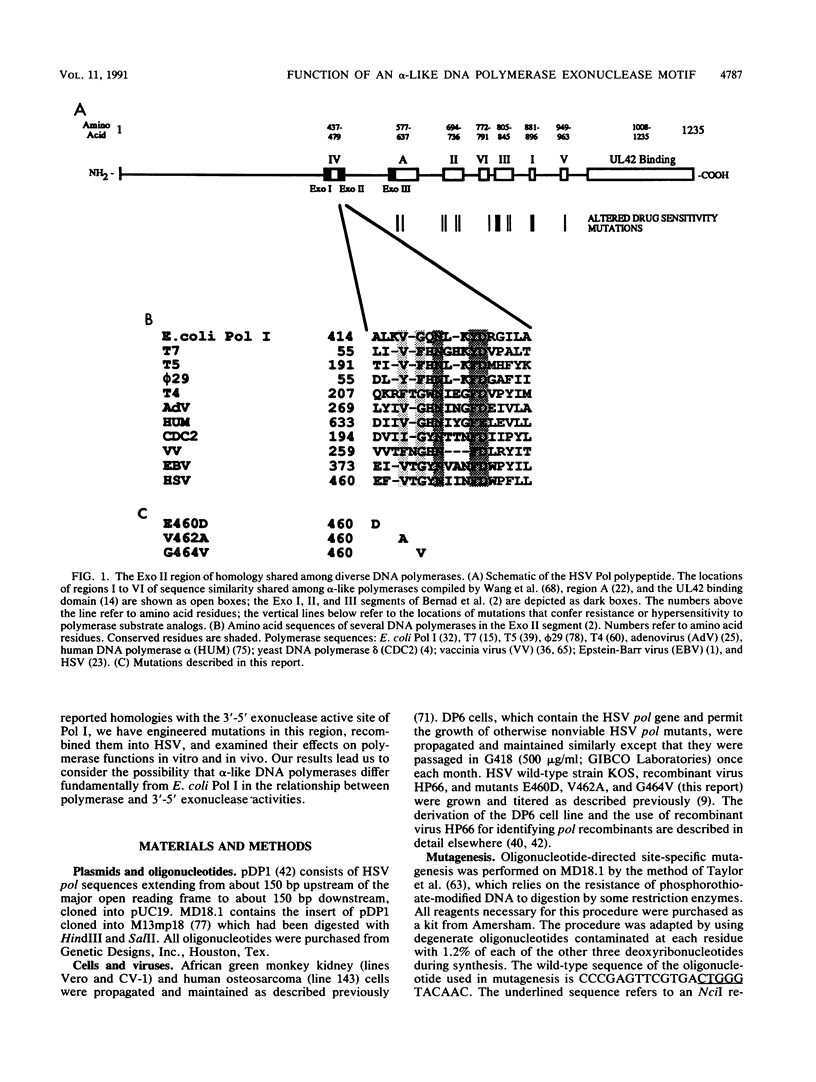
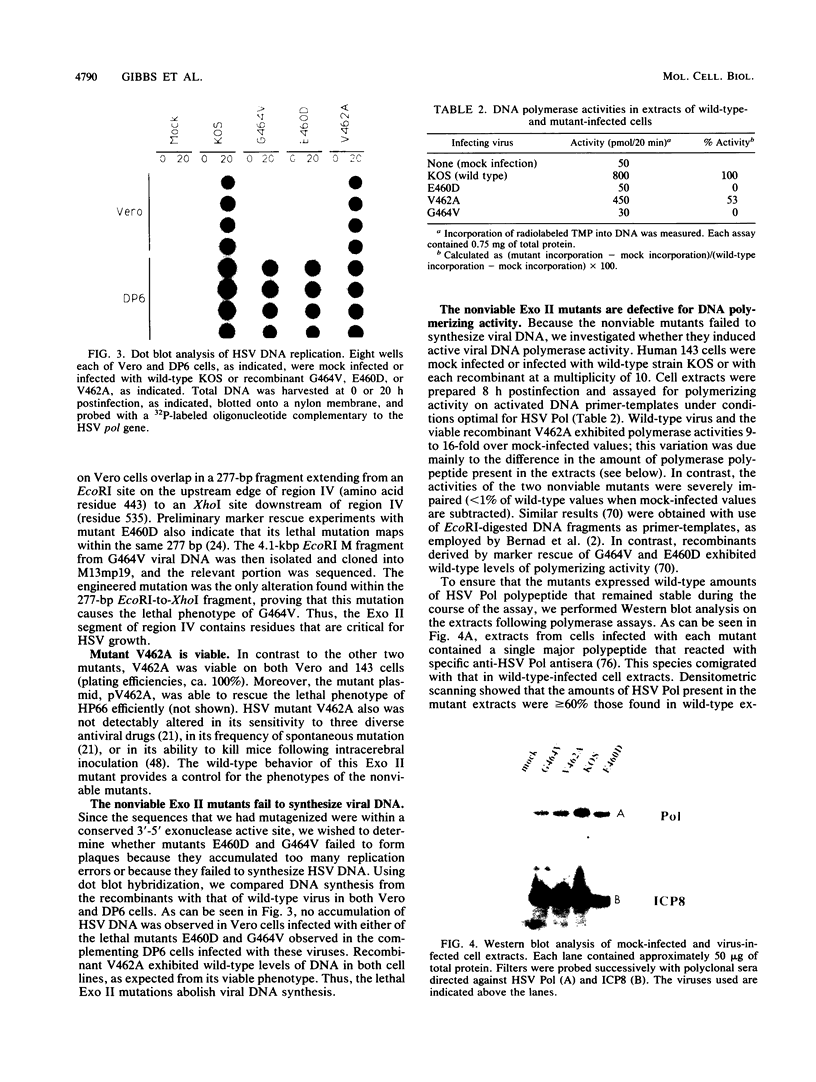
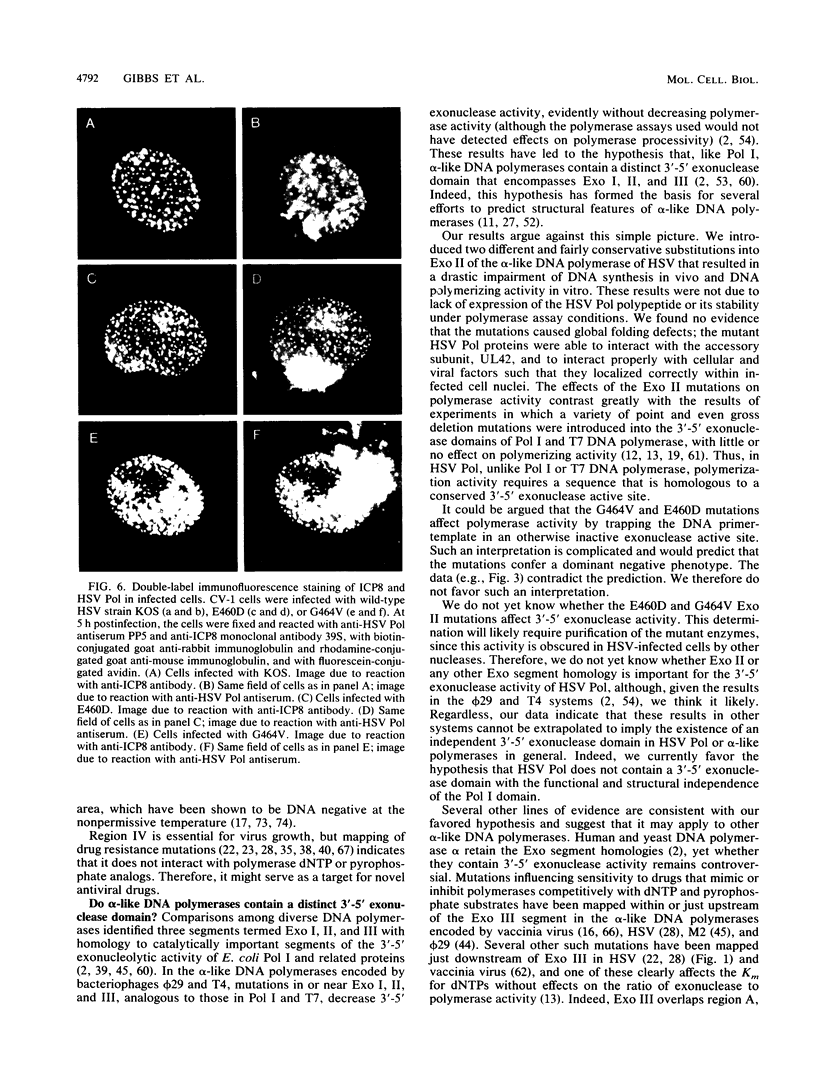
Abstract
Most DNA polymerases are multifunctional proteins that possess both polymerizing and exonucleolytic activities. For Escherichia coli DNA polymerase I and its relatives, polymerase and exonuclease activities reside on distinct, separable domains of the same polypeptide. The catalytic subunits of the alpha-like DNA polymerase family share regions of sequence homology with the 3'-5' exonuclease active site of DNA polymerase I; in certain alpha-like DNA polymerases, these regions of homology have been shown to be important for exonuclease activity. This finding has led to the hypothesis that alpha-like DNA polymerases also contain a distinct 3'-5' exonuclease domain. We have introduced conservative substitutions into a 3'-5' exonuclease active site homology in the gene encoding herpes simplex virus DNA polymerase, an alpha-like polymerase. Two mutants were severely impaired for viral DNA replication and polymerase activity. The mutants were not detectably affected in the ability of the polymerase to interact with its accessory protein, UL42, or to colocalize in infected cell nuclei with the major viral DNA-binding protein, ICP8, suggesting that the mutation did not exert global effects on protein folding. The results raise the possibility that there is a fundamental difference between alpha-like DNA polymerases and E. coli DNA polymerase I, with less distinction between 3'-5' exonuclease and polymerase functions in alpha-like DNA polymerases.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baer R., Bankier A. T., Biggin M. D., Deininger P. L., Farrell P. J., Gibson T. J., Hatfull G., Hudson G. S., Satchwell S. C., Séguin C. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984 Jul 19;310(5974):207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- Bernad A., Blanco L., Lázaro J. M., Martín G., Salas M. A conserved 3'----5' exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 1989 Oct 6;59(1):219–228. doi: 10.1016/0092-8674(89)90883-0. [DOI] [PubMed] [Google Scholar]
- Bernad A., Zaballos A., Salas M., Blanco L. Structural and functional relationships between prokaryotic and eukaryotic DNA polymerases. EMBO J. 1987 Dec 20;6(13):4219–4225. doi: 10.1002/j.1460-2075.1987.tb02770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulet A., Simon M., Faye G., Bauer G. A., Burgers P. M. Structure and function of the Saccharomyces cerevisiae CDC2 gene encoding the large subunit of DNA polymerase III. EMBO J. 1989 Jun;8(6):1849–1854. doi: 10.1002/j.1460-2075.1989.tb03580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush M., Yager D. R., Gao M., Weisshart K., Marcy A. I., Coen D. M., Knipe D. M. Correct intranuclear localization of herpes simplex virus DNA polymerase requires the viral ICP8 DNA-binding protein. J Virol. 1991 Mar;65(3):1082–1089. doi: 10.1128/jvi.65.3.1082-1089.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Horwitz M. S. Dissection of functional domains of adenovirus DNA polymerase by linker-insertion mutagenesis. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6116–6120. doi: 10.1073/pnas.86.16.6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou H. C., Weller S. K., Coen D. M. Mutations in the herpes simplex virus major DNA-binding protein gene leading to altered sensitivity to DNA polymerase inhibitors. Virology. 1985 Sep;145(2):213–226. doi: 10.1016/0042-6822(85)90155-2. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen D. M., Fleming H. E., Jr, Leslie L. K., Retondo M. J. Sensitivity of arabinosyladenine-resistant mutants of herpes simplex virus to other antiviral drugs and mapping of drug hypersensitivity mutations to the DNA polymerase locus. J Virol. 1985 Feb;53(2):477–488. doi: 10.1128/jvi.53.2.477-488.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue M., Poch O., Tordo N., Moras D., Argos P. An attempt to unify the structure of polymerases. Protein Eng. 1990 May;3(6):461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- Derbyshire V., Freemont P. S., Sanderson M. R., Beese L., Friedman J. M., Joyce C. M., Steitz T. A. Genetic and crystallographic studies of the 3',5'-exonucleolytic site of DNA polymerase I. Science. 1988 Apr 8;240(4849):199–201. doi: 10.1126/science.2832946. [DOI] [PubMed] [Google Scholar]
- Derse D., Bastow K. F., Cheng Y. Characterization of the DNA polymerases induced by a group of herpes simplex virus type I variants selected for growth in the presence of phosphonoformic acid. J Biol Chem. 1982 Sep 10;257(17):10251–10260. [PubMed] [Google Scholar]
- Digard P., Coen D. M. A novel functional domain of an alpha-like DNA polymerase. The binding site on the herpes simplex virus polymerase for the viral UL42 protein. J Biol Chem. 1990 Oct 15;265(29):17393–17396. [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Earl P. L., Jones E. V., Moss B. Homology between DNA polymerases of poxviruses, herpesviruses, and adenoviruses: nucleotide sequence of the vaccinia virus DNA polymerase gene. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3659–3663. doi: 10.1073/pnas.83.11.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank K. B., Cheng Y. C. Inhibition of herpes simplex virus DNA polymerase by purine ribonucleoside monophosphates. J Biol Chem. 1986 Feb 5;261(4):1510–1513. [PubMed] [Google Scholar]
- Freemont P. S., Ollis D. L., Steitz T. A., Joyce C. M. A domain of the Klenow fragment of Escherichia coli DNA polymerase I has polymerase but no exonuclease activity. Proteins. 1986 Sep;1(1):66–73. doi: 10.1002/prot.340010111. [DOI] [PubMed] [Google Scholar]
- Gadler H., Larsson A., Sølver E. Nucleic acid hybridization, a method to determine effects of antiviral compounds on herpes simplex virus type 1 DNA synthesis. Antiviral Res. 1984 Apr;4(1-2):63–70. doi: 10.1016/0166-3542(84)90026-3. [DOI] [PubMed] [Google Scholar]
- Gibbs J. S., Chiou H. C., Bastow K. F., Cheng Y. C., Coen D. M. Identification of amino acids in herpes simplex virus DNA polymerase involved in substrate and drug recognition. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6672–6676. doi: 10.1073/pnas.85.18.6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingeras T. R., Sciaky D., Gelinas R. E., Bing-Dong J., Yen C. E., Kelly M. M., Bullock P. A., Parsons B. L., O'Neill K. E., Roberts R. J. Nucleotide sequences from the adenovirus-2 genome. J Biol Chem. 1982 Nov 25;257(22):13475–13491. [PubMed] [Google Scholar]
- Gottlieb J., Marcy A. I., Coen D. M., Challberg M. D. The herpes simplex virus type 1 UL42 gene product: a subunit of DNA polymerase that functions to increase processivity. J Virol. 1990 Dec;64(12):5976–5987. doi: 10.1128/jvi.64.12.5976-5987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffey M. L., Novotny J., Bruccoleri R. E., Carroll R. D., Stevens J. T., Matthews J. T. Structure-function studies of the herpes simplex virus type 1 DNA polymerase. J Virol. 1990 Oct;64(10):5008–5018. doi: 10.1128/jvi.64.10.5008-5018.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J. D., Wang Y. S., Pierpont J., Berlin M. S., Rundlett S. E., Woodward S. Aphidicolin resistance in herpes simplex virus type I reveals features of the DNA polymerase dNTP binding site. Nucleic Acids Res. 1989 Nov 25;17(22):9231–9244. doi: 10.1093/nar/17.22.9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez T. R., Lehman I. R. Functional interaction between the herpes simplex-1 DNA polymerase and UL42 protein. J Biol Chem. 1990 Jul 5;265(19):11227–11232. [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Joyce C. M., Kelley W. S., Grindley N. D. Nucleotide sequence of the Escherichia coli polA gene and primary structure of DNA polymerase I. J Biol Chem. 1982 Feb 25;257(4):1958–1964. [PubMed] [Google Scholar]
- Knipe D. M., Senechek D., Rice S. A., Smith J. L. Stages in the nuclear association of the herpes simplex virus transcriptional activator protein ICP4. J Virol. 1987 Feb;61(2):276–284. doi: 10.1128/jvi.61.2.276-284.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopf C. W. Nucleotide sequence of the DNA polymerase gene of herpes simplex virus type 1 strain Angelotti. Nucleic Acids Res. 1986 Oct 24;14(20):8225–8226. doi: 10.1093/nar/14.20.8225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopf C. W. The herpes simplex virus type 1 DNA polymerase gene: site of phosphonoacetic acid resistance mutation in strain Angelotti is highly conserved. J Gen Virol. 1987 May;68(Pt 5):1429–1433. doi: 10.1099/0022-1317-68-5-1429. [DOI] [PubMed] [Google Scholar]
- Larder B. A., Kemp S. D., Darby G. Related functional domains in virus DNA polymerases. EMBO J. 1987 Jan;6(1):169–175. doi: 10.1002/j.1460-2075.1987.tb04735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt M. C., Ito J. T5 DNA polymerase: structural--functional relationships to other DNA polymerases. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4465–4469. doi: 10.1073/pnas.86.12.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcy A. I., Hwang C. B., Ruffner K. L., Coen D. M. Engineered herpes simplex virus DNA polymerase point mutants: the most highly conserved region shared among alpha-like DNA polymerases is involved in substrate recognition. J Virol. 1990 Dec;64(12):5883–5890. doi: 10.1128/jvi.64.12.5883-5890.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcy A. I., Olivo P. D., Challberg M. D., Coen D. M. Enzymatic activities of overexpressed herpes simplex virus DNA polymerase purified from recombinant baculovirus-infected insect cells. Nucleic Acids Res. 1990 Mar 11;18(5):1207–1215. doi: 10.1093/nar/18.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcy A. I., Yager D. R., Coen D. M. Isolation and characterization of herpes simplex virus mutants containing engineered mutations at the DNA polymerase locus. J Virol. 1990 May;64(5):2208–2216. doi: 10.1128/jvi.64.5.2208-2216.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden H. S., Campbell M. E., Haarr L., Frame M. C., Parris D. S., Murphy M., Hope R. G., Muller M. T., Preston C. M. The 65,000-Mr DNA-binding and virion trans-inducing proteins of herpes simplex virus type 1. J Virol. 1987 Aug;61(8):2428–2437. doi: 10.1128/jvi.61.8.2428-2437.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K., Kim C. I., Kobayashi H., Kanehiro H., Hirokawa H. Aphidicolin-resistant DNA polymerase of bacteriophage phi 29 APHr71 mutant is hypersensitive to phosphonoacetic acid and butylphenyldeoxyguanosine 5'-triphosphate. Virology. 1990 Sep;178(1):337–339. doi: 10.1016/0042-6822(90)90416-o. [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Takano H., Kim C. I., Hirokawa H. Primary structure of bacteriophage M2 DNA polymerase: conserved segments within protein-priming DNA polymerases and DNA polymerase I of Escherichia coli. Gene. 1989 Dec 14;84(2):247–255. doi: 10.1016/0378-1119(89)90498-8. [DOI] [PubMed] [Google Scholar]
- Miller B. W., Williams J. Cellular transformation by adenovirus type 5 is influenced by the viral DNA polymerase. J Virol. 1987 Nov;61(11):3630–3634. doi: 10.1128/jvi.61.11.3630-3634.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollis D. L., Brick P., Hamlin R., Xuong N. G., Steitz T. A. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. 1985 Feb 28-Mar 6Nature. 313(6005):762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- Que B. G., Downey K. M., So A. G. Mechanisms of selective inhibition of 3' to 5' exonuclease activity of Escherichia coli DNA polymerase I by nucleoside 5'-monophosphates. Biochemistry. 1978 May 2;17(9):1603–1606. doi: 10.1021/bi00602a004. [DOI] [PubMed] [Google Scholar]
- Quinlan M. P., Chen L. B., Knipe D. M. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell. 1984 Apr;36(4):857–868. doi: 10.1016/0092-8674(84)90035-7. [DOI] [PubMed] [Google Scholar]
- Reha-Krantz L. J. Amino acid changes coded by bacteriophage T4 DNA polymerase mutator mutants. Relating structure to function. J Mol Biol. 1988 Aug 20;202(4):711–724. doi: 10.1016/0022-2836(88)90552-9. [DOI] [PubMed] [Google Scholar]
- Reha-Krantz L. J. Locations of amino acid substitutions in bacteriophage T4 tsL56 DNA polymerase predict an N-terminal exonuclease domain. J Virol. 1989 Nov;63(11):4762–4766. doi: 10.1128/jvi.63.11.4762-4766.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reha-Krantz L. J., Stocki S., Nonay R. L., Dimayuga E., Goodrich L. D., Konigsberg W. H., Spicer E. K. DNA polymerization in the absence of exonucleolytic proofreading: in vivo and in vitro studies. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2417–2421. doi: 10.1073/pnas.88.6.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roovers D. J., Overman P. F., Chen X. Q., Sussenbach J. S. Linker mutation scanning of the genes encoding the adenovirus type 5 terminal protein precursor and DNA polymerase. Virology. 1991 Jan;180(1):273–284. doi: 10.1016/0042-6822(91)90032-7. [DOI] [PubMed] [Google Scholar]
- Roovers D. J., Young C. S., Vos H. L., Sussenbach J. S. Physical mapping of two temperature-sensitive adenovirus mutants affected in the DNA polymerase and DNA binding protein. Virus Genes. 1990 Jun;4(1):53–61. doi: 10.1007/BF00308565. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter S. D., Zweig M., Hampar B. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP 4. Infect Immun. 1981 Dec;34(3):684–692. doi: 10.1128/iai.34.3.684-692.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spicer E. K., Rush J., Fung C., Reha-Krantz L. J., Karam J. D., Konigsberg W. H. Primary structure of T4 DNA polymerase. Evolutionary relatedness to eucaryotic and other procaryotic DNA polymerases. J Biol Chem. 1988 Jun 5;263(16):7478–7486. [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. Selective inactivation of the exonuclease activity of bacteriophage T7 DNA polymerase by in vitro mutagenesis. J Biol Chem. 1989 Apr 15;264(11):6447–6458. [PubMed] [Google Scholar]
- Taddie J. A., Traktman P. Genetic characterization of the vaccinia virus DNA polymerase: identification of point mutations conferring altered drug sensitivities and reduced fidelity. J Virol. 1991 Feb;65(2):869–879. doi: 10.1128/jvi.65.2.869-879.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. W., Ott J., Eckstein F. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8765–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traktman P. The enzymology of poxvirus DNA replication. Curr Top Microbiol Immunol. 1990;163:93–123. doi: 10.1007/978-3-642-75605-4_4. [DOI] [PubMed] [Google Scholar]
- Tsurumi T., Maeno K., Nishiyama Y. A single-base change within the DNA polymerase locus of herpes simplex virus type 2 can confer resistance to aphidicolin. J Virol. 1987 Feb;61(2):388–394. doi: 10.1128/jvi.61.2.388-394.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. S., Wong S. W., Korn D. Human DNA polymerase alpha: predicted functional domains and relationships with viral DNA polymerases. FASEB J. 1989 Jan;3(1):14–21. doi: 10.1096/fasebj.3.1.2642867. [DOI] [PubMed] [Google Scholar]
- Weissbach A., Hong S. C., Aucker J., Muller R. Characterization of herpes simplex virus-induced deoxyribonucleic acid polymerase. J Biol Chem. 1973 Sep 25;248(18):6270–6277. [PubMed] [Google Scholar]
- Weller S. K., Aschman D. P., Sacks W. R., Coen D. M., Schaffer P. A. Genetic analysis of temperature-sensitive mutants of HSV-1: the combined use of complementation and physical mapping for cistron assignment. Virology. 1983 Oct 30;130(2):290–305. doi: 10.1016/0042-6822(83)90084-3. [DOI] [PubMed] [Google Scholar]
- Wilcock D., Lane D. P. Localization of p53, retinoblastoma and host replication proteins at sites of viral replication in herpes-infected cells. Nature. 1991 Jan 31;349(6308):429–431. doi: 10.1038/349429a0. [DOI] [PubMed] [Google Scholar]
- Williams J., Galos R. S., Binger M. H., Flint S. J. Location of additional early regions within the left quarter of the adenoviral genome. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):353–365. doi: 10.1101/sqb.1980.044.01.040. [DOI] [PubMed] [Google Scholar]
- Willians J. F., Young C. S., Austin P. E. Genetic analysis of human adenovirus type 5 in permissive and nonpermissive cells. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):427–437. doi: 10.1101/sqb.1974.039.01.055. [DOI] [PubMed] [Google Scholar]
- Wong S. W., Wahl A. F., Yuan P. M., Arai N., Pearson B. E., Arai K., Korn D., Hunkapiller M. W., Wang T. S. Human DNA polymerase alpha gene expression is cell proliferation dependent and its primary structure is similar to both prokaryotic and eukaryotic replicative DNA polymerases. EMBO J. 1988 Jan;7(1):37–47. doi: 10.1002/j.1460-2075.1988.tb02781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yager D. R., Marcy A. I., Coen D. M. Translational regulation of herpes simplex virus DNA polymerase. J Virol. 1990 May;64(5):2217–2225. doi: 10.1128/jvi.64.5.2217-2225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yoshikawa H., Ito J. Nucleotide sequence of the major early region of bacteriophage phi 29. Gene. 1982 Mar;17(3):323–335. doi: 10.1016/0378-1119(82)90149-4. [DOI] [PubMed] [Google Scholar]
- de Bruyn Kops A., Knipe D. M. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell. 1988 Dec 2;55(5):857–868. doi: 10.1016/0092-8674(88)90141-9. [DOI] [PubMed] [Google Scholar]