Abstract
Insects rely on innate immune responses controlled by the immune deficiency (IMD), Toll and other immune signaling pathways, to combat infection by a broad spectrum of pathogens. These pathways signal to downstream NF-κB family transcription factors which control specific anti-pathogen action via direct transcriptional control of immune effectors, hematopoiesis and melanization. Here we show that in the Anopheles malaria vector, IMD and Toll pathways mediate species-specific defenses against Plasmodium and bacteria through the transcriptional regulation of splicing factors Caper and IRSF1 that, in turn, determine the production of pathogen-specific splice variant repertoires of the hypervariable pattern recognition receptor AgDscam. This mechanism represents an additional level of immune response regulation that may provide a previously unrecognized level of plasticity to the insect immune pathway-regulated anti-pathogen defenses.
INTRODUCTION
Lacking the vast repertoire of antibodies or variable lymphocyte receptors provided by vertebrate adaptive immunity, insects largely rely on a finite number of germ line-encoded innate immune genes for their anti-pathogen defenses (Boehm et al., 2012; Kurtz and Armitage, 2006). While the RNAi defense system can provide large defense diversity it acts exclusively against viruses (Blair, 2011). Despite this limitation, insects can control infection by a great variety of parasites and microbes through deployment of anti-pathogen effector molecules, hemocyte proliferation and melanization that are largely mediated by NF-κB and other immune signaling pathways.
According to the established model of NF-κB signaling pathway –mediated immune responses, pattern recognition receptors either directly or indirectly activate the IMD or Toll pathways upon interaction with pathogen associated molecular patterns. This leads to the nuclear translocation of NF-κB transcription factors that control transcription of distinct sets of immune effector genes, hematopoiesis and melanization, thereby providing a certain degree of pathogen defense specificity (Cirimotich et al., 2010; Eleftherianos and Schneider, 2011; Hetru and Hoffmann, 2009; Valanne et al., 2011). The total humoral and cellular immune responses to pathogen challenge are also influenced by past infections (immune priming or trained immunity), environmental parameters, the circadian system, and diet (Chambers and Schneider, 2012). The Anopheles mosquito defenses against the human Plasmodium falciparum and the rodent P. berghei malaria parasites are largely controlled by the IMD and Toll pathways, respectively. The pathways regulate the expression of diverse anti-Plasmodium effectors that block the ookinete parasite stages during invasion of the midgut epithelium at around 18–30 hours after ingestion of an infectious blood meal (Cirimotich et al., 2010).
The gene encoding the A. gambiae Down syndrome cell adhesion molecule (AgDscam) is composed of 16 constant exons present in all splice forms and three immunoglobulin (Ig) domain-encoding exon cassettes (cassettes 4, 6, and 10) that are each alternatively spliced to contribute a single Ig exon in individual mature mRNAs (Yang et al., 2011). Each AgDscam protein contains ten Ig domains, six fibronectin repeat domains and a transmembrane domain (Fig. 1A) (Dong et al., 2006b). Invertebrate Down syndrome cell adhesion molecule (Dscam) was first identified as a highly diverse axon guidance molecule in Drosophila (Schmucker and Chen, 2009). More recent studies in mosquitoes and other arthropods have established it as an essential hypervariable pattern recognition receptor of the immune system, with the potential of generating over 31,000 alternative spliceforms with different pathogen-interaction and inhibition specificities (Brites et al., 2008; Dong et al., 2006b; Watson et al., 2005; Watthanasurorot et al., 2011). We have previously shown that bacterial infection-responsive AgDscam spliceform repertoires strongly correlate with their bacterial species-specific interaction and inhibition, and that this hypervariable pattern recognition receptor can influence the mosquito’s resistance to midgut infection with the rodent malaria parasite Plasmodium berghei (Dong et al., 2006b). This feature suggests that alternative splicing may play a role in providing an expanded insect immune response diversity and specificity. Here our goal was to address the regulation of immune responsive alternative splicing of AgDscam and its spliceform specificity in the defense against malaria parasite infection and microbiota of the mosquito midgut.
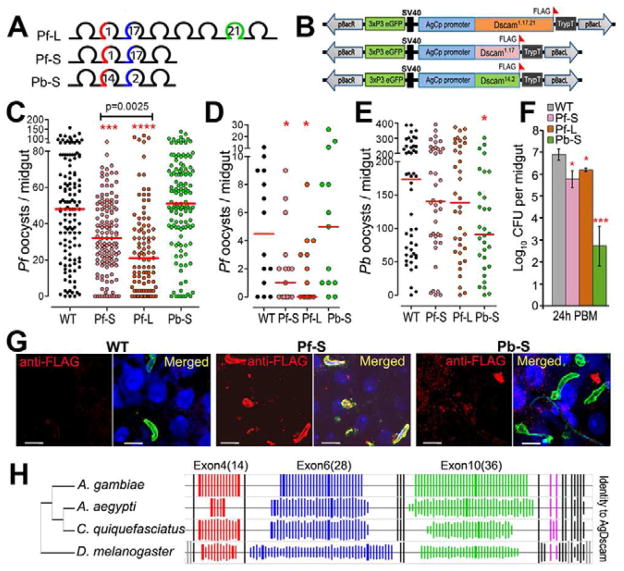
Figure 1. AgDscam gene, transcript and protein organization, and anti-P. falciparum activity.
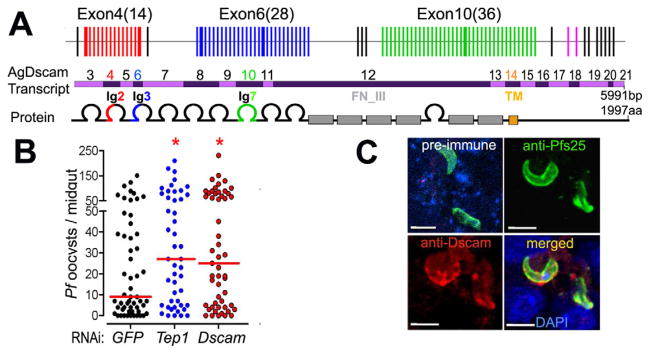
(A) Schematic presentation of AgDscam gene, transcript and protein organization. Upper panel shows the premature transcript with variable (colored) and constant (black) exons, and the three Ig exon cassettes. Middle panel shows the mRNA with indicated exon numbers. Lower panel shows AgDscam protein with 10 Ig-like domains (semicircles), six fibronectin type III domains (grey boxes, FN_III), and a trans-membrane domain (orange box, TM). Splice variants generated in the Ig2 (cassette 4), Ig3 (cassette 6), and Ig7 (cassette 10), domains are colored as red, blue, and green respectively. (B) Effect of total AgDscam silencing on A. gambiae susceptibility to P. falciparum infection. Mosquito cohorts were treated with dsRNAs of GFP, Tep1, or AgDscam. Points indicate the absolute value of oocyst counts in individual mosquitoes from 3 replicates, and horizontal red bars indicate the median value of oocysts, and the statistical numbers are presented in Supplementary Table S1. (C) Association of AgDscam with P. falciparum ookinetes during midgut invasion. Anti-Pfs 25 recognized the P. falciparum ookinetes (green), and anti-Dscam recognizes AgDscam total protein (red) and DAPI stains the nucleus blue. Scale bars: 20 μm. Pre-immune serum from AgDscam antibody generation was used for control with a red staining secondary antibody. See also Table S1.
RESULTS
AgDscam suppresses P. falciparum development and associates with the ookinete stage parasite in the mosquito midgut epithelium
While we have previously shown that AgDscam acts as an antagonist of the rodent malaria parasite (Dong et al., 2006b), here we show that RNAi-mediated depletion of total AgDscam, by targeting the constant exon 7, resulted in a 2.8 fold (278% of control) increase in A. gambiae susceptibility to the human malaria parasite P. falciparum by (p<0.05), as measured by median oocyst-stage parasite load on the midgut (p<0.05) (Fig. 1B, Table S1). The magnitude of this effect was similar to that observed when mosquitoes were depleted of the extensively studied anti-Plasmodium immune factor Tep1 (Dong et al., 2006a; Levashina et al., 2001). AgDscam-mediated inhibition of the malaria parasite most likely occurs while ookinete stages of the parasite are traversing the midgut epithelium, prior to forming oocysts on the basal side of this tissue (Cirimotich et al., 2010). To provide a basic mechanistic insight into AgDscam’s anti-Plasmodium function at the cellular level, we performed immunohistochemical analyses of infected midguts at 24 to 28 h post-ingestion of an infectious bloodmeal, when the ookinetes invade the midgut epithelium. A prominent and specific co-localization of AgDscam staining with the parasites was observed using Pfs25- and Dscam- specific antibodies (Dong et al., 2006b), suggesting a direct anti-Plasmodium activity that involves association, but not necessarily direct interaction, of the protein with the ookinete (Fig. 1C).
IMD and Toll pathways regulate alternative splicing of AgDscam
We and others have previously shown that the mosquito’s immune responses are significantly regulated by the IMD and Toll pathways, which control the nuclear translocation of the NF-κB-like transcription factors, Rel2 and Rel1, respectively. We have also shown that induction of IMD pathway-mediated immune responses more potently suppresses P. falciparum infection, while induction of the Toll pathway results in a more specific defense against P. berghei (Cirimotich et al., 2010; Garver et al., 2009). Based on our previous work showing that A. gambiae responds with differential production of AgDscam spliceform repertoires upon challenge with at least eight different pathogens and pathogen associated molecular patterns (PAMPs) (Dong et al., 2006b), we hypothesized that alternative splicing of this gene is likely to be regulated by the IMD and Toll immune signaling pathways. To test this hypothesis, we first investigated whether the activation or repression of these two immune signaling pathways could influence AgDscam spliceform repertoire production. We analyzed the transcript abundance of all 80 variable AgDscam exons, using an established microarray approach (Wojtowicz et al., 2004) in the A. gambiae Sua5B immune-competent cells that have previously been used to study mosquito immune pathways (Luna et al., 2006; Meister et al., 2005). We analyzed transcript levels following pathway activation without additional immune stimulus or inhibition upon immune-activation by two PAMPs, lipopolysaccharide (LPS) and peptidoglycan (PGN). Activation of either the IMD or Toll pathway significantly altered mRNA representations of variable AgDscam Ig domain-encoding exons as compared to representations of cells treated with GFP dsRNA (Figs. 2, S1A, Table S2). IMD pathway activation resulted in changes in the mRNA abundance of cassette 4 Ig exons with a pattern similar to that previously observed in P. falciparum-infected midguts of A. gambiae (Pearson correlation coefficient (R) =0.73) (Dong et al., 2006b). These changes included up-regulation of spliceforms containing the P. falciparum infection-inducible exon 4.1, corroborating the role of splice forms containing this Ig exon in the defense against the human malaria parasite P. falciparum (Fig. S1A, S1B, Table S2). In contrast, Toll pathway activation resulted in exon 4 transcript abundances that correlated (R=0.63) with those observed in midguts infected with the rodent malaria parasite P. berghei (Dong et al., 2006b). Transcript abundance of spliceforms containing the P. berghei infection-inducible exon 4.14 increased in Toll pathway-activated cells (Figs. 2, S1B, Table S2), further indicating a possible role of splice forms containing this exon in the defense against the rodent malaria parasite. Some overlap of Ig exon transcript abundance was also observed between IMD and Toll pathway-activated cells (R=0.54) (Fig. S1B), indicating that these two immune pathways also direct the production of common spliceform repertoires and therefore defenses, as has been indicated previously (Garver et al., 2009). Total AgDscam transcript analyses in mosquitoes show that expression levels are similar in the naïve gut, abdomen and thorax tissues, while they are elevated in hemocytes (Fig. S1C).
Figure 2. IMD and Toll pathway –regulated alternative splicing of AgDscam.
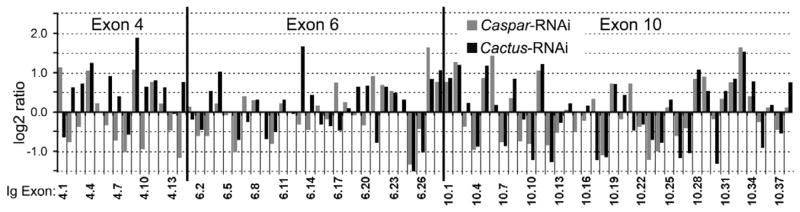
Differential transcript abundance of AgDscam’s 80 spliced Ig exons in A. gambiae Sua5B cells upon IMD or Toll pathway activation (Caspar- or Cactus- RNAi) compared to GFP dsRNA treated control Sua5B cells. Log2 transformed signal ratios of gene silenced samples (Cactus, Caspar-RNAi) versus GFP dsRNA treated control cells, through CombiMatrix microarray analysis. See also Figure S1 and Table S2.
IMD pathway activation enhances AgDscam association with P. falciparum ookinetes
Because pathogen infection induces AgDscam spliceform repertoires with a greater association affinity for the eliciting pathogen (Dong et al., 2006b) and the IMD pathway induces P. falciparum infection-responsive AgDscam spliceforms, we hypothesized that these spliceforms would have a greater association affinity for P. falciparum ookinetes. To test this, we used confocal microscopy to observe the colocalization of AgDscam with P. falciparum ookinetes in the mosquito midgut after IMD pathway activation or inhibition. Quantitative analysis of fluorescence intensities showed that the levels of AgDscam protein associated with P. falciparum ookinetes increased by 6.4 fold following IMD pathway activation, compared to intensity levels following inhibition of this pathway (Fig. 3, Table S3). We have previously shown that the abundance of constant AgDscam exons does not significantly change upon infection (Dong et al., 2006b), suggesting that the total AgDscam population is determined by immune pathway-regulated abundance of specific spliceform repertoires.
Figure 3. IMD pathway activation enhances AgDscam association to P. falciparum ookinetes.
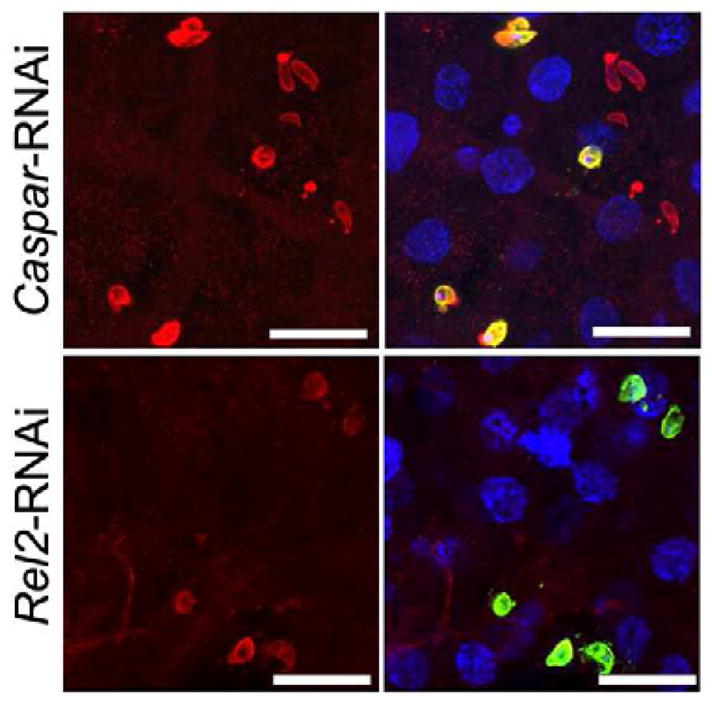
Immunohistochemical analysis of P. falciparum ookinetes during ookinete invasion of the midgut of Caspar dsRNA treated and Rel2 dsRNA treated mosquitoes. DAPI stains of nuclei blue, anti-Pfs25 stains P. falciparum ookinetes green and anti-AgDscam stains Dscam protein red. The right panels are merged immunofluorescence and DAPI signals. Scale bars: 10 μm. See also Table S3.
IMD pathway-controlled immune responsive splicing factors regulate anti-Plasmodium defense
AgDscam defense activity involves an association with the pathogen, suggesting that the IMD pathway-induced and P. falciparum infection-responsive AgDscam spliceforms would have a greater inhibitory activity against this parasite species. Therefore, we hypothesized that the production of P. falciparum-specific AgDscam isoforms is mediated by Rel2-regulated transcription of splicing factors that carry out alternative splicing of premature AgDscam. To test this hypothesis we performed experiments to identify and test putative splicing factors for implication in immune pathway-mediated alternative splicing of AgDscam. Through literature and blast homology searches to known D. melanogaster pre-messenger RNA processing factors and putative alternative splicing regulators we identified 88 putative A. gambiae splicing factors, including some that have been linked to Dscam splicing (Mount and Salz, 2000; Park et al., 2004). Of these, 9 were transcriptionally regulated by microbe or parasite infection in previous analyses (Dong et al., 2006a; Dong et al., 2009; Garver et al., 2009) (Table S4). We confirmed the infection-responsive nature of these 9 genes by profiling the changes in their transcript abundance in Sua5B cells upon challenge with either PGN or LPS, two immune elicitors that are less likely to induce alternative splicing through non-immune-related mechanisms compared to challenge with whole bacteria. Transcript abundances of YT521, Caper, and KHD were increased, while those of U2AF and immune responsive splicing factor 1 (IRSF1) decreased in response to challenge with either one or both of the immune elicitors (Fig. 4A). We also investigated whether the IMD pathway regulated these splicing factors. IMD pathway inactivation in PGN and LPS challenged cells (through Rel2 silencing) modulated the abundance of these transcripts in an opposite direction (Fig. 4B), further supporting the hypothesis that alternative splicing can be controlled through the IMD pathway. To provide evidence for the involvement of these splicing factors in regulating the anti-Plasmodium defense we profiled the changes of their transcript abundance upon P. falciparum infection at 24 h post-ingestion of an infectious bloodmeal, and found that Caper transcripts were enriched by 1.8 fold while IRSF1 transcripts decreased by a 2.1 fold. Transcript abundances of both Caper and IRSF1 showed no change upon P. berghei infection (Fig. 4C).
Figure 4. Immune responsive and IMD pathway –controlled splicing factors.
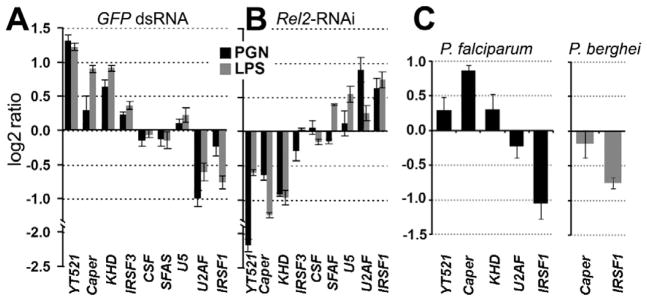
qRT-PCR –based differential transcript abundance analysis of 9 putative splicing factors in the Sua5B cells upon peptidoglycan (PGN) and lipopolysaccharide (LPS) challenge compared to naïve controls (A), or upon Rel2 depletion of PGN and LPS challenged cells compared to GFP dsRNA treated controls (B). (C) qRT-PCR-based differential transcript abundance analysis of 5 or 2 putative splicing factors in P. falciparum infected (left) or P. berghei infected (right) total female mosquitoes compared to naïve controls. Three biological replicates were included and data are represented as mean +/− SEM. See also Table S4.
Immune responsive splicing factors Caper and IRSF1 regulate AgDscam splicing and anti-Plasmodium defense–specificity
Because activation of the IMD pathway significantly alters transcription of Caper and IRSF1 and alternative splicing of AgDscam, we hypothesized that the two putative splicing regulators might be involved in regulating the alternative splicing of AgDscam. We tested this hypothesis by investigating AgDscam Ig exon transcript representation upon RNAi-mediated silencing of Caper and IRSF1 in Sua5B cells. Silencing of either splicing factor resulted in the production of a distinctly different AgDscam spliceform transcript representation in comparison to control cells treated with GFP dsRNA (Figs. 5, S2, Table S5). Silencing of Caper resulted in a transcript abundance pattern similar to that of IMD pathway inhibited (Rel2 depleted) cells (R= 0.66) (Fig. S2), including a decreased abundance of the P. falciparum infection-responsive exon 4.1 transcripts. Conversely, IRSF1 silencing resulted in an increased abundance of exon 4.1 transcripts. This result suggests that the IMD pathway directs AgDscam splicing through Caper and/or IRSF1 (and possibly other splicing factors), leading to the production of AgDscam spliceform repertoires with specific anti-pathogen activities. To confirm the involvement of these infection responsive splicing factors in the anti-Plasmodium defense, we assessed the impact of their RNAi-mediated depletion on mosquito resistance to the human malaria parasite. Caper silencing increased the susceptibility of mosquitoes to P. falciparum infection 1.7 fold (174% of control) (p<0.0001), whereas IRSF1 silencing resulted in a 5.2 fold (19.4% of control) (p<0.0001) greater resistance (Fig. 6A, Table S6). Our data suggests that Caper and IRSF1 play opposite roles in modulating the anti-Plasmodium defense, through the regulation of AgDscam and possibly other alternatively spliced parasite agonists and/or antagonists (Garver et al., 2008; Meister et al., 2005).
Figure 5. Caper and IRSF1 -regulate alternative splicing of AgDscam.
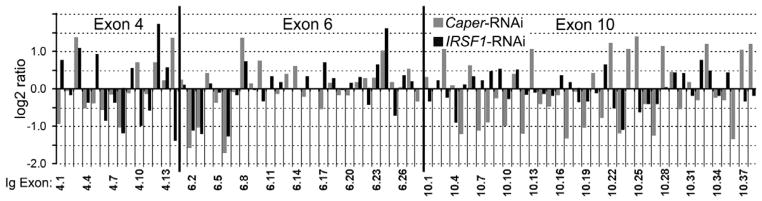
Differential transcript abundance of AgDscam’s 80 spliced Ig exons in A. gambiae Sua5B cells upon Caper or IRSF1 depletion compared to GFP dsRNA treated control Sua5B cells. Log2 transformed signal ratios of gene silenced Caper- and IRSF1-RNAi) versus GFP dsRNA treated control cells, through CombiMatrix microarray analysis. See also Figure S2 and Table S5.
Figure 6. Caper and IRSF1 regulate resistance to P. falciparum infection and AgDscam Ig exon specific implication in anti-P. falciparum defense.
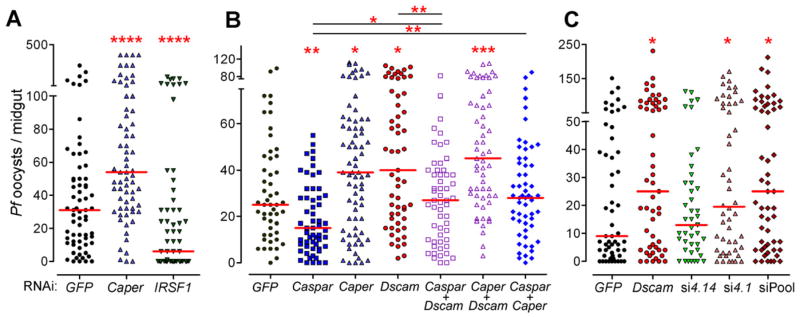
(A) P. falciparum oocyst infection intensity in Caper and IRSF1 depleted mosquitoes. Graphic presentation is as described in Figure 1 and the statistical numbers are presented in Supplementary Table S6. ****: p<0.0001. (B) P. falciparum oocyst infection intensity in independent Caspar, Caper and AgDscam silenced, or co-silenced silenced (Caspar+AgDscam, Caper+AgDscam, Caspar+Caper) mosquitoes. *: p<0.05; **: p<0.01; ***: p<0.001; ****: p<0.0001. (C) Effect of total AgDscam or specific spliceform repertoire silencing on A. gambiae susceptibility to P. falciparum infection. Mosquito cohorts were treated with dsRNAs of GFP, AgDscam or AgDscam siRNAs for Ig exons 4.1 and 4.14, or a siRNA pool targeting Ig exons 4.1, 6.17 and 10.21. Points indicate the absolute value of oocyst counts in individual mosquitoes from 3 replicates, and horizontal red bars indicate the median value of oocysts. See also Table S6.
Next we performed experiments to provide more detailed insight on the roles of Caper and Caspar in regulating anti-P. falciparum defense through AgDscam. We reasoned that, if AgDscam is the main anti-parasite factor spliced by Caper, then silencing transcripts of both genes would have a similar impact on infection to silencing either factor singly (Fig. 6B, Table S6). Indeed, co-silencing the two genes significantly increased parasite infection levels to a similar degree as when they were silenced independently. Co-silencing AgDscam or Caper with Caspar reverted the Caspar knockdown phenotype, but only to control levels, suggesting that Caspar is not exerting its anti-P. falciparum activity exclusively through Caper and AgDscam. Caspar is upstream of the other two factors and is likely exerting its anti-parasitic effect through multiple independent effectors as we have shown previously (Garver et al., 2009).
AgDscam exerts spliceform-specific defense against human and rodent malaria parasite species
Here we have shown that IMD pathway -controlled splicing factors regulate the production of AgDscam spliceform repertoires that correlate with pathogen-stimulated repertoires and show a stronger association with P. falciparum ookinetes. We have previously shown that AgDscam spliceforms containing Ig exons 4.1, 6.17, and 10.21 are induced upon challenge with P. falciparum, while spliceforms containing exon 4.14 are induced upon challenge with P. berghei (Dong et al., 2006a; Dong et al., 2006b). Because of this, we investigated the possible Plasmodium species-specificity of these specific exon-containing AgDscam spliceforms by using a siRNA-based approach to specifically deplete transcripts containing these P. falciparum infection-inducible Ig exons upon parasite challenge. Specific depletion of spliceforms containing the P. falciparum infection-responsive Ig exon 4.1 resulted in a 2.2 fold (217% of control) increase in median P. falciparum oocyst loads (p=0.045), an effect that was comparable to silencing of total AgDscam transcripts (Fig. 6C, Table S6). Depletion of spliceforms containing exons 4.1, 6.17 and 10.21 resulted in a 2.8-fold (278% of control) (p=0.040) increased susceptibility to P. falciparum infection to a similar degree as specifically depleting exon 4.1 (p=0.876). In contrast, selective depletion of spliceforms containing the P. berghei infection-responsive exon 4.14 did not influence mosquito susceptibility to P. falciparum infection (p=0.682). These data do not exclude the possibility that spliceforms representing other variable Ig exons are also involved in the anti-P. falciparum defense but we do show that spliceforms containing exon 4.1 are involved and confirm the correlation between P. falciparum infection-responsive spliceforms and defense specificity against this parasite species.
To provide more robust evidence and further mechanistic insight into AgDscam infection-responsive spliceform involvement in the defense against Plasmodium and also bacteria, we used a transgenic approach with A. stephensi mosquitoes. Previous recombinant protein expression studies of Drosophila Dscam have shown that the first four Ig domains of the extracellular protein domain are necessary and sufficient for binding to bacterial surfaces (Meijers et al., 2007; Watson et al., 2005). To examine whether these first 4 Ig domains of AgDscam (including cassettes 4 and 6) are implicated in anti-Plasmodium defense, we generated two separate transgenic mosquito lines. These lines, called Pf-S and Pf-L, express P. falciparum-inducible short (Pf-S, or Dscam1.17) and long (Pf-L, or Dscam1.17.21). AgDscam spliceforms containing the constant exons and the first four Ig domains (including exon 4.1 and 6.17) or the first eight Ig domains (including exon 4.1, 6.17, 10.21), respectively (Figs. 1A, 7A, 7B, Fig. S3A, S3B, Table S7). We also generated a third transgenic line, Pb-S, expressing a P. berghei and bacteria challenge-inducible short spliceform (Pb-S, or Dscam14.2) containing exons 4.14 and 6.2. The carboxypeptidase-1 promoter was used to drive over-expression of the recombinant genes in the female mosquito midgut upon a bloodmeal to effectively target invading ookinetes (Dong et al., 2009) (see Experimental Procedures for details). AgDscam spliceform-specific defenses against the two parasite species were confirmed through Plasmodium infection experiments with the transgenic and wt mosquitoes (Fig. S3C, S3D). P. falciparum infection intensity decreased 1.5 fold (66.7% of control) (p<0.0001) and 2.4 fold (41.7% of control) (p<0.0001) in the Pf-S and Pf-L transgenic lines, respectively (Fig. 7C, Table S7b). In contrast, over-expression of the P. berghei and bacteria challenge-inducible Pb-S spliceform did not impact P. falciparum infection in comparison to the wt mosquitoes (p=0.423). These data suggest that the inclusion of exon 4.1 in the first four Ig domains is sufficient for anti-P. falciparum defense, while the inclusion of exon 10.21 may provide additional anti-P. falciparum activity (Fig. 7C; Pf-L vs. Pf-S: p=0.0025). This observation corroborates a previous study showing that the variable splicing of cassette 4 in response to Plasmodium is more important than the variability conferred by cassettes 6 and 10 (Smith et al., 2011). Because our laboratory-based experimental system produces unnaturally high parasite infection intensities, we fed the Pf-S and Pf-L transgenic mosquitoes with a low-gametocytemic P. falciparum culture to obtain an infection intensity that more closely resembles mosquito infections in the field (Dong et al., 2011; Garver et al., 2012; Sinden et al., 2004). Under these conditions, over-expression of the Dscam1.17.21 transgene in Pf-L mosquitoes decreased the median oocyst intensity from 4.5 to 0, rendering the mosquitoes almost entirely refractory to the human malaria parasite (Fig. 7D, Table S7b). This implies that AgDscam will play a potent role in regulating vectorial capacity in the field. In contrast, the over-expression of the Dscam14.2transgene in the Pb-S transgenic mosquitoes did not confer elevated resistance to P. falciparum (p>0.05), though these mosquitoes were 1.9 fold (52.3% of control) more resistant to infection with the rodent malaria parasite (p<0.05) (Fig. 7E, Table S7b). Over-expression of transgenes Dscam1.17 (Pf-S) and Dscam1.17.21 (Pf-L) did not significantly inhibit P. berghei development (p=0.9). These results confirm the correlation between defense specificity of AgDscam spliceform repertoires and parasite species-specific immune activation.
Figure 7. Dscam phylogenetic analysis and Pf-S, Pb-S, and Pf-L –expressing transgenic mosquitoes.
(A) PCR fragments containing Pf-S, Pb-S, and Pf-L were separately cloned and used for transgenic constructs. The letter inside the Ig domain denotes the splice variant. The short isoform and long isoform induced upon P. falciparum challenge are designated as Dscam1.17 and Dscam1.17.21, and the short isoform fragment cloned from a septic blood fed midgut cDNA sample is designated as Dscam14.2. (B) Schematic representation of the pBac-3xP3eGFP- DscamPf-S-TrypT (pPf-S), pPb-S, and pPf-L plasmids used for germ-line transformation. (C, D) P. falciparum oocyst infection intensities in wild type and transgenic A. stephensi mosquitoes at 8 days after feeding on blood with a 0.3% (C) and 0.05% (D) gametocytemia. The p-value from comparison of Pf-S and Pf-L was calculated through a Mann-Whitney test. *: p<0.05; ****: p<0.0001. (E) P. berghei oocyst infection intensities of the wild type and transgenic mosquitoes. The statistical numbers are presented in Supplementary Table S7. (F) The midgut microbial flora (total bacterial load) of female transgenic and wt control (WT) mosquitoes at 24 h PBM, data are represented as mean +/− SEM. A Student’s t-test was used to calculate p-values and determine significance. (G) Pf-S –specific association to P. falciparum ookinetes. Anti-FLAG antibody (Sigma) recognizes recombinant Dscam (red), and P. falciparum ookinetes during invasion of the midgut in Pf-S and Pb-S transgenic mosquitoes are stained in green, DAPI stains of nuclei blue. Scale bars: 5 μm. (H) Phylogenetic analysis of Dscam across 3 mosquito species and Drosophila. The thicker lines indicate the Ig exons used for generating transgenic mosquitoes. Bar heights represent amino acid identity (%) to AgDscam with 100% as maximum identity. See also Figure S3 and Table S7.
AgDscam-mediated suppression of the midgut microbiota is spliceform-specific
The mosquito midgut contains a variety of mainly Gram-negative bacteria as part of its natural microbiota, which greatly proliferate after a bloodmeal (Cirimotich et al., 2011). Because the Dscam14.2 isoform was identified in septic (bacteria containing) bloodfed mosquito midguts and exon 4.14-containing spliceforms are induced upon infection with two tested Gram-negative bacteria (Dong et al., 2006b), we hypothesized that Dscam14.2 might be involved in regulating the bacterial load of the mosquito gut. Indeed, at 24 to 48 h after a blood meal, the bacterial loads in the Pb-S mosquito midguts was at least 500 fold lower (Student’s t-test, p<0.01) than those of Pf-S, Pf-L and wt mosquitoes (Figs. 7F and S3E), implicating exon 4.14-containing spliceforms in the suppression of midgut microbiota. The small effect of the Pf-S and Pf-L on bacteria proliferation may indicate weaker affinities of these recombinant proteins to bacterial surfaces. Tissue-specific cassette 4 exon transcript abundance analyses also showed that exon 4.14 was expressed at elevated levels in hemocytes which are known to be implicated in the cellular defenses against bacteria (Fig. S3E).
Anti-P. falciparum AgDscam spliceforms associate specifically with P. falciparum ookinetes
Our studies and those of others have shown that Dscam’s spliceform-specific anti-pathogen activity is mediated through an affinity for, and association with, the pathogen (Dong et al., 2006b; Watson et al., 2005; Watthanasurorot et al., 2011). To determine whether the P. falciparum infection-responsive Dscam1.17.21 (or Dscam1.17) and P. berghei infection-responsive Dscam14.2 recombinant spliceforms have different affinities for P. falciparum ookinetes in vivo, we used immunohistochemical assays. An anti-FLAG epitope antibody was used to discriminate recombinant from endogenous AgDscam molecules in infected transgenic mosquito midguts and a P. falciparum surface-specific antibody was used to investigate colocalization. Confocal microscopy studies of whole-mount mosquito midguts at 24 to 28 h after a P. falciparum-infected blood meal showed a 8.3 fold greater association of AgDscam with P. falciparum ookinetes (measured by fluorescence intensity) in the Pf-S transgenic mosquitoes than in the Pb-S transgenic mosquitoes (Fig. 7G, Table S7c), confirming the parasite association-dependent anti-Plasmodium activity of specific AgDscam spliceforms. Though the specific mode of association and inhibition is unknown it likely involves complexes with other effectors.
Dscam Ig exon conservation is variable amongst dipteran species
Ig exon sequence and phylogenetic analyses between Dscam molecules from the genomes of A. gambiae, Culex pipiens, Aedes aegypti and D. melanogaster showed that the amino acid sequences of most constant exons and the gene organization of immune-responsive exon variants were highly conserved across all four species, while the number of potential Ig exons per cassette were different and alternatively spliced exons showed different levels of conservation (Fig. 7H, Table S7d). Interestingly, while both exon 4.1 and 4.14 showed moderately conserved amino acid sequences between all three mosquito species, the ratio of synonymous to non-synonymous nucleotide substitutions differed (Table S6), indicating a reduced selective pressure on the sequence. This may reflect the continuous exposure of all dipterans to bacteria, while P. falciparum only infects a small fraction of A. gambiae, reducing the selective pressure on exon 4.1. The genome sequence of A. stephensi was not available and its Dscam gene could therefore not be included in this study.
DISCUSSION
The complex life cycle, diverse breeding habitats, and hematophagy of mosquitoes expose them to a variety of bacteria, fungi, and viruses, including many of medical and veterinary importance. Along with pathogenic microorganisms, there are also commensal and transient bacteria and fungi associated with the midgut and other tissues of the mosquito (Cirimotich et al., 2010). While the vertebrate immune surveillance system uses recombination and somatic hyper-mutation of antibody immunoglobulin (Ig) domains and assembly of variable lymphocyte receptors to discriminate between a similarly broad microbial spectrum, insects have to rely on approximately 150 pattern recognition receptors and the differential activation of a few immune signaling pathways that control the production of downstream effectors and defense mechanisms ( Boehm et al., 2012; Kurtz and Armitage, 2006). Activation of pathogen species- or class-specific antimicrobial peptides and other immune factors and defense mechanisms are to a significant extent controlled by immune signaling pathways, such as IMD and Toll (Chambers and Schneider, 2012; Cirimotich et al., 2010; Eleftherianos and Schneider, 2011; Hetru and Hoffmann, 2009). Here we show that these well-characterized insect NF-κB signaling pathways regulate defenses through transcriptional regulation of splicing factors that control alternative splicing of the hypervariable pattern recognition receptor AgDscam, and possibly other immune genes. AgDscam can produce different spliceform repertoires in response to challenge with each of at least eight different immune elicitors (two PAMPs and two malaria parasite species shown in this report and three different bacterial species and a fungus shown previously (Dong et al., 2006b)) and the induction of parasite- and bacteria-specific AgDscam spliceform repertoires correlates with the affinity for and activity against the eliciting pathogens (Dong et al., 2006b).
The single germ-line encoded gene AgDscam provides the mosquito with a remarkable degree of pathogen interaction and defense variability. Specificity for the eliciting pathogen or molecule is conferred through different Ig domain combinations that are generated by activation or suppression of the splicing factors Caper and IRSF1. The IMD, Toll and likely other immune signaling pathways regulate expression of these splicing factors in response to immune challenge. While the invertebrate splicing machinery has not been studied with respect to the regulation of immune responses, Caper has previously been implicated in the alternative splicing of a vascular endothelial growth factor gene (Huang et al., 2012).
Our study reveals an additional level of immune signaling pathway-regulated defense diversity and specificity, and supports a model by which the combined action of multiple immune pathways on the transcriptional regulation of splicing factors can generate diversity of pathogen-specific defenses, either through a continuous range or a large number of distinct AgDscam splice form repertoires that exert their activity by associating with the pathogens.
AgDscam is one of some 85 infection-responsive A. gambiae Ig domain superfamily genes, and numerous other mosquito immune genes are alternatively spliced (Christophides et al., 2002; Dong et al., 2006b; Garver et al., 2008; Meister et al., 2005). It is likely that splicing factors not investigated here, possibly also regulated by other immune pathways, play a role in insect anti-pathogen defenses. The understanding of insect innate immunity has experienced a remarkable progress over the past decade that has proven it to be much more complex and sophisticated than initially believed, including memory and adaptive characteristics (Chambers and Schneider, 2012; Eleftherianos and Schneider, 2011; Pham et al., 2007; Rodrigues et al., 2010). AgDscam’s plasticity renders it a likely component of such mechanisms. The potent anti-P. falciparum activity of immune pathway-regulated AgDscam spliceform repertoires underscores the value for further studies of this gene towards the development of a malaria control strategy. Such strategies could be based on the manipulation of the mosquito’s splicing machinery or overexpression of a Plasmodium sp. –specific splice variant in a way that would enable production of a broad-range of specificity against malaria parasites, which can show great genotypic and antigenic variability in the field.
EXPERIMENTAL PROCEDURES
Mosquito rearing, genomic DNA and total RNA isolation
The A. gambiae Keele and transgenic A. stephensi Liston strain (India strain) were maintained at 27°C and 80% humidity with a 12 hour day-night cycle. Adult mosquitoes were maintained on 10% sucrose and were fed on mice blood for egg production. RNA was extracted using RNeasy kit (Qiagen) and gDNA was isolated according to an established protocol (Dong et al., 2006a; Dong et al., 2006b; Dong et al., 2011).
dsRNA and siRNA -mediated gene silencing and real-time PCR (qRT-PCR)
Target gene mRNA was selectively depleted from adult female mosquitoes and Sua5B cells using established RNAi methodology (Table S1a) (Dong et al., 2006a). For the activation of the immune signaling pathways, 4 days post dsRNA treatment, lipopolysaccharide (LPS from Pseudomonas aeruginosa (Sigma Aldrich) or peptidoglycan (PGN from Staphylococcus aureus (Sigma Aldrich)) was added to the cells for 12 hrs at the final concentration of 10 μg/ml. RNAi assays were repeated at least three times with at least 60 mosquitoes for each group or 4 cell cultures, using GFP dsRNA treatment as control. Verification of gene silencing was performed 4 days after injection or incubation by quantitative real-time PCR (qRT-PCR) using ribosomal protein S7 gene as a normalization standard. The fold-change in transcript abundance levels after silencing was calculated by the standard EΔΔCt method (Pfaffl, 2001) (Table S1a).
AgDscam exon transcript abundnace analysis with oligonucleotide microarrays
Three different 21 to 23 -mere probe sequences targeting the specific sequences of AgDscam 99 exons were designed using Array Designer 4 software, and represented on Combimatrix microarrays that were used for assays as previously described (Dong et al., 2006a; Dong et al., 2009). A protocol developed by Watson et al., 2005 and Wojtowicz et al., 2004 was used for sample preparation. AgDscam fragments were PCR amplified from cDNA using PlatinumTaq Hifi polymerase (Invitrogen) with a forward primer specific for exon3 (5′-GGA ACC AAC GAA CCG TAT TGA TTT - 3′) and a reverse primers specific for exon7 (5′-TGT GCA CTT TCC TGA TCG TTG CGA AT - 3′) or with a forward primer specific for exon8 (5′-CGC GGT ACA CTG GAA GTG CAA GTA - 3′) and a reverse primer specific for exon12 (5′-GTA GTA GCC TTC GTT CGA TTT TTG G - 3′). Products of at least 3 PCR reactions were purified using the PCR purification kit (Qiagen) and pooled from 3 replicates. PCR products were labeled using the ArrayIt labeling kit (Mirus Bio) and hybridized according to manufacture instruction (CombiMatrix). The probe hybridization signal and local background fluorescence values for each spot were determined with the GenePix Pro 5.1 software. Since the labeled hybridized sample DNA was obtained from separate PCR reactions (exons 3- to7 and exons 8 to12), spots for variable exons in the Ig exon cassettes 4 and 6, and 10 were analyzed separately. Because only one dye (Cy5) was used across control and treatment samples, the data was first normalized by using total intensity method to obtain a sum ratio of samples versus GFP dsRNA treated controls equal to 1. Constant exons 5 and 7 were then used to normalize signals for variable Ig exon cassette 4 and 6 exons. Constant exons 9 and 11 signals were used to normalize signals for variable Ig exon cassette 10 exons. Corrected fluorescence values for each spot were exported to Microsoft Excel for further analysis. The signal ratios of RNAi silenced samples versus GFP dsRNA treated controls from three replicates were averaged and log2 transformed and presented as bar graphs (Fig. 2, Table S2). The total hybridization intensities of each exon from GFP dsRNA treated cells are also presented in Fig. S1A. The correlations of the transcript abundance of all 80 variable exons between different treatments are presented in Fig. S1B.
Immunofluorescence assay (IFA) and confocal microscopy
Immunostaining for confocal microscopy was performed as previously described (Dong and Dimopoulos, 2009). The anti-AgDscam antibody has previously been shown to bind specifically to AgDscam in adult mosquitoes and the Sua5B cell line and the protein is depleted when AgDscam is silenced via RNAi (Dong et al., 2006b). The anti-Pfs25 (MR4, Cat# MRA-28) and anti-FLAG (Sigma) primary antibodies were used at a 1:400 dilution while the Dscam peptide antibody (Dong et al., 2006b) was used at a 1:150 dilution. The secondary antibodies were the AlexaFluor 488-conjugated (green) goat anti-mouse antibody (1:500 dilution) for the parasite-specific antibodies, and AlexaFluor 568-conjugated (red) goat anti-rabbit antibody (1:500 dilution) for the anti-FLAG or anti-Dscam antibodies. The samples were examined with a Zeiss LSM 510 confocal microscope, collecting 0.2- to 1- μm optical sections. For the comparison across different slides, the confocal microscopy settings were kept the same and DAPI staining intensity from each treatment was used for standardization. The fluorescent intensities from the colocalization of total Dscam protein with P. falciarpum ookinetes were measured using the Image J and Volocity software. Total 4 midguts (with at least 4 parasites in each gut) were assayed for each treatment. The intensities of the green fluorescence of P. falciparum ookinetes samples were used as internal references for normalization. Mean ratio of intensities of Caspar-RNAi and Rel2-RNAi samples was first obtained by dividing Dscam intensities with Pfs25 intensities. Thereafter fold change of AgDscam mean intensity was calculated by dividing the mean ratio of Capar-RNAi with that of Rel2-RNAi samples (Table S3). The absolute intensities of 4 representative ookinetes (green) and Dscam stained protein (red) are presented in Table S4. Similarly, the fold difference of fluorescent intensity of the recombinant Dscam protein (with FLAG-tag) associated with the invading P. falciparum ookinetes between the transgenic Pf-S and Pb-S mosquitoes was determined (Table S7c).
Generation of transgenic mosquitoes
The primers used for generation and verification of transgenic mosquitoes are listed in Table S1b. Amplified AgDscam Ig exons fragments were separately cloned and fused to the carboxypeptidase promoter followed by the A. gambiae trypsin terminator sequence (antryp1T, TrypT) (Fig. 7B) and cloned into a piggyBac based plasmid (pBac[3xP3-eGFPafm]) that also included the 3xP3 eye specific promoter driven eGFP as selection marker to make the constructs for germline embryo microinjection, according to a previously described strategy (Dong et al., 2011). 1,200 A. stephensi eggs were injected with each construct along with helper plasmid, and the hatched larval survivors were screened for transient expression of the 3xP3- GFP marker. 38% of larvae showed transient expression of GFP and all the adult mosquitoes (247 for Pf-S, 118 for Pb-S, 221 for Pf-L) from the surviving larvae were crossed to wild-type mosquitoes. The offspring of these crosses were divided into 12, 6, and 12 groups, for Pf-S, Pb-S, and Pf-L, respectively, and crossed to wild type (wt) mosquitoes to give rise to 5, 4, and 3 stable trangenic lines for pPf-L, pPf-S, and pPb-S, respectively. For the out-crossing, about 20 G1 female mosquitoes were crossed with 7 wild-type male mosquitoes (3:1 sex ratio), while 20 G1 male mosquitoes were crossed with 100 wild-type female mosquitoes (1:5 sex ratio). Discrimination based on green fluorescence in the eyes was used to screen transgenic larvae and adult mosquitoes as shown in Fig. S3A. Confirmation of transgene insertions was performed according to an established protocol (Dong et al., 2011) and the sequences for the verification primers are presented in Table S7a (Fig. S3B). As shown in Supplementary Fig. S3, the Pf-L#3, Pf-S#7 and Pb-S#2 transgenic lines were selected and used throughout the study based on anti-Plasmodium activities; thereafter referred to as Pf-L, Pf-S, and Pb-S, respectively. Homozygous transgenic mosquitoes at generation 4, 5 and 6 were fed on either P. falciparum infected blood or P. berghei infected mice. The detailed statistical analysis of the infection assays are presented in Supplementary Table S7b.
P. falciparum and P. berghei infection assays
Mosquito infection assays and oocyst enumeration with the NF54 P. falciparum (provided by the Johns Hopkins Malaria Institute core facility and Sanaria) and P. berghei (ANKA strain) were performed according to standard procedures (Dong et al., 2006a; Dong et al., 2006b; Dong et al., 2009; Garver et al., 2009). At least three biological replicates were performed for each experiment (with each replicate including at least 80 mosquitoes) and the median number of oocysts per midgut was determined. The prevalence is the percentage of mosquitoes with at least one oocyst on its midgut. The dot plots of the oocysts within each treatment were produced using GraphPad Prism5 software with the median value indicated. The p value from non-parametric Mann-Whitney test and/or Kruskal-Wallis (KW) ANOVA on ranks was used to determine the significance of oocysts numbers. The detailed statistical analysis of the infection assays are presented in Supplementary Table S1b, S6, S7b.
Enumeration of endogenous midgut bacteria
Isolation and colony-forming unit (CFU) enumeration of endogenous midgut bacteria from the Pf-S, Pf-L, Pb-S and wt female mosquitoes were performed as described previously (Dong et al., 2011; Dong et al., 2009). Each experiment was performed with an individual midgut and results are representative of 8 individual experiments. CFU per midgut were calculated by multiplying the number of colonies on the plate with the dilution factor. The CFUs of midguts at 0-, 24-, 48-, 72- hrs were assayed (Fig. S3E). The significance of the differences in bacterial loads between mosquito groups was determined by student’s t-test using GraphPad Prism5.
Phylogenetic analysis of alternatively spliced Dscam exons
tblastn was used to search the putative genes of C. quinquefasciatus, A. aegypti and Drosophila melanogaster using protein sequences for each exon in A. gambiae (Fig. 7G). For each exon in each species, the A. gambiae exon with highest identity at the amino acid level was identified using blastp, and the percent identity at both the amino acid and nucleotide levels were calculated, as was the ratio of synonymous to non-synonymous substitutions, as shown in Table S7d.
Supplementary Material
HIGHLIGHTS.
AgDscam associates with and suppresses Plasmodium falciparum development in the mosquito
IMD and Toll pathway responsive splicing factors regulate AgDscam alternative splicing
AgDscam exerts spliceform-specific defense against different Plasmodium species
AgDscam-mediated suppression of the mosquito midgut microbiota is spliceform-specific
Acknowledgments
This work has been supported by National Institutes of Health/National Institute of Allergy and Infectious Disease grants R01AI061576 and R01AI081877 and the Calvin A. and Helen H. Lang fellowship (to C.M.C.). We thank Drs. Silverman, Jacobs-Lorena, Dr. Yoshida and the Johns Hopkins Malaria Research Institute Insectary and Parasitology core facilities.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Blair CD. Mosquito RNAi is the major innate immune pathway controlling arbovirus infection and transmission. Future Microbiol. 2011;6:265–277. doi: 10.2217/fmb.11.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm T, McCurley N, Sutoh Y, Schorpp M, Kasahara M, Cooper MD. VLR-based adaptive immunity. Annu Rev Immunol. 2012;30:203–220. doi: 10.1146/annurev-immunol-020711-075038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brites D, McTaggart S, Morris K, Anderson J, Thomas K, Colson I, Fabbro T, Little TJ, Ebert D, Du Pasquier L. The Dscam homologue of the crustacean Daphnia is diversified by alternative splicing like in insects. Mol Biol Evol. 2008;25:1429–1439. doi: 10.1093/molbev/msn087. [DOI] [PubMed] [Google Scholar]
- Chambers MC, Schneider DS. Pioneering immunology: insect style. Curr Opin Immunol. 2012;24:10–14. doi: 10.1016/j.coi.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, Blass C, Brey PT, Collins FH, Danielli A, Dimopoulos G, et al. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298:159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- Cirimotich CM, Dong Y, Clayton AM, Sandiford SL, Souza-Neto JA, Mulenga M, Dimopoulos G. Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science. 2011;332:855–858. doi: 10.1126/science.1201618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirimotich CM, Dong Y, Garver LS, Sim S, Dimopoulos G. Mosquito immune defenses against Plasmodium infection. Dev Comp Immunol. 2010;34:387–395. doi: 10.1016/j.dci.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Aguilar R, Xi Z, Warr E, Mongin E, Dimopoulos G. Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. PLoS Pathog. 2006a;2:e52. doi: 10.1371/journal.ppat.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Taylor HE, Dimopoulos G. AgDscam, a hypervariable immunoglobulin domain-containing receptor of the Anopheles gambiae innate immune system. PLoS Biol. 2006b;4:e229. doi: 10.1371/journal.pbio.0040229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Dimopoulos G. Anopheles fibrinogen-related proteins provide expanded pattern recognition capacity against bacteria and malaria parasites. J Biol Chem. 2009;284:9835–9844. doi: 10.1074/jbc.M807084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Manfredini F, Dimopoulos G. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 2009;5:e1000423. doi: 10.1371/journal.ppat.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Das S, Cirimotich C, Souza-Neto JA, McLean KJ, Dimopoulos G. Engineered anopheles immunity to Plasmodium infection. PLoS Pathog. 2011;7:e1002458. doi: 10.1371/journal.ppat.1002458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eleftherianos I, Schneider D. Drosophila immunity research on the move. Fly (Austin) 2011;5:247–254. doi: 10.4161/fly.5.3.17028. [DOI] [PubMed] [Google Scholar]
- Garver LS, Bahia AC, Das S, Souza-Neto JA, Shiao J, Dong Y, Dimopoulos G. Anopheles imd pathway factors and effectors in infection intensity-dependent anti-Plasmodium action. PLoS Pathog. 2012;8:e1002737. doi: 10.1371/journal.ppat.1002737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garver LS, Dong Y, Dimopoulos G. Caspar controls resistance to Plasmodium falciparum in diverse anopheline species. PLoS Pathog. 2009;5:e1000335. doi: 10.1371/journal.ppat.1000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garver LS, Xi Z, Dimopoulos G. Immunoglobulin superfamily members play an important role in the mosquito immune system. Dev Comp Immunol. 2008;32:519–531. doi: 10.1016/j.dci.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetru C, Hoffmann JA. NF-kappaB in the immune response of Drosophila. Cold Spring Harb Perspect Biol. 2009;1:a000232. doi: 10.1101/cshperspect.a000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Zhou Z, Wang H, Kleinerman ES. CAPER-alpha alternative splicing regulates the expression of vascular endothelial growth factor(165) in Ewing sarcoma cells. Cancer. 2012;118:2106–2116. doi: 10.1002/cncr.26488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz J, Armitage SA. Alternative adaptive immunity in invertebrates. Trends Immunol. 2006;27:493–496. doi: 10.1016/j.it.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Levashina EA, Moita LF, Blandin S, Vriend G, Lagueux M, Kafatos FC. Conserved role of a complement-like protein in phagocytosis revealed by dsRNA knockout in cultured cells of the mosquito, Anopheles gambiae. Cell. 2001;104:709–718. doi: 10.1016/s0092-8674(01)00267-7. [DOI] [PubMed] [Google Scholar]
- Luna C, Hoa NT, Lin H, Zhang L, Nguyen HL, Kanzok SM, Zheng L. Expression of immune responsive genes in cell lines from two different Anopheline species. Insect Mol Biol. 2006;15:721–729. doi: 10.1111/j.1365-2583.2006.00661.x. [DOI] [PubMed] [Google Scholar]
- Meijers R, Puettmann-Holgado R, Skiniotis G, Liu JH, Walz T, Wang JH, Schmucker D. Structural basis of Dscam isoform specificity. Nature. 2007;449:487–491. doi: 10.1038/nature06147. [DOI] [PubMed] [Google Scholar]
- Meister S, Kanzok SM, Zheng XL, Luna C, Li TR, Hoa NT, Clayton JR, White KP, Kafatos FC, Christophides GK, et al. Immune signaling pathways regulating bacterial and malaria parasite infection of the mosquito Anopheles gambiae. Proc Natl Acad Sci U S A. 2005;102:11420–11425. doi: 10.1073/pnas.0504950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount SM, Salz HK. Pre-messenger RNA processing factors in the Drosophila genome. J Cell Biol. 2000;150:F37–44. doi: 10.1083/jcb.150.2.f37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JW, Parisky K, Celotto AM, Reenan RA, Graveley BR. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc Natl Acad Sci U S A. 2004;101:15974–15979. doi: 10.1073/pnas.0407004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham LN, Dionne MS, Shirasu-Hiza M, Schneider DS. A specific primed immune response in Drosophila is dependent on phagocytes. PLoS Pathog. 2007;3:e26. doi: 10.1371/journal.ppat.0030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues J, Brayner FA, Alves LC, Dixit R, Barillas-Mury C. Hemocyte differentiation mediates innate immune memory in Anopheles gambiae mosquitoes. Science. 2010;329:1353–1355. doi: 10.1126/science.1190689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucker D, Chen B. Dscam and DSCAM: complex genes in simple animals, complex animals yet simple genes. Genes Dev. 2009;23:147–156. doi: 10.1101/gad.1752909. [DOI] [PubMed] [Google Scholar]
- Sinden RE, Alavi Y, Raine JD. Mosquito--malaria interactions: a reappraisal of the concepts of susceptibility and refractoriness. Insect Biochem Mol Biol. 2004;34:625–629. doi: 10.1016/j.ibmb.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Smith PH, Mwangi JM, Afrane YA, Yan G, Obbard DJ, Ranford-Cartwright LC, Little TJ. Alternative splicing of the Anopheles gambiae Dscam gene in diverse Plasmodium falciparum infections. Malar J. 2011;10:156. doi: 10.1186/1475-2875-10-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valanne S, Wang JH, Ramet M. The Drosophila Toll signaling pathway. J Immunol. 2011;186:649–656. doi: 10.4049/jimmunol.1002302. [DOI] [PubMed] [Google Scholar]
- Watson FL, Puttmann-Holgado R, Thomas F, Lamar DL, Hughes M, Kondo M, Rebel VI, Schmucker D. Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science. 2005;309:1874–1878. doi: 10.1126/science.1116887. [DOI] [PubMed] [Google Scholar]
- Watthanasurorot A, Jiravanichpaisal P, Liu H, Soderhall I, Soderhall K. Bacteria-Induced Dscam Isoforms of the Crustacean, Pacifastacus leniusculus. PLoS Pathog. 2011;7:e1002062. doi: 10.1371/journal.ppat.1002062. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wojtowicz WM, Flanagan JJ, Millard SS, Zipursky SL, Clemens JC. Alternative splicing of Drosophila Dscam generates axon guidance receptors that exhibit isoform-specific homophilic binding. Cell. 2004;118:619–633. doi: 10.1016/j.cell.2004.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhan L, Zhang W, Sun F, Wang W, Tian N, Bi J, Wang H, Shi D, Jiang Y, et al. RNA secondary structure in mutually exclusive splicing. Nat Struct Mol Biol. 2011;18:159–168. doi: 10.1038/nsmb.1959. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.