Abstract
Stem cells are characterized by their unique ability of self-renewal to maintain the so-called stem cell pool. Over the past decades, reactive oxygen species (ROS) have been recognized as toxic aerobic metabolism byproducts that are harmful to stem cells, leading to DNA damage, senescence or cell death. Recently, a growing body of literature has shown that stem cells reside in redox niches with low ROS levels. The balance of Redox homeostasis facilitates stem cell self-renewal by an intricate network. Thus, to fully decipher the underlying molecular mechanisms involved in the maintenance of stem cell self-renewal, it is critical to address the important role of redox homeostasis in the regulation of self-renewal and differentiation of stem cells. In this regard, we will discuss the regulatory mechanisms involved in the subtly orchestrated balance of redox status in stem cells by scavenger antioxidant enzyme systems that are well monitored by the hypoxia niches and crucial redox regulators including forkhead homeobox type O family (FoxOs), apurinic/apyrimidinic (AP) endonuclease1/redox factor-1 (APE1/Ref-1), nuclear factor erythroid-2-related factor 2 (Nrf2) and ataxia telangiectasia mutated (ATM). We will also introduce several pivotal ROS-sensitive molecules, such as hypoxia-inducible factors, p38 mitogen-activated protein kinase (p38) and p53, involved in the redox-regulated stem cell self-renewal. Specifically, all the aforementioned molecules can act as ‘redox sensors' by virtue of redox modifications of their cysteine residues, which are critically important in the control of protein function. Given the importance of redox homeostasis in the regulation of stem cell self-renewal, understanding the underlying molecular mechanisms involved will provide important new insights into stem cell biology.
Keywords: stem cell, self-renewal, ROS, redox homeostasis, redox modification
Facts
Stem cells reside in redox niches with low ROS levels.
Signaling molecules such as FoxOs, APE1/Ref-1, Nrf2, ATM, HIFs, p38 and p53 are involved in the regulation of stem cell self-renewal and differentiation.
FoxOs, APE1/Ref-1, Nrf2 and ATM can modulate the antioxidant enzyme systems and act upstream of ROS production, while HIFs, p38 and p53 are downstream molecules of ROS signaling in stem cells.
Certain proteins can act as ‘redox sensors' due to the redox modifications of their cysteine residues, which are critically important in the control of protein function.
Open questions
Whether ROS homeostasis is an important modulator in stem cell self-renewal and differentiation?
What roles do these signaling molecules (FoxOs, APE1/Ref-1, Nrf2, ATM, HIFs, p38 and p53) play in ROS-regulated stem cell self-renewal and differentiation?
Are the redox modifications of signaling molecules (FoxOs, APE1/Ref-1, Nrf2, ATM, HIFs, p38 and p53) involved in the regulation stem cell self-renewal and differentiation?
Self-renewal is the process by which a stem cell asymmetrically or symmetrically divides to propagate one or two daughter stem cells with similar developmental potential as the mother cells.1 The capability of self-renewal is essential for stem cells to expand their numbers during development and to maintain the stem cell pool.2 Abnormal self-renewal mechanisms can destroy the maintenance of the undifferentiated state and alter the balance of self-renewal versus differentiation.3 Thus, deciphering the molecular mechanisms behind stem cell self-renewal is of significant importance. However, it still remains enigmatic as to how exactly the self-renewal of stem cells is achieved.
Reactive oxygen species (ROS), initially implicated in stress and disease, have recently been revisited as influential new players in stem cell biology.4 High levels of ROS have long been suggested to be detrimental to mediate oxygen toxicity, while physiological low levels of ROS have been reported to operate as intracellular signaling molecules, a function that, although has been widely documented, is still controversial.5 A general movement towards the concept of ‘homeostatic ROS levels' versus ‘pathologic ROS levels' is gaining support and is replacing the older dogma that ROS are always ‘bad' for cells.6
Until recently, the focus in stem cell biology has been on the damaging effects of ROS accumulation, and various anti-oxidative and anti-stress mechanisms of stem cells have been characterized.7, 8 However, increasing evidence is now supporting the notion that, in some cases, ROS in the redox homeostasis play pivotal roles in the maintenance of stem cell self-renewal.9 Indeed, stem cells reside in niches characterized by low levels of ROS, which are critical for maintaining the potential for self-renewal and stemness, while high levels of ROS effectively shut down self-renewal and confer potent capacity for stem cell differentiation.10, 11 However, understanding of the myriad potential mechanisms whereby homeostatic ROS levels regulate stem cell self-renewal is still in a state of flux. In this review, we attempt to highlight the molecular mechanisms concerning the maintenance of stem cell self-renewal regulated by intracellular redox status. In addition, we will discuss several crucial ‘redox sensors' involved in the regulation of stem cell self-renewal and differentiation.
Balance of the redox status in stem cells
Stem cells undergoing the self-renewal process are believed to possess low levels of intracellular ROS.12 To balance the redox status, stem cells engage scavenger antioxidant enzyme systems to eliminate the intracellular ROS (Figure 1), which are well regulated by the hypoxia niches as well as several critical transcription factors including the forkhead homeobox type O (FoxO) family and nuclear factor erythroid-2-related factor 2 (Nrf2) that both activate the transcription of antioxidant enzymes.10, 11 Several other critical redox regulators such as apurinic/apyrimidinic (AP) endonuclease1/redox factor-1 (APE1/Ref-1) and ataxia telangiectasia mutated (ATM) are also involved in the elimination of intracellular ROS11, 13 (Figure 2).
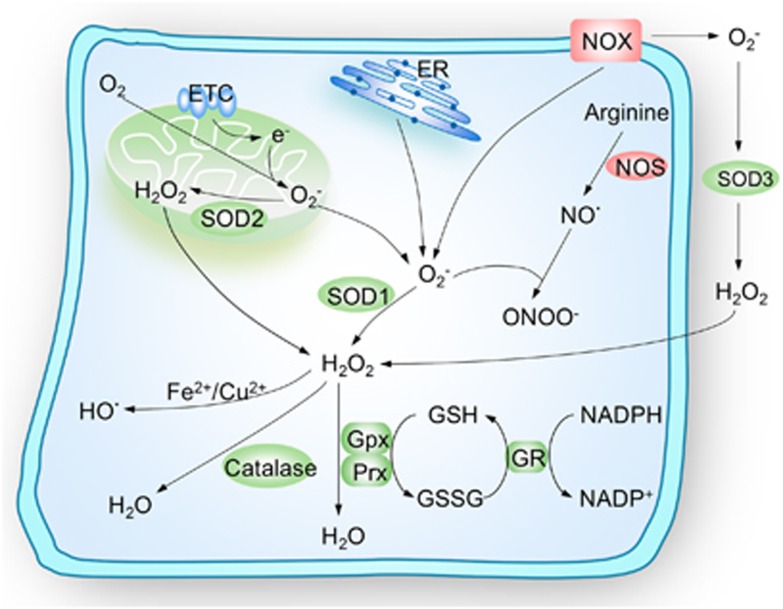
Figure 1.
Schematic illustration of cellular maintenance of redox homeostasis. Mitochondria electron-transport chain (ETC), membrane-bound NADPH oxidase (NOX) complex and endoplasmic reticulum (ER) are the three major intracellular sources of reactive oxygen species (ROS). Anion superoxide (O2−) is the principal form of ROS and can be rapidly converted into hydrogen peroxide (H2O2) by superoxide dismutases (SODs) or can alternatively, form peroxynitrite (ONOO−) through reacting with the nitric oxide (NO·). H2O2 can be catalyzed to HO· in the presence of Fe2+ or Cu2+ ions or be converted to H2O and O2 catalyzed by catalase, glutathione peroxidase (Gpx) or peroxiredoxins (Prx). To maintain the redox homeostasis, the living cells engage powerful scavenger antioxidant enzyme systems to eliminate the intracellular ROS, major ROS-scavenging enzymes are shown in green. NOS, nitric oxide synthase; GR, glutathione reductase; GSH, reduced glutathione; GSSG, oxidized glutathione
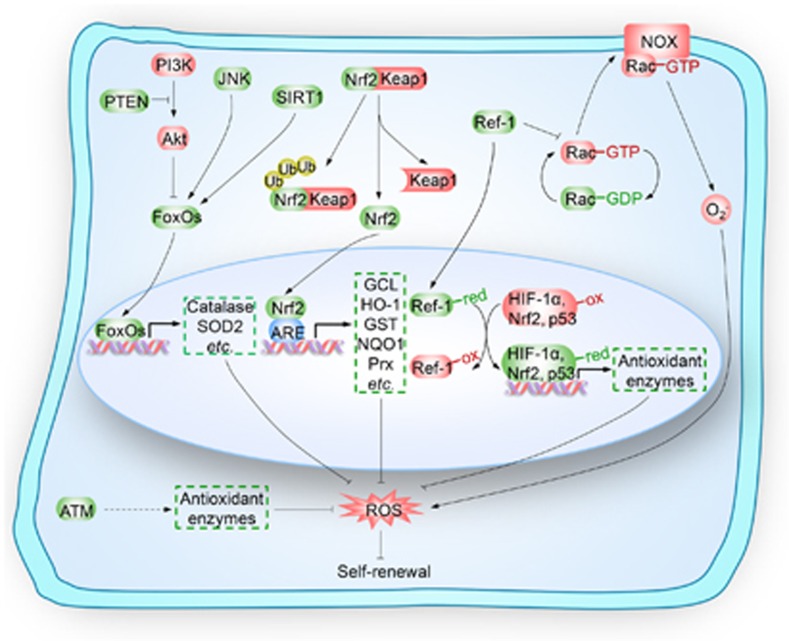
Figure 2.
Key regulators upstream of ROS in the self-renewal of stem cells. Key regulators, including forkhead homeobox type O family (FoxOs), nuclear factor erythroid-2-related factor 2 (Nrf2), apurinic/apyrimidinic (AP) endonuclease1/redox factor-1 (APE1/Ref-1) and ataxia telangiectasia mutated (ATM), play pivotal roles in the regulation of self-renewal of stem cells by modulating redox homeostasis. FoxOs, including FoxO1, FoxO3, FoxO4 and FoxO6, are negatively regulated by phosphoinositide 3-kinase (PI3K)/Akt kinase signaling cascades and positively regulated by c-Jun N-terminal kinase (JNK) phosphorylation and Sirtuin 1 (SIRT1) deacetylation. Activated FoxOs translocate into the nucleus and initiate the transcription of target antioxidant genes, such as catalase and superoxide dismutase 2 (SOD2). Nrf2 is sequestered in the cytoplasm by binding with Kelch-like ECH-associated protein 1 (Keap1), which causes proteasomal degradation of Nrf2. Nrf2 is also a transcription factor that combats intracellular detrimental reactive oxygen species (ROS) by transcribing genes encoding key antioxidant molecules, such as glutamate cysteine ligase (GCL), heme oxygenase-1 (HO-1), glutathione transferase (GST), NAD(P)H quinine oxidoreductase-1 (NQO1) and peroxiredoxins (Prx). APE1/Ref-1 inhibits Rac1-regulated membrane-bound NADPH oxidase (NOX) to decrease ROS production or by binding with oxidized transcription factors (such as hypoxia-inducible factor 1α (HIF-1α), Nrf2 and p53) that transcribing genes encoding antioxidant enzymes, to maintain the cysteine residues of these transcription factors in the reduced state. ATM controls the intracellular levels of ROS by regulating the expression of antioxidant enzymes. FoxOs, Nrf2, APE1/Ref1 and ATM modulate the self-renewal of stem cells by monitoring the redox homeostasis. PTEN, phosphatase and tensin homolog; Ub, ubiquitin; ARE, antioxidant responsive element
Intracellular ROS production and elimination
The three major intracellular sources of ROS are the mitochondria electron-transport chain (ETC), the membrane-bound NADPH oxidase (NOX) complex and the endoplasmic reticulum14 (Figure 1). Of these, the byproducts of electron leakage of mitochondrial ETC are the principal generators of ROS in mammalian cells. These electrons can then be captured by O2, forming anion superoxide (O2−).11 Then, O2− can be rapidly converted into hydrogen peroxide (H2O2) by superoxide dismutases (SODs).10 During steady-state cellular conditions, the intracellular ROS levels are tightly regulated to maintain redox homeostasis by scavenger antioxidant enzymatic defense systems, including SODs, glutathione peroxidase (Gpx), glutathione reductase (GR), peroxiredoxins (Prx), thioredoxin (Trx) and catalase10 (Figure 1). Obviously, these intracellular antioxidant enzymatic defense systems need to be subtly regulated by the hypoxia niche and critical regulators such as FoxO family, APE1/Ref-1, Nrf2 and ATM, all of which have been recently reported as key regulators of stem cell self-renewal by modulating redox homeostasis (Figure 2).
Hypoxia-mediated redox regulation in the maintenance of stem cell self-renewal
Stem cells have been shown to live in niches with low oxygen tension (hypoxia), and recent studies have underscored the importance of hypoxia in human bone marrow mesenchymal stem cells (MSCs) and induced pluripotent stem cells (iPSCs), suggesting that low oxygen tension promotes an undifferentiated state of the stem cells.15, 16, 17 In principle, a hypoxic milieu could be useful for maintaining cells in a non-dividing, or at least slow cycling stage protecting cells from detrimental accumulation of ROS. In other words, ROS would be reduced to secure the maintenance of stem cells in the case of hypoxia.18 Paradoxically, evidence suggests that acute hypoxia contributes to the generation of ROS.19 Although still controversial, it appears that there must be a tight balance between hypoxia and ROS levels. Recent studies indicated that hypoxia significantly induced proliferation and migration of ASCs by increasing the intracellular levels of ROS,20, 21 however, the underlying mechanisms are still not clear. Until now, hypoxia-induced ROS accumulation has received little attention in other types of stem cells.
FoxOs-mediated redox regulation in the maintenance of stem cell self-renewal
The forkhead box O (FoxO) family is a large family of transcription factors and has diverse physiological functions. FoxOs can be phosphorylated by the phosphoinositide 3-kinase (PI3K)/Akt kinase, leading to subsequent nuclear export and ultimately transcriptional inactivation.22 By contrast, c-Jun N-terminal kinase (JNK) phosphorylates FoxOs at a distinct set to mediate nuclear import and transcriptional activation.23 In addition, deacetylation of FoxOs by Sirtuin 1 (SIRT1) also promotes FoxOs-mediated transcription of target genes.24 Among the FoxO family, FoxO1, FoxO3a and FoxO4 have been implicated in many of the ROS-regulated processes including maintenance of stem cell self-renewal.25 For example, FoxO3a regulates hematopoietic homeostasis and is essential for maintenance of the HSC pool,26, 27 FoxO1/3/4 governs neural stem cell (NSC) homeostasis and regulates the NSC pool.28, 29 And FoxO1 is reported to be required for the maintenance of human ESC pluripotency.30, 31
In spite of the increasing reports on regulation of stem cell self-renewal by FoxOs, little is known about the underlying molecular mechanisms. Since target genes of the FoxO family, such as SOD2 and catalase, are involved in cellular detoxification of ROS,25 it is plausible that FoxOs might regulate stem cell self-renewal by maintaining the homeostatic ROS levels. Indeed, this hypothesis has been recently substantiated. Tothova et al.32 found a marked increase of ROS in FoxO-deficient HSCs that correlated with changes in expression of SOD1/3. Thus, FoxO proteins play essential roles in response to physiologic oxidative stress and thereby enhance survival in the HSC compartment. In another report, loss of FoxO3 decreases SOD2 and catalase expression, leading to over-accumulation of ROS and HSCs defects.27, 33 Taken together, these experiments imply that repression of ROS levels by FoxOs is required for maintenance of the stem cell self-renewal.
APE1/Ref-1-mediated redox regulation in the maintenance of stem cell self-renewal
APE1/Ref-1 is a multifunctional protein possessing both DNA repair and transcriptional regulatory activities.34 APE1/Ref-1 also has a pleiotropic role in controlling cellular response to oxidative stress by decreasing ROS production, or by binding with oxidized transcription factors (hypoxia-inducible factor 1α (HIF-1α), Nrf2 and p53 etc.).35 Recent studies have addressed the important role of APE1/Ref-1-medidated ROS in the regulation of stem cell self-renewal. For instance, the redox function of Ref-1, but not the repair endonuclease activity, was required in normal embryonic hematopoietic development.36 Similarly, in cardiac stem cells, Ref-1 inhibition followed by H2O2 treatment extensively induced the levels of intracellular ROS, resulting in significant elevation in cardiac differentiation and apoptosis.37 In another observation on human bone-marrow-derived MSCs, over-expression of APE1/Ref-1 further suppressed superoxide production and alleviated senescence.38 In summary, these important findings suggest that APE1/Ref-1 is implicated in the maintenance of the stem cell pool by modulating intracellular redox homeostasis.
Nrf2-mediated redox regulation in the maintenance of stem cell self-renewal
Nrf2 activates antioxidant responsive element (ARE)-dependent gene expression to maintain cellular redox homeostasis.39 Nrf2 is sequestered in the cytoplasm by binding to Kelch-like ECH-associated protein 1 (Keap1), which causes proteasomal degradation of Nrf2.40 Nrf2 combats detrimental intracellular ROS by transcribing genes encoding several key antioxidant molecules.41 Nrf2 stabilization has been found to profoundly attenuate H2O2-induced cell death of human NSCs (hNSCs), suggesting that Nrf2 might be recognized as a regulatory factor that controls stem cell fate by modulating redox homeostasis.42 Takahata et al.43 showed a similar effect in MSCs where Nrf2 protected MSCs from oxidative stress damage. Further studies on intestinal stem cells (ISCs) of Drosophila indicated that repression of Nrf2 caused accumulation of ROS and accelerated age-related degeneration of the intestinal epithelium.44 However, intriguingly, Nrf2 was also found to regulate mouse HSC survival independently of ROS levels.45 Collectively, these reports imply that Nrf2 could be a potent regulator of stem cell self-renewal and differentiation due, at least in part, to its antioxidant capacity.
ATM-mediated redox regulation in the maintenance of stem cell self-renewal
The ATM protein kinase is best known for its important role in monitoring genomic integrity in response to DNA damage.46 Recent findings suggest that ATM protein kinase also controls the intracellular levels of ROS,47 and that ATM-regulated ROS levels are critical for stem cell self-renewal. This is evidenced by the finding that ATM-mediated BID phosphorylation led to quiescence of HSCs due to BID's ability to regulate oxidative stress, resulting in maintenance of the HSC pool and HSC repopulating potential in vivo.48 Moreover, recent advances from several groups have also underscored the importance of ROS in ATM-regulated stem cell self-renewal. For example, Ito's group found that Atm−/− mice showed progressive bone marrow failure, resulting from a defect in HSC function that was associated with elevated ROS.49 In another study, Atm−/− NSCs showed impaired proliferation by intrinsic elevation of ROS levels.50 These reports suggest that the self-renewal capacity of stem cells depends on ATM-mediated redox homeostasis.
Regulation of stem cell self-renewal by redox homeostasis
As reviewed above, stem cells are capable of coping with oxidative stress to maintain redox homeostasis through unique regulatory mechanisms whereby they upregulate their own antioxidant defense systems. Additionally, a growing body of literature supports the notion that redox homeostasis is an important modulator in the self-renewal and differentiation of stem cells51, 52, 53, 54 (Figure 3). However, most of these studies did not clarify the underlying molecular mechanisms. Here, we will introduce several potential or well-documented ROS-sensitive molecules that participate in the regulation of stem cell self-renewal.
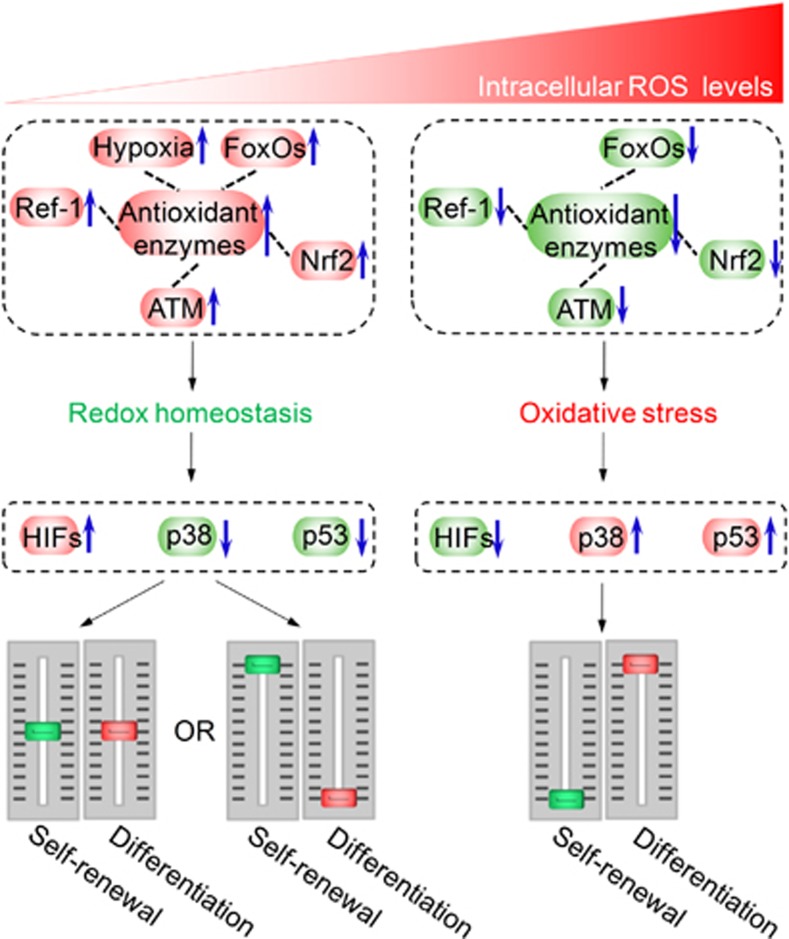
Figure 3.
Redox homeostasis in stem cell self-renewal and differentiation. Low reactive oxygen species (ROS) levels maintain the self-renewal of stem cells by monitoring the redox homeostasis, which is well regulated by the antioxidant enzymatic defense systems and hypoxia niches as well as several key redox regulators such as forkhead homeobox type O family (FoxOs), nuclear factor erythroid-2-related factor 2 (Nrf2), apurinic/apyrimidinic (AP) endonuclease1/redox factor-1 (APE1/Ref-1) and ataxia telangiectasia mutated (ATM); high ROS levels cause abnormal differentiation, apoptosis or senescence of stem cells by the ROS-sensitive molecules, including hypoxia-inducible factors (HIFs), p38 and p53
HIFs might be required in ROS-regulated stem cell self-renewal
HIFs are the most commonly known transcription factors in response to hypoxia.55 Increasing studies support the notion that HIFs play an important role in the maintenance of the stem cell pool. For example, in HSCs, HIF-1α was stabilized and precisely regulated to maintain cell-cycle quiescence and unique metabolic characteristics.56, 57 HIF-2α has been shown to increase the expression of Oct-4 in the maintenance of a stem cell state in HSCs.58 However, whether HIFs-regulated maintenance of stem cell self-renewal is due to the ROS-modulated HIFs still remains unclear.
p38 is a key mediator of ROS-regulated stem cell self-renewal
p38 plays a critical role in cell proliferation, differentiation, survival and migration.59 A plethora of evidence suggests that p38 plays a primordial role in response to changes in the cellular redox balance.60, 61 p38α was recently identified as a modulator of ex vivo stem cell proliferation in human hematopoietic stem and progenitor cells.62 Indeed, p38 activation status can determine proliferation/differentiation fates in human MSCs.63
According to the important role of ROS homeostasis in the self-renewal and differentiation of stem cells, it can be speculated that p38 might be the executor of ROS-regulated stem cell self-renewal. For instance, Ito's group showed that elevation of ROS levels induced HSC-specific phosphorylation of p38 concomitant with a defect in the maintenance of HSC quiescence, and inactivation of p38 protects HSCs against loss of self-renewal capacity.49 Consistent with these results, HSCs with low levels of ROS had a higher self-renewal potential, while with high levels of ROS, HSC exhaustion was observed accompanied by higher expression of activated p38.64 In addition to HSCs, ROS-induced p38 also regulated self-renewal in other types of stem cells, including ESCs and NSCs.50, 65 Taken together, these intriguing findings supported the notion that p38 is a key mediator of ROS-regulated stem cell self-renewal.
p53 is a critical regulator of ROS-mediated stem cell self-renewal
As the guardian of the genome, p53 prevents the accumulation of genetic mutations by inducing cell-cycle arrest, apoptosis or senescence of somatic cells after genotoxic and oncogenic stresses.66 Although p53 is probably best characterized among the hundreds of oncogenes and tumor suppressors, recent advances have highlighted novel emerging roles of p53 in the regulation of stem cell self-renewal and homeostasis.67, 68 Disruption of the p53 network improved the efficacy of stem cell production, catapulting p53 into the center of stem cell research.69, 70, 71, 72 Additionally, a whole-genome study of p53-mediated DNA damage signaling was carried out in mouse ESCs.73 Unexpectedly, they found that many ESC-enriched core transcription factors, including Oct-4, Sox273 and Nanog,74 were repressed by p53, while p53-activated genes were linked to differentiation.73 Further studies showed that disabling p53 signaling enhanced the stemness and regenerative potential of human ESCs.75 Moreover, studies have also confirmed that p53 negatively regulates proliferation, survival, and thereby self-renewal, of NSCs.76
Emerging evidence suggests that p53 also acts as a finely tuned regulator of redox-dependent physiological processes.77 It was shown that ROS can control the activity of p53 by the modification of redox state of p53.78 Accordingly, since p53 is a key regulator in stem cell self-renewal, it is plausible that ROS-regulated p53 might play a critical role in stem cell self-renewal. Indeed, endogenous ROS have been shown to cause p53 translocation into mitochondria and trigger apoptosis in wild-type mouse ESCs, but induce p53 translocation into the nucleus and inhibit Nanog expression in SIRT1−/− mouse ESCs.79 Moreover, ROS-induced p53 activity has also been found to be important in the control of survival of HSCs.80 Taken together, these results suggest that p53 might be a critical regulator of ROS-mediated stem cell self-renewal, which warrants further studies in the future.
Redox modifications might be involved in stem cell self-renewal
Post-translational modifications (PTMs) of proteins play central roles in creating a continuously dynamic fine-tuned regulatory network implicated in a wide variety of cellular processes.81 However, such modifications failed to clarify ROS-initiated cellular signaling. Recently, oxidative cysteine modifications have emerged as a central mechanism for dynamic post-translational regulation of almost all major protein classes, and correlate with many disease states.82 Certain proteins in which the redox state of cysteine residues is modified (termed ‘redox sensors') seem to be involved in the initial and direct regulation of signaling molecules in response to ROS.83 Such ‘redox sensors' commonly possess highly conserved free cysteine (Cys) residues of which the thiol (RSH) functional groups are the most important direct cellular targets or ‘sensors' of ROS.84, 85 These active RSH groups are easily oxidized to a sulfenic oxoform (RSOH), the initial oxidative product of cysteine.86 The transient sulfenic acids can also result from the hydrolysis of S-nitrosothiols (RSNO), which are the oxidative product of the RSH group in response to reactive nitrogen species (RNS).87 The highly reactive nature of sulfenic acids often leads to reaction with nearby thiols to form disulfide bonds (RS-SR or RS-SR') or condense with GSH resulting in S-glutathionylation (RS-SG).88 Alternatively, sulfenic acids can be further oxidized to sulfinic acids (RSO2H) and, under more severe oxidizing conditions, to sulfonic acids (RSO3H)82 (Figure 4). Such oxidative cysteine modifications have been found to constitute a facile switch for modulating the function of proteins involved in many different biological functions.85 Recently, a cornucopia of ‘redox sensors' have been identified that participate in many important biological functions, some of which are crucial molecules modulating stem cell self-renewal and differentiation, including HIF-1α, FoxOs, APE1/Ref-1, Nrf2, ATM, p38 and p53 (Figure 5).
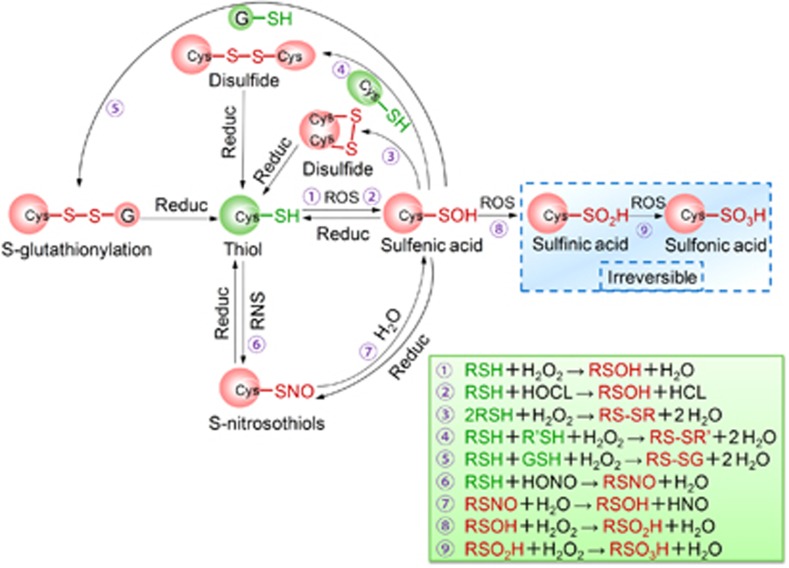
Figure 4.
Oxidative thiol modifications in redox-sensitive cysteine residues. Active thiol groups are easily oxidized to sulfenic acids (RSOH), the initial oxidation product. These transient sulfenic acids can also result from the hydrolysis of S-nitrosothiols (RSNO), which are the oxidative products of thiol groups in response to RNS. Sulfenic acids often condense with nearby thiols to form intermolecular or intramolecular disulfide bonds (RS-SR' or RS-SR), or with GSH resulting in S-glutathionylation (RS-SG). These oxoforms are reversible and can be restored to free thiols through the action of cellular reductants. Alternatively, sulfenic acids can be further oxidized to sulfinic acids (RSO2H) and, under more severe oxidizing conditions, to sulfonic acids (RSO3H), both of which are irreversible modifications. Cys, cysteine; ROS, reactive oxygen species; RNS, reactive nitrogen species; GSH, reduced glutathione; Reduc, Reductants
Figure 5.
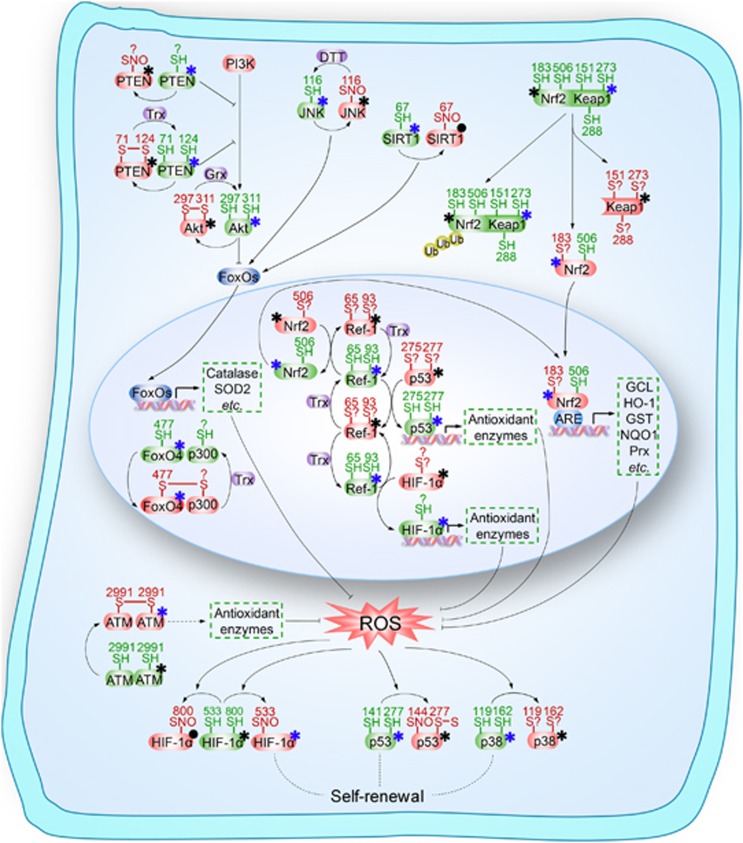
Schematic illustration of key ‘redox sensors' that might be involved in the self-renewal of stem cells. All key molecules shown are involved in the regulation of stem cell self-renewal by modulating redox homeostasis, including forkhead homeobox type O family (FoxOs), nuclear factor erythroid-2-related factor 2 (Nrf2), apurinic/apyrimidinic (AP) endonuclease1/redox factor-1 (APE1/Ref-1) and ataxia telangiectasia mutated (ATM), hypoxia-inducible factor 1α (HIF-1α), p38 and p53, as well as several upstream signaling molecules including phosphatase and tensin homolog (PTEN), Sirtuin 1 (SIRT1), c-Jun N-terminal kinase (JNK) and Kelch-like ECH-associated protein 1 (Keap1), are subject to redox modifications, or named ‘redox sensors.' These redox modifications might play an important role in the regulation of redox homeostasis thereby modulating the self-renewal of stem cells. SH, thiol; SNO, S-nitrosothiols; PI3K, phosphoinositide 3-kinase; Grx, glutaredoxin; Trx, thioredoxin; DTT, Dithiothreitol; SOD2, superoxide dismutase 2; Ub, ubiquitin; ARE, antioxidant responsive element; GCL, glutamate cysteine ligase; HO-1, heme oxygenase-1; GST glutathione transferase; NQO1, NAD(P)H quinine oxidoreductase-1; Prx, peroxiredoxins; ROS, reactive oxygen species. The blue asterisk denotes activated state of ‘redox sensors', while the black asterisk denotes inactivated state. The black solid dot indicates that the redox modification has no effect on the turnover of activated/inactivated state of ‘redox sensors', or this effect is not affected or has not been clarified; ‘S?' indicates that the types of oxidative modification are not clear. ‘?' denotes that the active cysteine site is undefined
HIF-1α as a target for S-nitrosylation
HIF-1α, containing 15 free thiol groups, was recently characterized as a potential target for S-nitrosylation that may facilitate transcriptional activity.89 It was demonstrated that HIF-1α can be subjected to S-nitrosylation, resulting in HIF-1α stabilization.90 Further, S-nitrosylation of thiol group of Cys800 under normoxia was found to be required for the transcriptional activity of HIF-1α by stimulating the recruitment of p300.91 Cys533 (equivalent to Cys520 in human) in normoxic murine HIF-1α is also an effective target of S-nitrosylation.92 These results indicate that the S-nitrosylation of free thiols in cysteines might be of key importance in the activation of HIF-1α. However paradoxically, recent studies demonstrated a significant disruption of the p300-HIF-1α interaction upon Cys800 S-nitrosylation.93 Moreover, H2O2 significantly potentiated HIF-1α inactivation in hypoxia cells.94 Thus, oxidative modification can both positively and negatively regulate stabilization of HIF-1α. Although this may seem contradictory, it is ipso facto reasonable. Indeed, oxidation controls HIF-1α activation depending on the differential oxygen content of the environment: oxidative modification stabilizes normoxic HIF-1α, while in the hypoxic scenario, oxidation of HIF-1α disrupts its DNA-binding capacity.
Redox modification involved in the modulation of FoxOs signaling
The transcriptional activity of FoxO family is known to be inactivated by the PI3K/Akt pathway, in which phosphatase and tensin homolog (PTEN) acts as a major gatekeeper that can reverse the action of PI3K.95 Increasing numbers of studies have reported oxidation of the catalytic cysteines within the active site of PTEN in response to exposure to ROS, and have shown that redox modification of PTEN potentiates Akt activation.96 The essential Cys124 residue in the active site of PTEN specifically forms a disulfide with Cys71 during oxidation, thereby inactivating the protein.97 In addition to disulfide formation, PTEN can also be subject to S-nitrosylation that stimulates the Akt activity.98 Thus, the activity of Akt is partly determined by the redox state of PTEN. Intriguingly, Akt can be also directly regulated by oxidation of its own cysteines. An intramolecular disulfide could be formed between Cys297 and Cys311 in Akt under oxidative stress, leading to enhanced dephosphrylation and the loss of activity.99 These results are consistent with analysis of the crystal structure of inactive Akt.100 Taken together, the oxidative modification of PTEN activates Akt while oxidation of Akt contributes to Akt inactivation, thereby negatively or positively regulating the transcriptional activity of FoxOs, respectively.
As mentioned above, JNK is a key player in the activation of FoxOs, which counteracts the Akt-mediated export of FoxOs from the nucleus.23 NO donors can inhibit JNK1 via S-nitrosylation at Cys116, leading to reduced JNK1 phosphorylation.101, 102 In this case, oxidized JNK decreased FoxOs activity. SIRT1, another crucial regulator implicated in the activation of FoxOs, is sensitive to S-glutathionylation at Cys67, but having no effect on its basal deacetylase activity. Thus, the S-glutathionylation of SIRT1 on Cys67 might not be required for the activation of FoxOs, but it cannot be completely excluded that oxidative modification of other cysteine residues in SIRT1 may be involved in the regulation of FoxOs.
In addition to the above-mentioned examples of cysteine redox modifications upstream of FoxOs that can result in sending a positive or negative signal to FoxOs, it has now become clear that FoxOs themselves can also act as ‘redox sensors.' It has been reported that an intermolecular disulfide bridge can be formed between FoxO4 Cys477 and p300/CBP acetyltransferase.103 The modulation of the switch of FoxO4 signaling from cell-cycle arrest to apoptosis by p300/CBP-mediated acetylation is completely dependent on the presence of this redox-sensitive cysteine modification.103 In humans, FoxO4 contains five cysteines, while FoxO1, FoxO3a, and FoxO6 have 7, 5 and 10 cysteines, respectively, among which two cysteines (including Cys477 in FoxO4) in these four FoxOs are highly conserved.104 It can be speculated that all these four FoxO family members are regulated by redox-dependent cysteine modification.
APE1/Ref-1 is recognized as a ‘redox sensor' that regulates the activity of certain transcription factors
The redox status of reactive cysteine residues, located within the DNA-binding domain of some transcription factors, may control transcriptional activity.105 APE1/Ref-1 has been identified to be a transcriptional regulator that directly modulates the redox status of transcription factors, including activator protein-1 (AP-1),106 HIF-1α,94 Nrf2107 and p53,108 by maintaining the cysteine residues of these transcription factors in the reduced state.105 Redox switches of transcription factors are accomplished by the redox activity of Cys65 (possibly along with Cys93) of APE1/Ref-1 that is required for reduction.109 It has not been completely determined whether this oxidation event results in the formation of an intramolecular disulfide bond or conversion to sulfenic acid, sulfinic acid or S-nitrosothiols.34 Unexpectedly, evidence has emerged that knock-in mice with a cysteine-to-alanine point mutation in the APE1/Ref-1 gene showed no evidence of aberrant redox regulation of AP-1 in vivo, questioning the role of APE1/Ref-1 in redox regulation.110 This contradiction might be due to the presence of compensatory control mechanisms in transgenic mice.105 Indeed, a recent study suggested that other cysteines (Cys93 and Cys310) in APE1/Ref-1 can be S-nitrosylated, potentially explaining the lack of a phenotype in the cysteine mutant mouse.84 The S-nitrosylation of Cys93 and Cys310 partly explained how NO triggers nuclear export of APE1/Ref-1, but the mechanisms involved still remain unknown.
Redox modification in the regulation of Nrf2/Keap1 signaling
Murine Keap1 contains 25 conserved cysteines, of which eight are in the Kelch domain through which Keap1 binds to Nrf2.111 These cysteine residues provide many opportunities for cysteine-dependent redox switches. Under homeostatic conditions, Keap1 interacts with the cullin-3 E3-ubiquitin ligase (Cul3) and serves as a platform for the ubiquitination and resultant proteasomal degradation of Nrf2. However, under conditions of oxidative stress, Keap1 acts as a redox-sensitive adaptor for Cul3, leading to the liberation of Nrf2 from Keap1 suppression.112 It has been reported that oxidative modifications in Cys151, Cys273 and Cys288 of Keap1, probably by the formation of intramolecular or intermolecular disulfide bridges, are of pivotal importance in reduced ubiquitination leading to subsequent accumulation of Nrf2.113, 114, 115, 116, 117, 118 Other possible thiol modifications of Keap1 include S-nitrosylation in response to NO or S-nitrosocysteine causing nuclear accumulation of Keap1.119 However, an increasing number of studies have provided evidence suggesting that these Keap1 redox modifications fail to disrupt the Nrf2/Keap1 complex.117, 120 Thus, additional mechanisms for sensing oxidant signals are believed to be critical for activation of Nrf2. It was shown that Nrf2 mediated the activation of ARE, which is regulated by APE1/Ref-1, suggesting the possibility of Nrf2 redox regulation directly by APE1/Ref-1.121 Indeed, Nrf2 contains a cysteine (Cys514) located in the DNA-binding domain that is the conserved site of APE1/Ref-1-mediated redox regulation. Mutation of murine Nrf2 at Cys506 (equivalent to Cys514 in human) to Ser506 lowered its affinity for the ARE enhancer, suggesting that reduced Cys506 was required for the interaction of Nrf2 and ARE.107 These results were further corroborated by the findings that other cysteine residues with redox modification potential, such as Cys119 and Cys235, play critical roles in the transcriptional activation of Nrf2.122 Additionally, Nrf2 nuclear export from the cell nucleus into the cytoplasm was also regulated by redox signals via Cys183 in Nrf2.123 These observations show that Nrf2 uses a dual sensor mechanism in which both Keap1 and Nrf2 are redox regulated via their cysteine thiols to activate Nrf2 through an integrated signaling cascade.
Cysteine redox modification of ATM
ATM is regarded as the major regulator of the cellular response to double-strand breaks (DSBs), in response to which ATM dimers dissociate to active monomers to orchestrate signaling cascades that initiate the DNA damage.124 However, it has recently been shown that ATM can also serve as a sensor to oxidative stress. In the presence of H2O2, the activation of ATM by DSBs was completely abolished by inhibiting the binding of MRN with DNA. In this situation, ATM was preferentially activated directly by cysteine oxidation. Further characterization demonstrated that the oxidized form of ATM was a disulfide-cross-linked dimer, and Cys2991, was primarily involved in the disulfide bond formation and oxidative activation.124, 125 The specific mechanisms directing redox modification will contribute significantly to the overall levels of ATM activity in the presence of both DSBs and oxidative stress.
p38 is identified as a ‘redox sensor'
p38 protein kinase contains four active cysteine residues, which have the potential to be oxidized under oxidative stress. Indeed, all cysteines in p38 were oxidized under H2O2 treatment, with Cys119 and Cys162 oxidized to a greater extent than Cys211 and Cys39.126 These results were consistent with observations based on the structural analysis of p38 that Cys119 and Cys162 might be more accessible for oxidation.127 Further studies showed that oxidative modification resulted in the loss of p38 kinase activity, and intramolecular disulfide bonds or disulfide bridges to another protein were likely not involved in p38 inhibition.126
Redox modification regulates the transcriptional activity of p53
The remarkable redox-sensitivity of p53 was first discovered by researchers attempting to purify p53. p53 shows high affinity binding to consensus DNA sequences under strong reducing conditions,128, 129, 130 while oxidizing agents can dramatically disrupt p53 conformation and inhibit DNA binding.131, 132 These observations are excellent and agree with evidence from the crystal structure of the DNA-binding core domain of p53.133 Further studies confirmed that Cys277 oxidation was critical for decreased DNA-binding capacity of p53.134 Several other cysteines, including Cys176, Cys238 and Cys242, may also be targets for redox regulation.135 Additionally, p53 is also a substrate for S-glutathionylation under oxidative stress. Molecular modeling studies showed that S-glutathionylation of Cys124 and Cys141 inhibited p53–DNA association and also interfered with protein dimerization.136, 137 It is noteworthy that the redox modification of p53 can be regulated by oxidative stress both directly and indirectly. In the indirect model, APE1/Ref-1 binds to p53 and maintains the thiol side chain of cysteine in the reduced state to impede the dissociation of p53 from DNA.138 In this scenario, redox status of Cys 275 and Cys277 in p53 was demonstrated to be responsible for DNA-binding capacity.108
Conclusions
Recent studies have provided a flood of information concerning the role of redox homeostasis in the regulation of stem cell behavior. Although great progresses have been made in the last two decades, the regulatory mechanisms of stem cell self-renewal mediated by ROS, especially for the downstream signaling mechanisms of ROS stimuli, are still not fully understood. Intriguingly, many of the molecules implicated in ROS-regulated stem cell self-renewal (FoxOs, APE1/Ref-1, Nrf2, ATM, HIF-1α, p38 and p53) have been found to be modified at redox-active cysteine residues, thereby acting as ‘redox sensors.' Understanding whether the ‘redox sensors' play important roles in the regulation of stem cell self-renewal will offer significant potential for stem cell biology in the foreseeable future.
Acknowledgments
This work was supported by grants from the National 973 Basic Research Program of China (2013CB911300, 2012CB518900), the National Science and Technology Major Project (2011ZX09302-001-01, 2012ZX09501001-003) and Chinese NSFC (81172173, 81225015).
Glossary
- APE1/Ref-1
apurinic/apyrimidinic (AP) endonuclease1/redox factor-1
- ARE
antioxidant responsive element
- ASCs
adipose-derived stem cells
- ATM
ataxia telangiectasia mutated
- Cul3
cullin-3
- Cys
cysteine
- DSBs
double-strand breaks
- ER
endoplasmic reticulum
- ESCs
embryonic stem cells
- ETC
mitochondria electron-transport chain
- FoxO
forkhead homeobox type O
- Gpx
glutathione peroxidase
- GR
glutathione reductase
- Grx
glutaredoxin
- H2O2
hydrogen peroxide
- HIF-1α
hypoxia-inducible factor 1α
- iPSC
induced pluripotent stem cell
- ISCs
intestinal stem cells
- JNK
c-Jun N-terminal kinase
- Keap1
kelch-like ECH-associated protein 1
- MSCs
mesenchymal stem cells
- NAC
N-acetyl-cysteine
- NO•
nitric oxide
- NOS
nitric oxide synthase
- NOX
NADPH oxidase
- Nrf2
nuclear factor erythroid-2-related factor 2
- NSC
neural stem cell
- O2−
anion superoxide
- PI3K
phosphoinositide 3-kinase
- Prx
peroxiredoxins
- PTEN
phosphatase and tensin homolog
- PTMs
post-translational modifications
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- RSH
thiol
- RSNO
S-nitrosothiols
- RSO2H
sulfinic acids
- RSO3H
sulfonic acids
- RSOH
sulfenic acids
- RS-SG
S-glutathionylation
- RS-SR
RS-SR', disulfide bonds
- SIRT1
sirtuin 1
- SOD
superoxide dismutase
- Trx
thioredoxin
- Ub
ubiquitin
The authors declare no conflict of interest.
Footnotes
Edited by A Finazzi-Agró
References
- He S, Nakada D, Morrison SJ. Mechanisms of stem cell self-renewal. Annu Rev Cell Dev Biol. 2009;25:377–406. doi: 10.1146/annurev.cellbio.042308.113248. [DOI] [PubMed] [Google Scholar]
- Simons BD, Clevers H. Strategies for homeostatic stem cell self-renewal in adult tissues. Cell. 2011;145:851–862. doi: 10.1016/j.cell.2011.05.033. [DOI] [PubMed] [Google Scholar]
- Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat Rev Genet. 2008;9:115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- Ogasawara MA, Zhang H. Redox regulation and its emerging roles in stem cells and stem-like cancer cells. Antioxid Redox Signal. 2009;11:1107–1122. doi: 10.1089/ars.2008.2308. [DOI] [PubMed] [Google Scholar]
- D'Autreaux B, Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- Mantel C, Messina-Graham SV, Broxmeyer HE. Superoxide flashes, reactive oxygen species, and the mitochondrial permeability transition pore: potential implications for hematopoietic stem cell function. Curr Opin Hematol. 2011;18:208–213. doi: 10.1097/MOH.0b013e3283475ffe. [DOI] [PubMed] [Google Scholar]
- Ghaffari S. Oxidative stress in the regulation of normal and neoplastic hematopoiesis. Antioxid Redox Signal. 2008;10:1923–1940. doi: 10.1089/ars.2008.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case J, Ingram DA, Haneline LS. Oxidative stress impairs endothelial progenitor cell function. Antioxid Redox Signal. 2008;10:1895–1907. doi: 10.1089/ars.2008.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi CI, Suda T. Regulation of reactive oxygen species in stem cells and cancer stem cells. J Cell Physiol. 2012;227:421–430. doi: 10.1002/jcp.22764. [DOI] [PubMed] [Google Scholar]
- Pervaiz S, Taneja R, Ghaffari S. Oxidative stress regulation of stem and progenitor cells. Antioxid Redox Signal. 2009;11:2777–2789. doi: 10.1089/ars.2009.2804. [DOI] [PubMed] [Google Scholar]
- Shi X, Zhang Y, Zheng J, Pan J. Reactive oxygen species in cancer stem cells. Antioxid Redox Signal. 2012;16:1215–1228. doi: 10.1089/ars.2012.4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haneline LS. Redox regulation of stem and progenitor cells. Antioxid Redox Signal. 2008;10:1849–1852. doi: 10.1089/ars.2008.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naka K, Muraguchi T, Hoshii T, Hirao A. Regulation of reactive oxygen species and genomic stability in hematopoietic stem cells. Antioxid Redox Signal. 2008;10:1883–1894. doi: 10.1089/ars.2008.2114. [DOI] [PubMed] [Google Scholar]
- Dickinson BC, Chang CJ. Chemistry and biology of reactive oxygen species in signaling or stress responses. Nat Chem Biol. 2011;7:504–511. doi: 10.1038/nchembio.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohyeldin A, Garzon-Muvdi T, Quinones-Hinojosa A. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell. 2010;7:150–161. doi: 10.1016/j.stem.2010.07.007. [DOI] [PubMed] [Google Scholar]
- Holzwarth C, Vaegler M, Gieseke F, Pfister SM, Handgretinger R, Kerst G, et al. Low physiologic oxygen tensions reduce proliferation and differentiation of human multipotent mesenchymal stromal cells. BMC Cell Biol. 2010;11:11. doi: 10.1186/1471-2121-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell. 2009;5:237–241. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Eliasson P, Jonsson JI. The hematopoietic stem cell niche: low in oxygen but a nice place to be. J Cell Physiol. 2010;222:17–22. doi: 10.1002/jcp.21908. [DOI] [PubMed] [Google Scholar]
- Vieira HL, Alves PM, Vercelli A. Modulation of neuronal stem cell differentiation by hypoxia and reactive oxygen species. Prog Neurobiol. 2011;93:444–455. doi: 10.1016/j.pneurobio.2011.01.007. [DOI] [PubMed] [Google Scholar]
- Lee EY, Xia Y, Kim WS, Kim MH, Kim TH, Kim KJ, et al. Hypoxia-enhanced wound-healing function of adipose-derived stem cells: increase in stem cell proliferation and up-regulation of VEGF and bFGF. Wound Repair Regen. 2009;17:540–547. doi: 10.1111/j.1524-475X.2009.00499.x. [DOI] [PubMed] [Google Scholar]
- Kim JH, Park SH, Park SG, Choi JS, Xia Y, Sung JH. The pivotal role of reactive oxygen species generation in the hypoxia-induced stimulation of adipose-derived stem cells. Stem Cells Dev. 2011;20:1753–1761. doi: 10.1089/scd.2010.0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dansen TB. Forkhead Box O transcription factors: key players in redox signaling. Antioxid Redox Signal. 2011;14:559–561. doi: 10.1089/ars.2010.3778. [DOI] [PubMed] [Google Scholar]
- Tothova Z, Gilliland DG. FoxO transcription factors and stem cell homeostasis: insights from the hematopoietic system. Cell Stem Cell. 2007;1:140–152. doi: 10.1016/j.stem.2007.07.017. [DOI] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Storz P. Forkhead homeobox type O transcription factors in the responses to oxidative stress. Antioxid Redox Signal. 2011;14:593–605. doi: 10.1089/ars.2010.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K, Araki KY, Naka K, Arai F, Takubo K, Yamazaki S, et al. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell. 2007;1:101–112. doi: 10.1016/j.stem.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Miyamoto K, Miyamoto T, Kato R, Yoshimura A, Motoyama N, Suda T. FoxO3a regulates hematopoietic homeostasis through a negative feedback pathway in conditions of stress or aging. Blood. 2008;112:4485–4493. doi: 10.1182/blood-2008-05-159848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault VM, Rafalski VA, Morgan AA, Salih DA, Brett JO, Webb AE, et al. FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell. 2009;5:527–539. doi: 10.1016/j.stem.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik JH, Ding Z, Narurkar R, Ramkissoon S, Muller F, Kamoun WS, et al. FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell Stem Cell. 2009;5:540–553. doi: 10.1016/j.stem.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yalcin S, Lee DF, Yeh TY, Lee SM, Su J, et al. FOXO1 is an essential regulator of pluripotency in human embryonic stem cells. Nat Cell Biol. 2011;13:1092–1099. doi: 10.1038/ncb2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales KA, Ng HH. FoxO: a new addition to the ESC cartel. Cell Stem Cell. 2011;9:181–183. doi: 10.1016/j.stem.2011.08.003. [DOI] [PubMed] [Google Scholar]
- Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Yalcin S, Zhang X, Luciano JP, Mungamuri SK, Marinkovic D, Vercherat C, et al. Foxo3 is essential for the regulation of ataxia telangiectasia mutated and oxidative stress-mediated homeostasis of hematopoietic stem cells. J Biol Chem. 2008;283:25692–25705. doi: 10.1074/jbc.M800517200. [DOI] [PubMed] [Google Scholar]
- Bhakat KK, Mantha AK, Mitra S. Transcriptional regulatory functions of mammalian AP-endonuclease (APE1/Ref-1), an essential multifunctional protein. Antioxid Redox Signal. 2009;11:621–638. doi: 10.1089/ars.2008.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angkeow P, Deshpande SS, Qi B, Liu YX, Park YC, Jeon BH, et al. Redox factor-1: an extra-nuclear role in the regulation of endothelial oxidative stress and apoptosis. Cell Death Differ. 2002;9:717–725. doi: 10.1038/sj.cdd.4401025. [DOI] [PubMed] [Google Scholar]
- Zou GM, Luo MH, Reed A, Kelley MR, Yoder MC. Ape1 regulates hematopoietic differentiation of embryonic stem cells through its redox functional domain. Blood. 2007;109:1917–1922. doi: 10.1182/blood-2006-08-044172. [DOI] [PubMed] [Google Scholar]
- Gurusamy N, Mukherjee S, Lekli I, Bearzi C, Bardelli S, Das DK. Inhibition of ref-1 stimulates the production of reactive oxygen species and induces differentiation in adult cardiac stem cells. Antioxid Redox Signal. 2009;11:589–600. doi: 10.1089/ars.2008.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JY, Jing K, Song KS, Seo KS, Park JH, Kim JS, et al. Downregulation of APE1/Ref-1 is involved in the senescence of mesenchymal stem cells. Stem Cells. 2009;27:1455–1462. doi: 10.1002/stem.54. [DOI] [PubMed] [Google Scholar]
- Sporn MB, Liby KT. NRF2 and cancer: the good, the bad and the importance of context. Nat Rev Cancer. 2012;12:564–571. doi: 10.1038/nrc3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004;10:549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Li J, Johnson D, Calkins M, Wright L, Svendsen C, Johnson J. Stabilization of Nrf2 by tBHQ confers protection against oxidative stress-induced cell death in human neural stem cells. Toxicol Sci. 2005;83:313–328. doi: 10.1093/toxsci/kfi027. [DOI] [PubMed] [Google Scholar]
- Takahata Y, Takarada T, Iemata M, Yamamoto T, Nakamura Y, Kodama A, et al. Functional expression of beta2 adrenergic receptors responsible for protection against oxidative stress through promotion of glutathione synthesis after Nrf2 upregulation in undifferentiated mesenchymal C3H10T1/2 stem cells. J Cell Physiol. 2009;218:268–275. doi: 10.1002/jcp.21594. [DOI] [PubMed] [Google Scholar]
- Hochmuth CE, Biteau B, Bohmann D, Jasper H. Redox regulation by Keap1 and Nrf2 controls intestinal stem cell proliferation in Drosophila. Cell Stem Cell. 2011;8:188–199. doi: 10.1016/j.stem.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant AA, Singh A, Matsui W, Biswal S. The redox-sensitive transcription factor Nrf2 regulates murine hematopoietic stem cell survival independently of ROS levels. Blood. 2011;118:6572–6579. doi: 10.1182/blood-2011-05-355362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatti S, Kozlov S, Farooqi AA, Naqi A, Lavin M, Khanna KK. ATM protein kinase: the linchpin of cellular defenses to stress. Cell Mol Life Sci. 2011;68:2977–3006. doi: 10.1007/s00018-011-0683-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditch S, Paull TT. The ATM protein kinase and cellular redox signaling: beyond the DNA damage response. Trends Biochem Sci. 2012;37:15–22. doi: 10.1016/j.tibs.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryanovich M, Oberkovitz G, Niv H, Vorobiyov L, Zaltsman Y, Brenner O, et al. The ATM-BID pathway regulates quiescence and survival of haematopoietic stem cells. Nat Cell Biol. 2012;14:535–541. doi: 10.1038/ncb2468. [DOI] [PubMed] [Google Scholar]
- Ito K, Hirao A, Arai F, Takubo K, Matsuoka S, Miyamoto K, et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat Med. 2006;12:446–451. doi: 10.1038/nm1388. [DOI] [PubMed] [Google Scholar]
- Kim J, Wong PK. Loss of ATM impairs proliferation of neural stem cells through oxidative stress-mediated p38 MAPK signaling. Stem Cells. 2009;27:1987–1998. doi: 10.1002/stem.125. [DOI] [PubMed] [Google Scholar]
- Chen C, Liu Y, Liu R, Ikenoue T, Guan KL, Zheng P. TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J Exp Med. 2008;205:2397–2408. doi: 10.1084/jem.20081297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntilla MM, Patil VD, Calamito M, Joshi RP, Birnbaum MJ, Koretzky GA. AKT1 and AKT2 maintain hematopoietic stem cell function by regulating reactive oxygen species. Blood. 2010;115:4030–4038. doi: 10.1182/blood-2009-09-241000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuikov S, Levi BP, Smith ML, Morrison SJ. Prdm16 promotes stem cell maintenance in multiple tissues, partly by regulating oxidative stress. Nat Cell Biol. 2010;12:999–1006. doi: 10.1038/ncb2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko E, Lee KY, Hwang DS. Human umbilical cord blood-derived mesenchymal stem cells undergo cellular senescence in response to oxidative stress. Stem Cells Dev. 2012;21:1877–1886. doi: 10.1089/scd.2011.0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson WR, Hay MP. Targeting hypoxia in cancer therapy. Nat Rev Cancer. 2011;11:393–410. doi: 10.1038/nrc3064. [DOI] [PubMed] [Google Scholar]
- Takubo K, Goda N, Yamada W, Iriuchishima H, Ikeda E, Kubota Y, et al. Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell. 2010;7:391–402. doi: 10.1016/j.stem.2010.06.020. [DOI] [PubMed] [Google Scholar]
- Simsek T, Kocabas F, Zheng J, Deberardinis RJ, Mahmoud AI, Olson EN, et al. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell. 2010;7:380–390. doi: 10.1016/j.stem.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covello KL, Kehler J, Yu H, Gordan JD, Arsham AM, Hu CJ, et al. HIF-2alpha regulates Oct-4: effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes Dev. 2006;20:557–570. doi: 10.1101/gad.1399906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulthard LR, White DE, Jones DL, McDermott MF, Burchill SA. p38(MAPK): stress responses from molecular mechanisms to therapeutics. Trends Mol Med. 2009;15:369–379. doi: 10.1016/j.molmed.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- Torres M, Forman HJ. Redox signaling and the MAP kinase pathways. Biofactors. 2003;17:287–296. doi: 10.1002/biof.5520170128. [DOI] [PubMed] [Google Scholar]
- Baudet A, Karlsson C, Safaee Talkhoncheh M, Galeev R, Magnusson M, Larsson J. RNAi screen identifies MAPK14 as a druggable suppressor of human hematopoietic stem cell expansion. Blood. 2012;119:6255–6258. doi: 10.1182/blood-2012-01-403949. [DOI] [PubMed] [Google Scholar]
- Bhandari DR, Seo KW, Roh KH, Jung JW, Kang SK, Kang KS. REX-1 expression and p38 MAPK activation status can determine proliferation/differentiation fates in human mesenchymal stem cells. PLoS One. 2010;5:e10493. doi: 10.1371/journal.pone.0010493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang YY, Sharkis SJ. A low level of reactive oxygen species selects for primitive hematopoietic stem cells that may reside in the low-oxygenic niche. Blood. 2007;110:3056–3063. doi: 10.1182/blood-2007-05-087759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Liang XG, Hu Y, Zhu DY, Lou YJ. Involvement of p38MAPK and reactive oxygen species in icariin-induced cardiomyocyte differentiation of murine embryonic stem cells in vitro. Stem Cells Dev. 2008;17:751–760. doi: 10.1089/scd.2007.0206. [DOI] [PubMed] [Google Scholar]
- Junttila MR, Evan GI. p53--a Jack of all trades but master of none. Nat Rev Cancer. 2009;9:821–829. doi: 10.1038/nrc2728. [DOI] [PubMed] [Google Scholar]
- Bonizzi G, Cicalese A, Insinga A, Pelicci PG. The emerging role of p53 in stem cells. Trends Mol Med. 2012;18:6–12. doi: 10.1016/j.molmed.2011.08.002. [DOI] [PubMed] [Google Scholar]
- Abbas HA, Pant V, Lozano G. The ups and downs of p53 regulation in hematopoietic stem cells. Cell Cycle. 2011;10:3257–3262. doi: 10.4161/cc.10.19.17721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, He Y, Dubois W, Wu X, Shi J, Huang J. Distinct regulatory mechanisms and functions for p53-activated and p53-repressed DNA damage response genes in embryonic stem cells. Mol Cell. 2012;46:30–42. doi: 10.1016/j.molcel.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E, et al. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol. 2005;7:165–171. doi: 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
- Das B, Bayat-Mokhtari R, Tsui M, Lotfi S, Tsuchida R, Felsher DW, et al. HIF-2alpha suppresses p53 to enhance the stemness and regenerative potential of human embryonic stem cells. Stem Cells. 2012;30:1685–1695. doi: 10.1002/stem.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao M, Campbell K, Burns K, Kuan CY, Trumpp A, Nakafuku M. Coordinated control of self-renewal and differentiation of neural stem cells by Myc and the p19ARF-p53 pathway. J Cell Biol. 2008;183:1243–1257. doi: 10.1083/jcb.200807130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafsi H, Hainaut P. Redox control and interplay between p53 isoforms: roles in the regulation of basal p53 levels, cell fate, and senescence. Antioxid Redox Signal. 2011;15:1655–1667. doi: 10.1089/ars.2010.3771. [DOI] [PubMed] [Google Scholar]
- Maillet A, Pervaiz S. Redox regulation of p53, redox effectors regulated by p53: a subtle balance. Antioxid Redox Signal. 2012;16:1285–1294. doi: 10.1089/ars.2011.4434. [DOI] [PubMed] [Google Scholar]
- Han MK, Song EK, Guo Y, Ou X, Mantel C, Broxmeyer HE. SIRT1 regulates apoptosis and Nanog expression in mouse embryonic stem cells by controlling p53 subcellular localization. Cell Stem Cell. 2008;2:241–251. doi: 10.1016/j.stem.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas HA, Maccio DR, Coskun S, Jackson JG, Hazen AL, Sills TM, et al. Mdm2 is required for survival of hematopoietic stem cells/progenitors via dampening of ROS-induced p53 activity. Cell Stem Cell. 2010;7:606–617. doi: 10.1016/j.stem.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deribe YL, Pawson T, Dikic I. Post-translational modifications in signal integration. Nat Struct Mol Biol. 2010;17:666–672. doi: 10.1038/nsmb.1842. [DOI] [PubMed] [Google Scholar]
- Leonard SE, Carroll KS. Chemical 'omics' approaches for understanding protein cysteine oxidation in biology. Curr Opin Chem Biol. 2011;15:88–102. doi: 10.1016/j.cbpa.2010.11.012. [DOI] [PubMed] [Google Scholar]
- Ray PD, Huang BW, Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal. 2012;24:981–990. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen-Heininger YM, Mossman BT, Heintz NH, Forman HJ, Kalyanaraman B, Finkel T, et al. Redox-based regulation of signal transduction: principles, pitfalls, and promises. Free Radic Biol Med. 2008;45:1–17. doi: 10.1016/j.freeradbiomed.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen CE, Carroll KS. Orchestrating redox signaling networks through regulatory cysteine switches. ACS Chem Biol. 2010;5:47–62. doi: 10.1021/cb900258z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T. Signal transduction by reactive oxygen species. J Cell Biol. 2011;194:7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddie KG, Carroll KS. Expanding the functional diversity of proteins through cysteine oxidation. Curr Opin Chem Biol. 2008;12:746–754. doi: 10.1016/j.cbpa.2008.07.028. [DOI] [PubMed] [Google Scholar]
- Thamsen M, Jakob U. The redoxome: proteomic analysis of cellular redox networks. Curr Opin Chem Biol. 2011;15:113–119. doi: 10.1016/j.cbpa.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisy K, Peet DJ. Turn me on: regulating HIF transcriptional activity. Cell Death Differ. 2008;15:642–649. doi: 10.1038/sj.cdd.4402315. [DOI] [PubMed] [Google Scholar]
- Sumbayev VV, Budde A, Zhou J, Brune B. HIF-1 alpha protein as a target for S-nitrosation. FEBS Lett. 2003;535:106–112. doi: 10.1016/s0014-5793(02)03887-5. [DOI] [PubMed] [Google Scholar]
- Yasinska IM, Sumbayev VV. S-nitrosation of Cys-800 of HIF-1alpha protein activates its interaction with p300 and stimulates its transcriptional activity. FEBS Lett. 2003;549:105–109. doi: 10.1016/s0014-5793(03)00807-x. [DOI] [PubMed] [Google Scholar]
- Li F, Sonveaux P, Rabbani ZN, Liu S, Yan B, Huang Q, et al. Regulation of HIF-1alpha stability through S-nitrosylation. Mol Cell. 2007;26:63–74. doi: 10.1016/j.molcel.2007.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Ahn DR, Park H, Yang EG. Modulation of p300 binding by posttranslational modifications of the C-terminal activation domain of hypoxia-inducible factor-1alpha. FEBS Lett. 2007;581:1542–1548. doi: 10.1016/j.febslet.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Huang LE, Arany Z, Livingston DM, Bunn HF. Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J Biol Chem. 1996;271:32253–32259. doi: 10.1074/jbc.271.50.32253. [DOI] [PubMed] [Google Scholar]
- Wrighton KH. Tumour suppressors: role of nuclear PTEN revealed. Nat Rev Cancer. 2011;11:154. doi: 10.1038/nrc3028. [DOI] [PubMed] [Google Scholar]
- Kwon J, Lee SR, Yang KS, Ahn Y, Kim YJ, Stadtman ER, et al. Reversible oxidation and inactivation of the tumor suppressor PTEN in cells stimulated with peptide growth factors. Proc Natl Acad Sci USA. 2004;101:16419–16424. doi: 10.1073/pnas.0407396101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Yang KS, Kwon J, Lee C, Jeong W, Rhee SG. Reversible inactivation of the tumor suppressor PTEN by H2O2. J Biol Chem. 2002;277:20336–20342. doi: 10.1074/jbc.M111899200. [DOI] [PubMed] [Google Scholar]
- Lim S, Clement MV. Phosphorylation of the survival kinase Akt by superoxide is dependent on an ascorbate-reversible oxidation of PTEN. Free Radic Biol Med. 2007;42:1178–1192. doi: 10.1016/j.freeradbiomed.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Murata H, Ihara Y, Nakamura H, Yodoi J, Sumikawa K, Kondo T. Glutaredoxin exerts an antiapoptotic effect by regulating the redox state of Akt. J Biol Chem. 2003;278:50226–50233. doi: 10.1074/jbc.M310171200. [DOI] [PubMed] [Google Scholar]
- Huang X, Begley M, Morgenstern KA, Gu Y, Rose P, Zhao H, et al. Crystal structure of an inactive Akt2 kinase domain. Structure. 2003;11:21–30. doi: 10.1016/s0969-2126(02)00937-1. [DOI] [PubMed] [Google Scholar]
- Park HS, Huh SH, Kim MS, Lee SH, Choi EJ. Nitric oxide negatively regulates c-Jun N-terminal kinase/stress-activated protein kinase by means of S-nitrosylation. Proc Natl Acad Sci USA. 2000;97:14382–14387. doi: 10.1073/pnas.97.26.14382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Korolchuk VI, Renna M, Imarisio S, Fleming A, Williams A, et al. Complex inhibitory effects of nitric oxide on autophagy. Mol Cell. 2011;43:19–32. doi: 10.1016/j.molcel.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dansen TB, Smits LM, van Triest MH, de Keizer PL, van Leenen D, Koerkamp MG, et al. Redox-sensitive cysteines bridge p300/CBP-mediated acetylation and FoxO4 activity. Nat Chem Biol. 2009;5:664–672. doi: 10.1038/nchembio.194. [DOI] [PubMed] [Google Scholar]
- de Keizer PL, Burgering BM, Dansen TB. Forkhead box o as a sensor, mediator, and regulator of redox signaling. Antioxid Redox Signal. 2011;14:1093–1106. doi: 10.1089/ars.2010.3403. [DOI] [PubMed] [Google Scholar]
- Tell G, Quadrifoglio F, Tiribelli C, Kelley MR. The many functions of APE1/Ref-1: not only a DNA repair enzyme. Antioxid Redox Signal. 2009;11:601–620. doi: 10.1089/ars.2008.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xanthoudakis S, Curran T. Identification and characterization of Ref-1, a nuclear protein that facilitates AP-1 DNA-binding activity. EMBO J. 1992;11:653–665. doi: 10.1002/j.1460-2075.1992.tb05097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom D, Dhakshinamoorthy S, Jaiswal AK. Site-directed mutagenesis of cysteine to serine in the DNA binding region of Nrf2 decreases its capacity to upregulate antioxidant response element-mediated expression and antioxidant induction of NAD(P)H:quinone oxidoreductase1 gene. Oncogene. 2002;21:2191–2200. doi: 10.1038/sj.onc.1205288. [DOI] [PubMed] [Google Scholar]
- Seo YR, Kelley MR, Smith ML. Selenomethionine regulation of p53 by a ref1-dependent redox mechanism. Proc Natl Acad Sci USA. 2002;99:14548–14553. doi: 10.1073/pnas.212319799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiadis MM, Luo M, Gaur RK, Delaplane S, Li X, Kelley MR. Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease. Mutat Res. 2008;643:54–63. doi: 10.1016/j.mrfmmm.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordway JM, Eberhart D, Curran T. Cysteine 64 of Ref-1 is not essential for redox regulation of AP-1 DNA binding. Mol Cell Biol. 2003;23:4257–4266. doi: 10.1128/MCB.23.12.4257-4266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang D, Hannink M, Beamer LJ. Crystal structure of the Kelch domain of human Keap1. J Biol Chem. 2004;279:54750–54758. doi: 10.1074/jbc.M410073200. [DOI] [PubMed] [Google Scholar]
- Villeneuve NF, Lau A, Zhang DD. Regulation of the Nrf2-Keap1 antioxidant response by the ubiquitin proteasome system: an insight into cullin-ring ubiquitin ligases. Antioxid Redox Signal. 2010;13:1699–1712. doi: 10.1089/ars.2010.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, et al. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci USA. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, Kang MI, Kobayashi A, Yamamoto M, et al. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci USA. 2004;101:2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Kang MI, Watai Y, Tong KI, Shibata T, Uchida K, et al. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol Cell Biol. 2006;26:221–229. doi: 10.1128/MCB.26.1.221-229.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Suzuki T, Kobayashi A, Wakabayashi J, Maher J, Motohashi H, et al. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol Cell Biol. 2008;28:2758–2770. doi: 10.1128/MCB.01704-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley BJ, Li S, Whorton AR. Keap1 modification and nuclear accumulation in response to S-nitrosocysteine. Free Radic Biol Med. 2008;44:692–698. doi: 10.1016/j.freeradbiomed.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Chen MG, Lin GX, Ma Q. Arsenic induces NAD(P)H-quinone oxidoreductase I by disrupting the Nrf2 x Keap1 x Cul3 complex and recruiting Nrf2 x Maf to the antioxidant response element enhancer. J Biol Chem. 2006;281:23620–23631. doi: 10.1074/jbc.M604120200. [DOI] [PubMed] [Google Scholar]
- Iwasaki K, Mackenzie EL, Hailemariam K, Sakamoto K, Tsuji Y. Hemin-mediated regulation of an antioxidant-responsive element of the human ferritin H gene and role of Ref-1 during erythroid differentiation of K562 cells. Mol Cell Biol. 2006;26:2845–2856. doi: 10.1128/MCB.26.7.2845-2856.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Ma Q. NRF2 cysteine residues are critical for oxidant/electrophile-sensing, Kelch-like ECH-associated protein-1-dependent ubiquitination-proteasomal degradation, and transcription activation. Mol Pharmacol. 2009;76:1265–1278. doi: 10.1124/mol.109.058453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Yu SW, Kong AN. Nrf2 possesses a redox-sensitive nuclear exporting signal in the Neh5 transactivation domain. J Biol Chem. 2006;281:27251–27263. doi: 10.1074/jbc.M602746200. [DOI] [PubMed] [Google Scholar]
- Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT. ATM activation by oxidative stress. Science. 2010;330:517–521. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- Guo Z, Deshpande R, Paull TT. ATM activation in the presence of oxidative stress. Cell Cycle. 2010;9:4805–4811. doi: 10.4161/cc.9.24.14323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton DJ, Aye MS, Rady J, Xu F, Cross JV. Purification of reversibly oxidized proteins (PROP) reveals a redox switch controlling p38 MAP kinase activity. PLoS One. 2010;5:e15012. doi: 10.1371/journal.pone.0015012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KP, Fitzgibbon MJ, Caron PR, Griffith JP, Chen W, McCaffrey PG, et al. Crystal structure of p38 mitogen-activated protein kinase. J Biol Chem. 1996;271:27696–27700. doi: 10.1074/jbc.271.44.27696. [DOI] [PubMed] [Google Scholar]
- Hupp TR, Meek DW, Midgley CA, Lane DP. Regulation of the specific DNA binding function of p53. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- Parks D, Bolinger R, Mann K. Redox state regulates binding of p53 to sequence-specific DNA, but not to non-specific or mismatched DNA. Nucleic Acids Res. 1997;25:1289–1295. doi: 10.1093/nar/25.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainaut P, Mann K. Zinc binding and redox control of p53 structure and function. Antioxid Redox Signal. 2001;3:611–623. doi: 10.1089/15230860152542961. [DOI] [PubMed] [Google Scholar]
- Hainaut P, Milner J. Redox modulation of p53 conformation and sequence-specific DNA binding in vitro. Cancer Res. 1993;53:4469–4473. [PubMed] [Google Scholar]
- Wu HH, Momand J. Pyrrolidine dithiocarbamate prevents p53 activation and promotes p53 cysteine residue oxidation. J Biol Chem. 1998;273:18898–18905. doi: 10.1074/jbc.273.30.18898. [DOI] [PubMed] [Google Scholar]
- Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- Buzek J, Latonen L, Kurki S, Peltonen K, Laiho M. Redox state of tumor suppressor p53 regulates its sequence-specific DNA binding in DNA-damaged cells by cysteine 277. Nucleic Acids Res. 2002;30:2340–2348. doi: 10.1093/nar/30.11.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainwater R, Parks D, Anderson ME, Tegtmeyer P, Mann K. Role of cysteine residues in regulation of p53 function. Mol Cell Biol. 1995;15:3892–3903. doi: 10.1128/mcb.15.7.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velu CS, Niture SK, Doneanu CE, Pattabiraman N, Srivenugopal KS. Human p53 is inhibited by glutathionylation of cysteines present in the proximal DNA-binding domain during oxidative stress. Biochemistry. 2007;46:7765–7780. doi: 10.1021/bi700425y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusuf MA, Chuang T, Bhat GJ, Srivenugopal KS. Cys-141 glutathionylation of human p53: Studies using specific polyclonal antibodies in cancer samples and cell lines. Free Radic Biol Med. 2010;49:908–917. doi: 10.1016/j.freeradbiomed.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann S, Hainaut P. Roles of thioredoxin reductase 1 and APE/Ref-1 in the control of basal p53 stability and activity. Oncogene. 2005;24:3853–3863. doi: 10.1038/sj.onc.1208549. [DOI] [PubMed] [Google Scholar]