Abstract
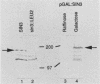
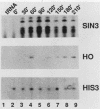
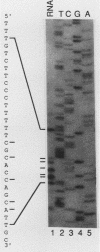
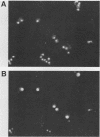
The SIN3 gene (also known as SDI1) is a negative regulator of the yeast HO gene. Mutations in SIN3 suppress the requirement for the SWI5 activator for expression of the yeast HO gene and change the normal asymmetric pattern of HO expression in mother and daughter cells. Furthermore, the in vitro DNA-binding activity of several DNA-binding proteins is reduced in extracts prepared from sin3 mutants. We have cloned the SIN3 gene and determined that a haploid strain with a SIN3 gene disruption is viable. We determined the sequence of the SIN3 gene, which is predicted to encode a 175-kDa polypeptide with four paired amphipathic helix motifs. These motifs have been identified in the myc family of helix-loop-helix DNA-binding proteins and in the TPR family of regulatory proteins. The SIN3 transcript was mapped, and it was determined that the SIN3 transcript was absent in stationary-phase cells. Immunofluorescence microscopy with anti-SIN3 antibody demonstrated that SIN3 protein was present in nuclei. A comparison of restriction map and sequence data revealed that SIN3 is the same as regulatory genes UME4 and RPD1.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams A. E., Pringle J. R. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J Cell Biol. 1984 Mar;98(3):934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Breeden L., Nasmyth K. Cell cycle control of the yeast HO gene: cis- and trans-acting regulators. Cell. 1987 Feb 13;48(3):389–397. doi: 10.1016/0092-8674(87)90190-5. [DOI] [PubMed] [Google Scholar]
- Carlson M., Osmond B. C., Neigeborn L., Botstein D. A suppressor of SNF1 mutations causes constitutive high-level invertase synthesis in yeast. Genetics. 1984 May;107(1):19–32. doi: 10.1093/genetics/107.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan A. M., Donahue T. F. Sequence and structural features associated with translational initiator regions in yeast--a review. Gene. 1987;59(1):1–18. doi: 10.1016/0378-1119(87)90261-7. [DOI] [PubMed] [Google Scholar]
- Davis R. L., Cheng P. F., Lassar A. B., Weintraub H. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell. 1990 Mar 9;60(5):733–746. doi: 10.1016/0092-8674(90)90088-v. [DOI] [PubMed] [Google Scholar]
- Dieckmann C. L., Tzagoloff A. Assembly of the mitochondrial membrane system. CBP6, a yeast nuclear gene necessary for synthesis of cytochrome b. J Biol Chem. 1985 Feb 10;260(3):1513–1520. [PubMed] [Google Scholar]
- Fassler J. S., Winston F. The Saccharomyces cerevisiae SPT13/GAL11 gene has both positive and negative regulatory roles in transcription. Mol Cell Biol. 1989 Dec;9(12):5602–5609. doi: 10.1128/mcb.9.12.5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink G. R., Styles C. A. Curing of a killer factor in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1972 Oct;69(10):2846–2849. doi: 10.1073/pnas.69.10.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyer M. S. The gamma delta sequence of F is an insertion sequence. J Mol Biol. 1978 Dec 15;126(3):347–365. doi: 10.1016/0022-2836(78)90045-1. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hirano T., Kinoshita N., Morikawa K., Yanagida M. Snap helix with knob and hole: essential repeats in S. pombe nuclear protein nuc2+. Cell. 1990 Jan 26;60(2):319–328. doi: 10.1016/0092-8674(90)90746-2. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R. E., Herskowitz I. Directionality and regulation of cassette substitution in yeast. Cold Spring Harb Symp Quant Biol. 1984;49:97–104. doi: 10.1101/sqb.1984.049.01.013. [DOI] [PubMed] [Google Scholar]
- Kukuruzinska M. A., Bergh M. L., Jackson B. J. Protein glycosylation in yeast. Annu Rev Biochem. 1987;56:915–944. doi: 10.1146/annurev.bi.56.070187.004411. [DOI] [PubMed] [Google Scholar]
- Marck C. 'DNA Strider': a 'C' program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1988 Mar 11;16(5):1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight S. L., Gavis E. R., Kingsbury R., Axel R. Analysis of transcriptional regulatory signals of the HSV thymidine kinase gene: identification of an upstream control region. Cell. 1981 Aug;25(2):385–398. doi: 10.1016/0092-8674(81)90057-x. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989 Mar 10;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Vaessin H., Caudy M., Jan L. Y., Jan Y. N., Cabrera C. V., Buskin J. N., Hauschka S. D., Lassar A. B. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989 Aug 11;58(3):537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Molecular analysis of a cell lineage. Nature. 1983 Apr 21;302(5910):670–676. doi: 10.1038/302670a0. [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Seddon A., Ammerer G. Cell cycle regulation of SW15 is required for mother-cell-specific HO transcription in yeast. Cell. 1987 May 22;49(4):549–558. doi: 10.1016/0092-8674(87)90457-0. [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Stillman D., Kipling D. Both positive and negative regulators of HO transcription are required for mother-cell-specific mating-type switching in yeast. Cell. 1987 Feb 27;48(4):579–587. doi: 10.1016/0092-8674(87)90236-4. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. The determination of mother cell-specific mating type switching in yeast by a specific regulator of HO transcription. EMBO J. 1987 Jan;6(1):243–248. doi: 10.1002/j.1460-2075.1987.tb04745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara P. J., Horowitz H., Eichinger G., Young E. T. The yeast ADR6 gene encodes homopolymeric amino acid sequences and a potential metal-binding domain. Nucleic Acids Res. 1988 Nov 11;16(21):10153–10169. doi: 10.1093/nar/16.21.10153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer K., Kim K. S., Kogan S., Guarente L. Functional dissection and sequence of yeast HAP1 activator. Cell. 1989 Jan 27;56(2):291–301. doi: 10.1016/0092-8674(89)90903-3. [DOI] [PubMed] [Google Scholar]
- Pinkham J. L., Olesen J. T., Guarente L. P. Sequence and nuclear localization of the Saccharomyces cerevisiae HAP2 protein, a transcriptional activator. Mol Cell Biol. 1987 Feb;7(2):578–585. doi: 10.1128/mcb.7.2.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V., Manet E., Hardon E., Bickel S. E., Benson M. Structure and sequence of the Drosophila zeste gene. EMBO J. 1987 Mar;6(3):791–799. doi: 10.1002/j.1460-2075.1987.tb04821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S., Wells R., Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986 Oct 17;234(4774):364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J., Sherman F. Genes affecting the expression of cytochrome c in yeast: genetic mapping and genetic interactions. Genetics. 1980 Apr;94(4):871–889. doi: 10.1093/genetics/94.4.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüther U., Müller-Hill B. Easy identification of cDNA clones. EMBO J. 1983;2(10):1791–1794. doi: 10.1002/j.1460-2075.1983.tb01659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J., Carlson M. Molecular analysis of SSN6, a gene functionally related to the SNF1 protein kinase of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Oct;7(10):3637–3645. doi: 10.1128/mcb.7.10.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengstag C., Hinnen A. The sequence of the Saccharomyces cerevisiae gene PHO2 codes for a regulatory protein with unusual aminoacid composition. Nucleic Acids Res. 1987 Jan 12;15(1):233–246. doi: 10.1093/nar/15.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987 Feb 11;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Boguski M. S., Goebl M., Hieter P. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell. 1990 Jan 26;60(2):307–317. doi: 10.1016/0092-8674(90)90745-z. [DOI] [PubMed] [Google Scholar]
- Stern M., Jensen R., Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. J Mol Biol. 1984 Oct 5;178(4):853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- Sternberg P. W., Stern M. J., Clark I., Herskowitz I. Activation of the yeast HO gene by release from multiple negative controls. Cell. 1987 Feb 27;48(4):567–577. doi: 10.1016/0092-8674(87)90235-2. [DOI] [PubMed] [Google Scholar]
- Stillman D. J., Bankier A. T., Seddon A., Groenhout E. G., Nasmyth K. A. Characterization of a transcription factor involved in mother cell specific transcription of the yeast HO gene. EMBO J. 1988 Feb;7(2):485–494. doi: 10.1002/j.1460-2075.1988.tb02836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern J. N., Herskowitz I. Asymmetry and directionality in production of new cell types during clonal growth: the switching pattern of homothallic yeast. Cell. 1979 Jun;17(2):371–381. doi: 10.1016/0092-8674(79)90163-6. [DOI] [PubMed] [Google Scholar]
- Strich R., Slater M. R., Esposito R. E. Identification of negative regulatory genes that govern the expression of early meiotic genes in yeast. Proc Natl Acad Sci U S A. 1989 Dec;86(24):10018–10022. doi: 10.1073/pnas.86.24.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Nogi Y., Abe A., Fukasawa T. GAL11 protein, an auxiliary transcription activator for genes encoding galactose-metabolizing enzymes in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Nov;8(11):4991–4999. doi: 10.1128/mcb.8.11.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumbly R. J. Cloning and characterization of the CYC8 gene mediating glucose repression in yeast. Gene. 1988 Dec 15;73(1):97–111. doi: 10.1016/0378-1119(88)90316-2. [DOI] [PubMed] [Google Scholar]
- Vidal M., Buckley A. M., Hilger F., Gaber R. F. Direct selection for mutants with increased K+ transport in Saccharomyces cerevisiae. Genetics. 1990 Jun;125(2):313–320. doi: 10.1093/genetics/125.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson C. R., Sigler P. B., McKnight S. L. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989 Nov 17;246(4932):911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- Wang H., Nicholson P. R., Stillman D. J. Identification of a Saccharomyces cerevisiae DNA-binding protein involved in transcriptional regulation. Mol Cell Biol. 1990 Apr;10(4):1743–1753. doi: 10.1128/mcb.10.4.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford J. L., Jr Nuclear pre-mRNA splicing in yeast. Yeast. 1989 Nov-Dec;5(6):439–457. doi: 10.1002/yea.320050604. [DOI] [PubMed] [Google Scholar]
- Wu C. T., Goldberg M. L. The Drosophila zeste gene and transvection. Trends Genet. 1989 Jun;5(6):189–194. doi: 10.1016/0168-9525(89)90074-7. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]