Abstract
Despite its key role in mediating a plethora of cellular signaling cascades pertinent to health and disease, little is known about the structural landscape of the proline-rich (PR) domain of Sos1 guanine nucleotide exchange factor. Herein, using a battery of biophysical tools, we provide evidence that the PR domain of Sos1 is structurally disordered and adopts an extended random coil-like conformation in solution. Of particular interest is the observation that while chemical denaturation of PR domain results in the formation of a significant amount of polyproline II (PPII) helices, it has little or negligible effect on its overall size as measured by its hydrodynamic radius. Our data also show that the PR domain displays a highly dynamic conformational basin in agreement with the knowledge that the intrinsically unstructured proteins rapidly interconvert between an ensemble of conformations. Collectively, our study provides new insights into the conformational equilibrium of a key signaling molecule with important consequences on its physiological function.
Keywords: Proline-rich proteins, Structural disorder, Random coil, Polyproline II helices, Conformational dynamics
INTRODUCTION
Sos1 guanine nucleotide exchange factor, comprised of the HF-DH-PH-REM-Cdc25-PR signaling cassette (Figure 1a), activates Ras and Rac GTPases that relay external signals from receptor tyrosine kinases (RTKs) such as EGFR at the cell surface to downstream effectors such as transcription factors within the nucleus (1–7). Notably, both Ras and Rac are tethered to the inner membrane surface via prenylation and act as molecular switches by virtue of their ability to cycle between active GTP-bound and inactive GDP-bound states. How does Sos1 activate Ras and Rac?
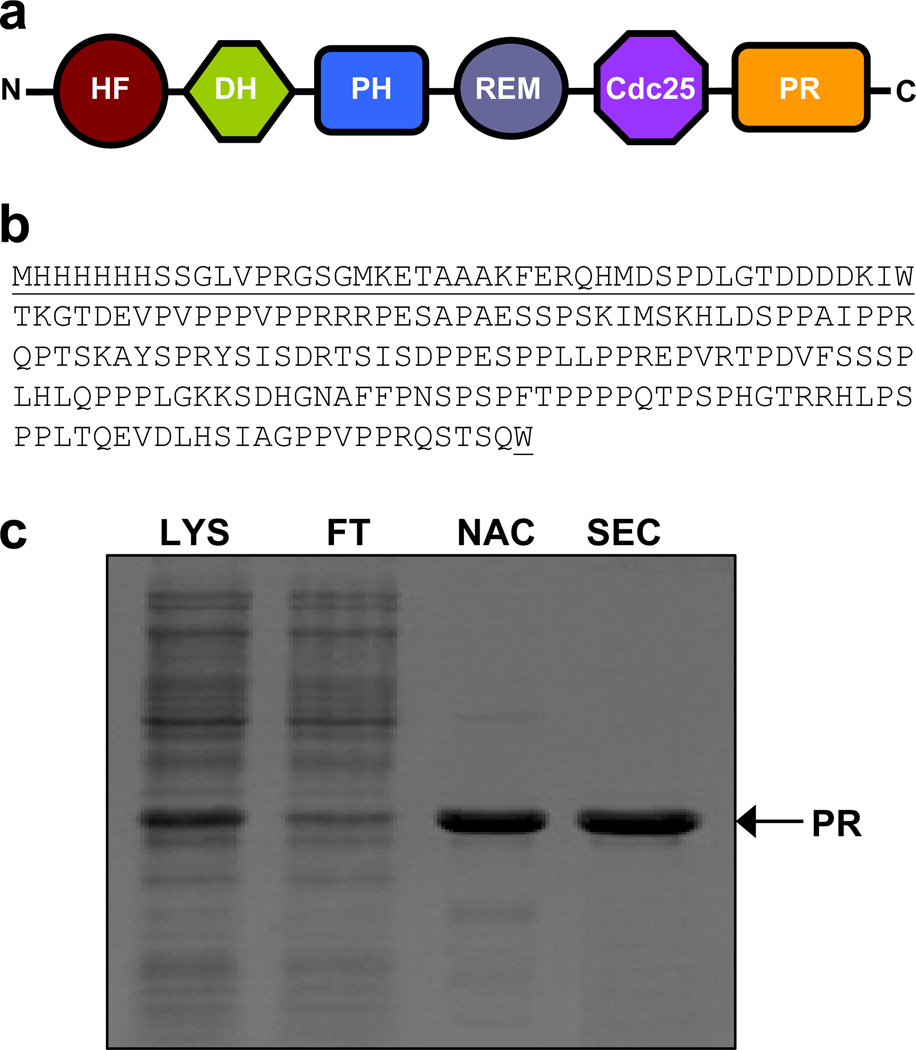
Figure 1.
Purification and characterization of the PR domain of Sos1. (a) Within Sos1 (residues 1–1333), the PR domain lies at the extreme C-terminal end. Other domains within Sos1 are HF (histone fold), DH (Dbl homology), PH (pleckstrin homology), REM (Ras exchange motif) and Cdc25 (cell division cycle 25). (b) Complete amino acid sequence of the recombinant PR domain (residues 1141–1300). The non-native amino acid residues introduced during cloning at both the N- and C-termini of the PR domain are underlined for clarity. (c) SDS-PAGE analysis of the PR domain. Briefly, total bacterial lysate (LYS) was loaded onto a Ni-NTA column, the flow-through (FT) was collected and after elution from the Ni-NTA affinity chromatography (NAC) column, the recombinant protein was further purified to apparent homogeneity by size-exclusion chromatography (SEC).
In the cytoplasm of quiescent cells, Sos1 exists in two functional pools in complex with Grb2 and Abi1 signaling adaptors—this association is mediated via the binding of the proline-rich (PR) domain of Sos1 to SH3 domains within Grb2 and Abi1. Upon stimulation of RTKs with growth factors and hormones, the Sos1-Grb2 complex becomes recruited to the inner membrane surface from the cytoplasm in a phosphotyrosine-dependent manner. Such translocation facilitates the Cdc25 domain of Sos1 to trigger GTP-GDP exchange within Ras and, in so doing, switches on a key signaling circuit that involves the activation of downstream MAP kinase cascade central to cellular growth and proliferation (8, 9). On the other hand, Sos1-Abi1 complex is recruited to actin filaments found within membrane ruffles in an Eps8-dependent manner. Given that Rac preferentially localizes to the confined areas of membrane ruffles, the recruitment of Sos1-Abi complex to the actin cytoskeleton network aids the DH domain of Sos1 to catalyze GTP-GDP exchange within Rac and, in so doing, plays a key role in actin remodeling central to cell invasion and migration (10, 11).
Interestingly, Sos1 can also be recruited to the inner membrane surface via the binding of its PH domain to phosphatidic acid, a component of phospholipids, in response to RTK stimulation with growth factors and hormones (12). Accordingly, recruitment of Sos1 to the inner membrane surface in a PH-dependent manner provides an alternative route for the activation of Ras. However, unlike the dispensability of Sos1-Grb2 complex for the activation of Ras, the Sos1-Abi1 complex is believed to be absolutely required for the activation of Rac. Importantly, the HF and REM domains within Sos1 play a regulatory role and fine tune the activity of Sos1 (13, 14). Briefly, the binding of Ras-GTP to REM domain serves as an allosteric switch to further stimulate the catalytic activity of Cdc25 domain. In contrast, the HF domain—comprised of a tandem copy of histone folds—associates in an intramolecular manner with the PH domain and, in so doing, down-regulates the PH-dependent activation of Ras.
In an attempt to understand the physical basis of how Sos1 activates Ras and Rac GTPases, the crystal structure of a Sos1 construct containing all contiguous domains but the C-terminal PR domain was recently solved to high resolution (15). However, structural insights into the ability of the PR domain to adopt a well-defined conformation, or lack thereof, would further our understanding of how Sos1 mediates RTK signaling. Herein, using a battery of biophysical tools, we provide evidence that the PR domain of Sos1 is structurally disordered and adopts an extended random coil-like conformation in solution. Given that many intrinsically unstructured proteins undergo folding in the presence of their cognate ligands (16–22), the possibility that the PR domain may also adopt a well-defined conformation upon binding to its ligands cannot be ruled out.
MATERIALS and METHODS
Sample preparation
The PR domain (residues 1141–1300) of human Sos1 was cloned into pET30 bacterial expression vector with an N-terminal sequence containing an His-tag (HHHHHH) and an enterokinase cleavage site (DDDDK) using Novagen LIC technology (Figures 1a and 1b). Additionally, a tryptophan (W) residue was added to both the N- and C-termini of the PR domain to aid in the quantification of protein concentration using spectrophotometry. The recombinant protein was expressed in Escherichia coli BL21* (DE3) bacterial strain and purified on a Ni-NTA affinity column followed by size-exclusion chromatography (SEC) on a Hiload Superdex 200 column using standard procedures as described previously (23). Final yield was typically between 5–10mg protein of apparent homogeneity, as judged by SDS-PAGE analysis (Figure 1c), per liter of bacterial culture. Notably, the molar mass of the PR domain was estimated to be around 26kD on the basis of SDS-PAGE analysis. This is in an excellent agreement with the theoretical molar mass of 22kD calculated from its amino acid sequence alone. Protein concentration was determined by the fluorescence-based Quant-It assay (Invitrogen) and spectrophotometrically on the basis of an extinction coefficient of 13,980 M−1cm−1 calculated for the recombinant PR domain using the online software ProtParam at ExPasy Server (24). Results from both methods were in an excellent agreement. The PR domain was dialyzed into a buffer of 50 mM Sodium phosphate at pH 8.0 in the presence or absence of appropriate concentrations of urea or GuHCl prior to the conduct of all biophysical measurements described hereinafter. All measurements were repeated at least three times. It should be noted that the treatment of recombinant PR domain with enterokinase substantially reduced the yield of the protein due to partial digestion. Accordingly, all experiments reported herein were conducted on the recombinant PR domain containing non-native residues at both the N- and C-termini (Figure 1b). Importantly, while control experiments were also carried out on the cleaved construct to check that the non-native residues did not alter the properties of the PR domain, the signal-to-noise ratio obtained for these measurements was relatively poor due to low yield of cleaved protein.
Circular dichroism
Far-UV circular dichroism (CD) measurements were conducted on a Jasco J-815 spectropolarimeter thermostatically controlled at 25 °C. All experiments were conducted on a 15 µM sample of PR domain alone and in the presence of varying concentrations of urea or GuHCl. Data were collected using a quartz cuvette with a 2-mm pathlength in the 190–250 nm wavelength range and with a slit bandwidth of 2 nm at a scan rate of 10 nm/min. Data were normalized against reference spectra to remove the contribution of appropriate buffers. Each data set represents an average of four scans acquired at 0.1 nm intervals. Data were converted to mean residue ellipticity, [θ], as a function of wavelength (λ) of electromagnetic radiation using the following equation:
| [1] |
where Δθ is the observed ellipticity in mdeg, N is the number of residues within the recombinant PR domain, c is the concentration of recombinant PR domain in µM, and l is the cuvette pathlength in cm.
Dynamic light scattering
Dynamic light scattering (DLS) experiments were conducted on a Wyatt miniDAWN TREOS instrument equipped with a QELS dynamic light scattering detector, positioned at 90° with respect to the incident laser beam at a wavelength of 658 nm. All measurements were carried out under steady-state conditions on a 50 µM sample of PR domain alone and in the presence of varying concentrations of urea or GuHCl at 10 °C. The time-dependence of DLS intensity fluctuation of PR domain under various conditions was collected for 30 min. The hydrodynamic radius (Rh) was calculated from the fit of DLS data using the regularization method according to the instructions embodied in the built-in ASTRA software. The Rh values expected for the PR domain resembling a fully-folded globular conformation or a fully-denatured state were calculated using the following expression based on polymer theory (25, 26):
| [2] |
where N is the number of amino acid residues within a polypeptide chain, while a and b are empirically-derived constants. For the fully-folded globular proteins, the constants a and b respectively take up values of 4.75Å and 0.29, while for the fully-denatured globular proteins, the constants a and b respectively take up values of 2.21Å and 0.57 (27, 28).
Small-angle x-ray scattering
Small-angle x-ray scattering (SAXS) analysis was conducted at the Beamline X9 of the National Synchrotron Light Source (NSLS) at the Brookhaven National Laboratory using the standard high throughput solution scattering setup at a photon energy of 13.5keV (29). Measurements were carried out on PR domain of Sos1 at protein concentrations of 100, 200 and 400µM. Wide-angle and small-angle data were collected simultaneously from two detectors. The data were subsequently converted to 1D scattering profiles, merged and the background scattering was subtracted using the integrated pyXS software.
Molecular dynamics
Molecular dynamics (MD) simulations were performed with the GROMACS software (30, 31) using the integrated GROMOS96-53A6 (32), AMBER99SB-ILDN (33, 34), CHARMM-27 (35, 36), or OPLS-AA (37, 38) force fields. Briefly, the PR domain of Sos1 was folded into a random-coil state using the QUARK server based on ab initio modeling. The QUARK server can be accessed online at http://zhanglab.ccmb.med.umich.edu/QUARK. Next, the random coil conformation of the PR domain was centered within a cubic box and hydrated with a water layer that extended 10Å (box size) from the protein surface along each orthogonal direction using the extended simple point charge (SPC/E) water model (39, 40). The total number of water molecules within the cubic box was ~11,000 and the ionic strength of solution was set to 100mM with Na+ (21 cations) and Cl− (25 anions) ions. Additionally, basic (Arg/Lys) and acidic (Asp/Glu) residues within the PR domain carried a net charge of +1 and −1, respectively. The hydrated structure was energy-minimized with the steepest descent algorithm prior to equilibration under the NPT ensemble conditions, wherein the number of atoms (N), pressure (P) and temperature (T) within the system were respectively kept constant at ~35,000, 1 bar and 300 K. The Particle-Mesh Ewald (PME) method was employed to compute long-range electrostatic interactions with a 10Å cut-off (41) and the Linear Constraint Solver (LINCS) algorithm to restrain bond lengths (42). All MD simulations were performed under periodic boundary conditions (PBC) using the leap-frog integrator with a time step of 2fs. For the final MD production runs, data were collected every 10ps over a time scale of 100ns. All MD simulations were run on a Linux workstation using parallel processors at the High Performance Computing facility within the Center for Computational Science of the University of Miami.
RESULTS and DISCUSSION
PR domain displays high propensity for structural disorder
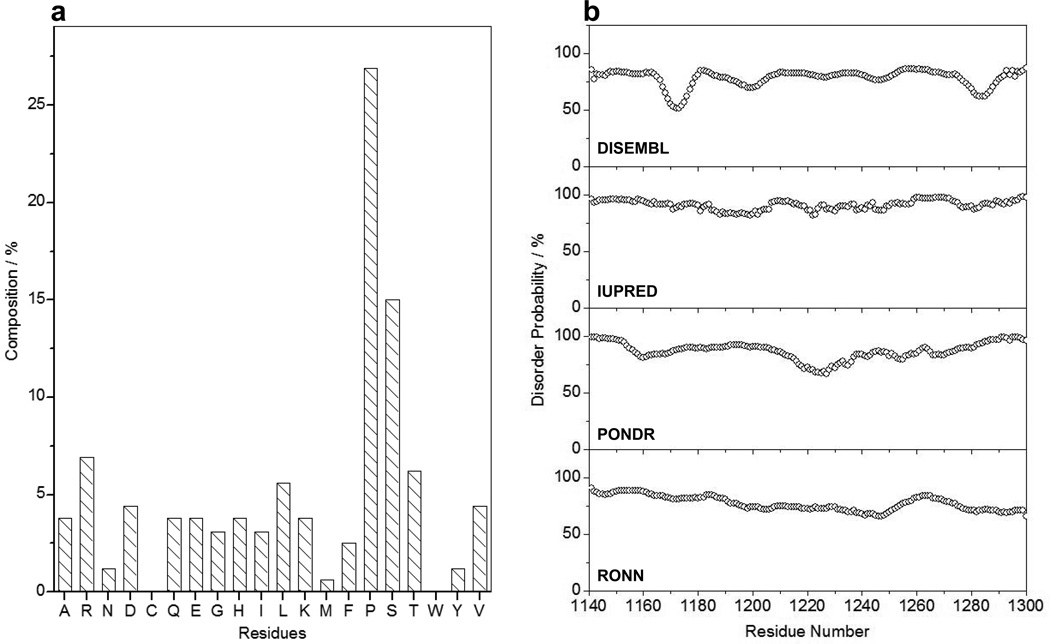
It is now well recognized that non-polar residues such as proline and glycine as well as polar and charged residues such as serine, threonine, glutamine, glutamate, lysine and arginine display high propensity for structural disorder within proteins (43–46). Analysis of amino acid composition of the PR domain indeed suggests that these aforementioned residues constitute close to 75% of its primary sequence (Figure 2a). Importantly, the charged residues alone (D/E/K/R) constitute about 20% of the total amino acid content of the PR domain. Taken together, these observations strongly argue that the PR domain apparently bears rather high propensity for structural disorder. This notion gains further credibility in view of the fact that the PR domain is largely depleted of hydrophobic residues such as valine, leucine, isoleucine, methionine, phenylalanine, tyrosine and tryptophan that are commonly found in globular proteins. Accordingly, we next subjected the PR domain to various online disorder predictors to quantify the degree of such structural disorder (Figure 2b). Our in silico analysis reveals that the PR domain is intrinsically disordered with approximately 75% probability.
Figure 2.
In silico analysis of the PR domain of Sos1. (a) The composition of amino acid residues within the PR domain (residues 1141–1300) as a percentage of its chain length. (b) Prediction of intrinsic disorder within the PR domain using DISEMBL (82), IUPRED (83), PONDR (43) and RONN (84). Note that these disorder predictors can be accessed online at http://www.disprot.org/predictors.php.
PR domain harbors structural features characteristic of a random coil
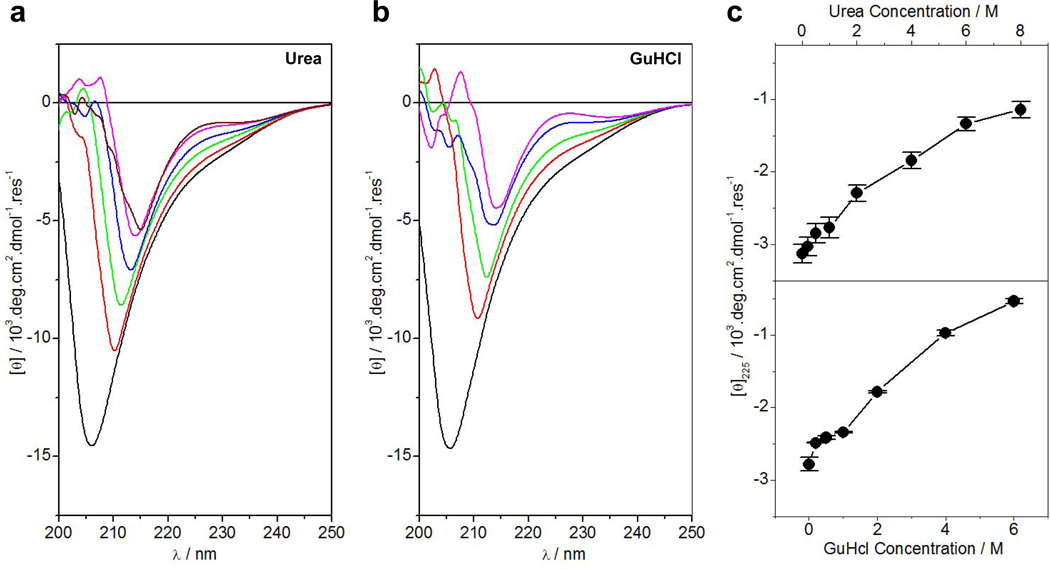
To experimentally test the notion that PR domain of Sos1 likely harbors structural disorder, we next conducted far-UV CD analysis on the PR domain alone and in the presence of chemical denaturants such as urea and GuHCl (Figure 3). Our data show that the far-UV CD spectrum of the PR domain is characterized by a negative band centered around 208nm in the absence of denaturants (Figures 3a and 3b). This strongly suggests that the PR domain predominantly adopts a random coil-like conformation in solution though the possibility of some degree of polyproline II (PPII) helices cannot be completely ruled out. Strikingly, the spectrum of the PR domain undergoes substantial changes with increasing concentration of denaturants. Firstly, the negative band not only undergoes a reduction in spectral intensity but also becomes red-shifted to around 215nm with increasing denaturant concentration. Secondly, increasing denaturant concentration results in the appearance of a positive band around 225nm. Such spectral features are frequently observed and well-documented for proline-rich peptides and proteins harboring PPII-helical conformation (47, 48). Notably, a plot of change in spectral intensity at 225nm, [θ]225, as a function of denaturant concentration follows a hyperbolic trend, increasing linearly at lower concentrations of denaturants and then slowly plateauing out at saturating concentrations (Figure 3c). Taken together, these salient observations suggest that the PR domain assumes PPII-helical conformation in the chemically-denatured state. This notion is further corroborated by the fact that the PPII-helices constitute a major component of proteins in their denatured states (49, 50). Importantly, we believe that the PPII-helices in the denatured state of PR domain may also be aided by the trans-cis isomerization about the X-Pro bonds in the presence of denaturants. It is noteworthy that the proline largely exists in cis-conformation in the denatured state of proteins and that cis-trans isomerization is a rate-limiting step in the folding of proline-containing proteins (51–53). In short, our far-UV CD data indicate that while the PR domain of Sos1 adopts a native random coil-like conformation in solution, it is highly malleable to chemical denaturants that trigger a structural transition to a denatured state harboring a substantial amount of PPII-helices. Importantly, while the PR domain of Sos1 may lack intrinsic structure, the possibility that it may also adopt a well-defined conformation upon binding to its ligands cannot be ruled out. It is indeed well-documented that many intrinsically unstructured proteins undergo folding upon ligand binding (16–22).
Figure 3.
CD analysis of the PR domain of Sos1 alone and in the presence of chemical denaturants. (a) Far-UV CD Spectra of the PR domain in 0M (black), 1M (red), 2M (green), 4M (blue), 6M (magenta) and 8M (brown) urea. (b) Far-UV CD Spectra of the PR domain in 0M (black), 1M (red), 2M (green), 4M (blue) and 6M (magenta) GuHCl. (c) Dependence of ellipticity of the positive band at 225nm, [θ]225, of the PR domain on urea (top panel) and GuHCl (bottom panel) concentrations. Error bars were calculated from three independent measurements to one standard deviation. The solid lines connect appropriate data points for clarity.
Chemical denaturants have little or no effect on the size of PR domain
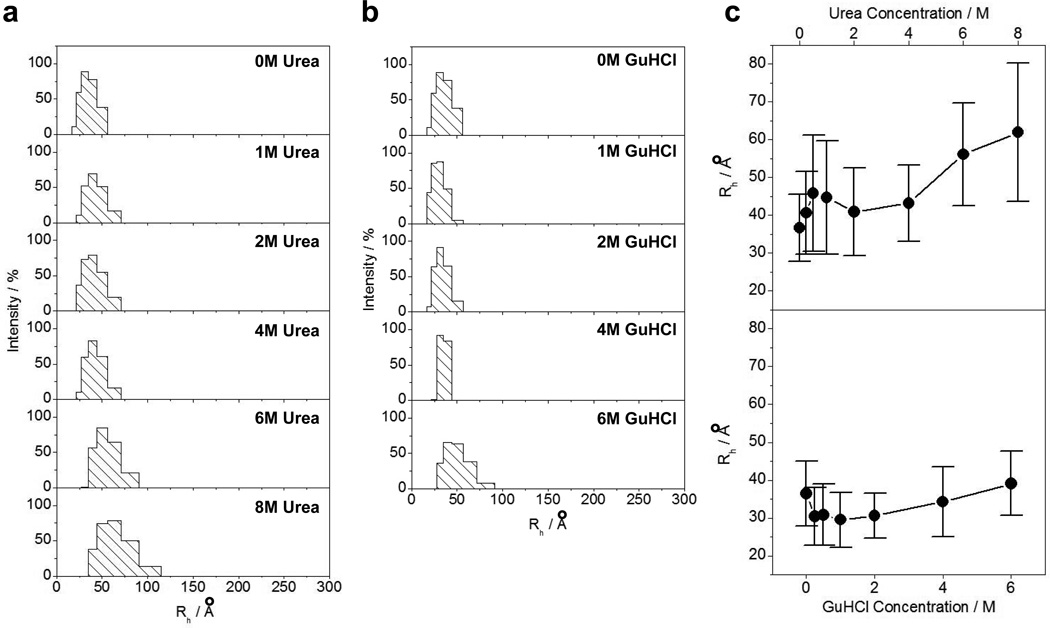
In order to test whether the overall size of the PR domain of Sos1 is consistent with a random coil conformation and to what extent chemical denaturation affects its overall size, we measured the hydrodynamic radius (Rh) of the PR domain alone and in the presence of urea and GuHCl using DLS (Figure 4). Strikingly, our analysis shows that the denaturants have little or no effect on the hydrodynamic radius of the PR domain. It is generally believed that chemical denaturants exert their actions via weakening the interior hydrophobic forces that hold the core of globular proteins together by virtue of their ability to form hydrogen bonds with polar residues, thereby altering the structure of surrounding water molecules and culminating in the breaking of the hydrophobic collapse (54–57). In agreement with this model, chemical denaturants should be expected to result in the expansion of globular proteins (58). However, the fact that the PR domain of Sos1 appears to lack a well-defined hydrophobic core reminiscent of globular proteins (Figure 2a), its ability to resist the action of chemical denaturants on its size should not therefore come as a surprise. Importantly, the expected hydrodynamic radii for the PR domain resembling a fully-folded globular conformation and a fully-denatured state were respectively calculated to be 21Å and 40Å using Eq [2]. Accordingly, our hydrodynamic data further substantiate the notion that the PR domain likely adopts a random coil-like conformation in solution.
Figure 4.
DLS analysis of the PR domain of Sos1 alone and in the presence of chemical denaturants. (a) Regularization plots of the PR domain in the absence and presence of varying concentrations of urea as indicated. (b) Regularization plots of the PR domain in the absence and presence of varying concentrations of GuHCl as indicated. (c) Comparison of the dependence of hydrodynamic radius (Rh) of the PR domain on urea (top panel) and GuHCl (bottom panel) concentrations. Error bars represent standard deviation in the fit of the regularization data. The solid lines connect appropriate data points for clarity.
PR domain adopts an extended random coil-like conformation
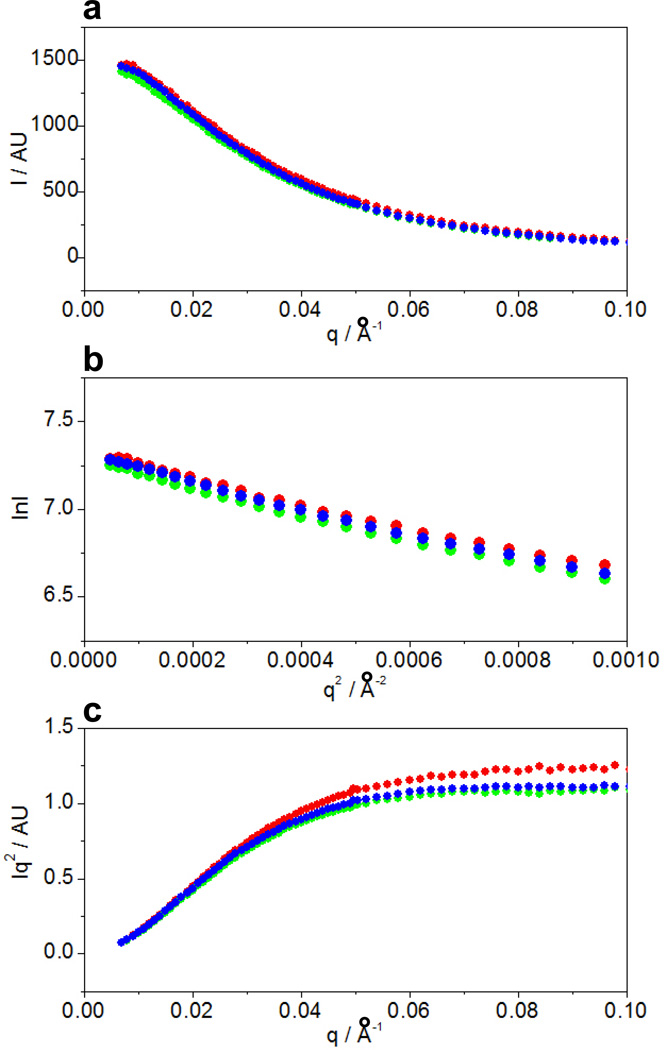
To shed light on the overall three-dimensional conformation of the PR domain of Sos1, we next conducted SAXS analysis at three different protein concentrations (Figure 5). Notably, the SAXS analysis correlates the scattering intensity (I) of atoms to the scattering vector (q), which is defined as q=(4πsinθ/λ), where θ is the scattering angle and λ is the wavelength of the x-ray incident beam. Our SAXS analysis shows that the scattering profiles follow similar trends for all three protein concentrations (Figure 5a), implying that the inter-particle scattering is similar within this concentration range. Consistent with this observation, the radius of gyration (Rg) calculated from the slopes of Guinier plots at low scattering angles is around 47Å at all three protein concentrations (Figure 5b). In light of the fact that hydrodynamic radius (Rh) of the PR domain calculated from our DLS analysis is 37Å (Figure 4), we obtained a shape factor (ρ=Rg/Rh) of 1.3. A value of ρ greater than unity implies that the PR domain most likely adopts an elongated shape in lieu of a compact conformation. In order to directly obtain shape information from our SAXS measurements, we also generated Kratky plots (Figure 5c). For a compact globular conformation, the Kratky plot initially rises at low scattering angles and then descends at higher scattering angles carving out a hump or bell-shaped curve (59). Strikingly, the Kratky plots for the PR domain at all three concentrations initially rise at low scattering angles but then plateau out at higher scattering angles. Such behavior is consistent with an extended random coil-like conformation characteristic of proteins devoid of a globular structure (59). Notably, the molar mass of the PR domain was estimated to be around 28kD from our SAXS data. Given that the theoretical molar mass of PR domain calculated from its amino acid sequence alone is 22kD, our SAXS analysis indicates that the PR domain largely exists as a monomer in solution in agreement with our previous static light scattering measurements (23). In short, our SAXS analysis suggests that the PR domain is likely to adopt an extended random coil-like conformation in solution.
Figure 5.
SAXS analysis of the PR domain of Sos1 at protein concentrations of 120µM ( ), 240µM (
), 240µM ( ) and 360µM (
) and 360µM ( ). (a) Scattering profiles. (b) Guinier plots. (c) Kratky plots.
). (a) Scattering profiles. (b) Guinier plots. (c) Kratky plots.
PR domain samples a highly dynamic conformational basin
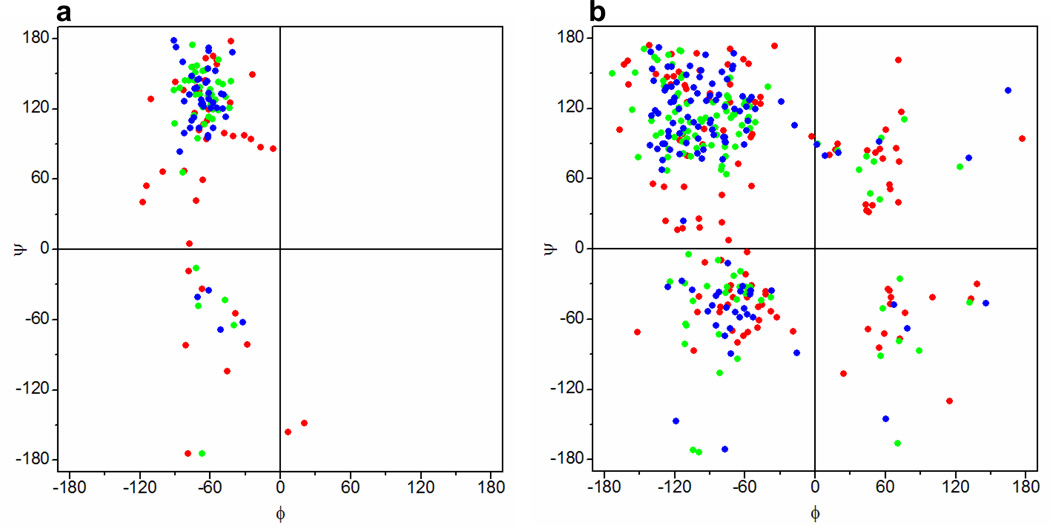
The data presented above are based on experimental techniques that probe the average behavior of large ensembles of the PR domain of Sos1 in lieu of any specific conformation. In particular, such bulk measurements do not provide information on the conformational space available to the PR domain. In an attempt to uncover the conformational dynamics in terms of the ϕ and ψ torsion angles respectively sampled by the backbone N-Cα and Cα-C bonds, we conducted MD simulations starting with a random coil state of the PR domain using GROMOS96-53A6 (32), AMBER99SB-ILDN (33, 34), CHARMM-27 (35, 36), or OPLS-AA (37, 38) force fields. Figure 6 shows Ramachandran plots obtained for proline and non-proline residues within the PR domain at various time points during such simulations using the GROMOS96-53A6 force field. Unsurprisingly, the ϕ and ψ torsion angles of both the proline and non-proline residues within the random coil state of the PR domain sample a large conformational basin within the Ramachandran plot (Figures 6a and 6b). Interestingly, such torsional space does not appear to become substantially restricted after 10-ns in the course of MD simulations. Moreover, this behavior does not substantially change as the simulation time is increased to 100ns, particularly for the non-proline residues. Importantly, the fact that the ϕ-ψ torsional space for proline residues within the PR domain becomes somewhat more restricted compared to non-proline residues with increasing simulation time should be expected due to the rigidity of the pyrrolidine ring of proline residues. It should be noted that the highly dynamic fluctuation of non-proline residues within the PR domain on the Ramachandran plot is also observed in the case of the three other force fields used (data not shown). In particular, our MD simulations suggest that the non-proline residues within the PR domain sample the ϕ-ψ torsional geometries reminiscent of all four major secondary structural elements: right-handed α-helix ((ϕ,ψ)≈(−60°,−45°)), β-sheet ((ϕ,ψ)≈(−135°,+135°)), PPII helix ((ϕ,ψ)≈(−75°,+150°)), and left-handed α-helix ((ϕ,ψ)≈(+60°,+60°)). Taken together, our MD analysis suggests that the PR domain displays backbone conformational dynamics expected of a highly fluctuating random coil-like conformation in agreement with the data from our bulk measurements. Importantly, such conformational characteristics of the PR domain are consistent with the highly fluctuating ensemble of rapidly interconverting conformations observed for structurally disordered proteins (60–63). Nonetheless, it should be borne in mind that we did not conduct MD analysis starting with a random coil conformation of an otherwise fully-folded globular protein of similar size to compare its backbone conformational dynamics to the PR domain. However, we anticipate that the conformational space available to a folded globular protein would be much more restricted than that observed for the PR domain.
Figure 6.
Ramachandran plots obtained for proline (a) and non-proline (b) residues within the random-coil state of the PR domain of Sos1 before ( ) and after being subjected to MD simulations for 10ns (
) and after being subjected to MD simulations for 10ns ( ) and 100ns (
) and 100ns ( ) using the GROMOS96-53A6 force field (32).
) using the GROMOS96-53A6 force field (32).
CONCLUSIONS
One of the basic tenets of structural biology is that all proteins must take up a three-dimensional structure in order to attain biological function. Indeed, structure-function relationships in proteins form the basis of specificity and fidelity in virtually all cellular processes central to driving life at molecular level. Yet, developments over the past decade or so indicate that a growing number of proteins, usually interspersed with proline-rich segments as well as polar and charged residues, are intrinsically unstructured or structurally disordered (64–73). Importantly, while such structurally disordered proteins orchestrate a plethora of signaling cascades driving cellular functions ranging from growth and motility to apoptosis and immunity (72), our understanding of the structural landscape that they sample remains hitherto poorly understood. Toward this goal, our data presented herein suggest that the PR domain of Sos1 is intrinsically unstructured and adopts an overall random coil-like conformation. Moreover, the PR domain appears to be conformationally dynamic in a manner akin to the conformational flexibility of structurally disordered proteins (60–63). We believe that the rapidly fluctuating nature of the PR domain is functionally advantageous in that it allows it to recognize a variety of different ligands such as Grb2 and Abi1 among others in the context of RTK signaling (1–7).
It would be fitting to add here that while the PR domain of Sos1 may lack intrinsic structure, the possibility that it may also adopt a well-defined conformation upon binding to its ligands cannot be ruled out. It is indeed well-documented that many intrinsically unstructured proteins undergo folding in the presence of their cognate ligands or in response to osmolytes. In the ligand-induced mechanism, the folding of intrinsically unstructured proteins to a stable globular structure is coupled to the binding of a ligand (16–22). The free energy released upon ligand binding essentially lowers the free energy of the folded state, thereby shifting the equilibrium in favor of the globular structure. Importantly, such energetic coupling is believed to augment the rate of ligand binding by virtue of the fact that the greater capture radius of intrinsically unstructured proteins enables them to sample a much larger space so as to reduce the dimensionality of the search for their partners (74). On the other hand, osmolyte-induced folding is driven by changes in the polarity of the surrounding solute molecules (75–79). This effectively raises the free energy of the unfolded state higher than that of the folded state, leading to thermodynamic stabilization of the latter. In a manner that hydrophobic effect drives the folding of hydrophobic regions in globular proteins, the osmophobic effect may also drive the folding of intrinsically unstructured proteins. Notably, the osmophobic effect arises from the unfavorable interaction of osmolytes with the backbone atoms of the protein so as to thermodynamically destabilize the unfolded state (80, 81, 76). Importantly, many of these osmolytes, such as TMAO, are found within living cells where they play a key role in the maintenance of native conformations of proteins against denaturing effects of environmental stresses such as urea (75, 81). Accordingly, the ability of these osmolytes to induce folding of what appear to be intrinsically unstructured proteins is physiologically relevant.
In sum, our study establishes that the PR domain of Sos1 lacks a well-defined intrinsic structure and that such virtue is likely to be of central importance to its physiological function.
ACKNOWLEDGMENTS
We are deeply indebted to Lin Yang at the National Synchrotron Light Source, Brookhaven National Laboratory, for his help with SAXS data collection and analysis. We thank the Sylvia Daunert Group for the use of its Jasco J-815 spectropolarimeter. This work was supported by the National Institutes of Health Grants R01-GM083897 (to AF) and R01-AG033719 (to IKL), and funds from the USylvester Braman Family Breast Cancer Institute (to AF). CBM is a recipient of a postdoctoral fellowship from the National Institutes of Health (Award# T32-CA119929). Use of the National Synchrotron Light Source, Brookhaven National Laboratory, was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-98CH10886.
ABBREVIATIONS
- Abi1
Abl interactor 1
- CD
Circular dichroism
- Cdc25
cell cycle division 25
- DH
Dbl homology
- DLS
Dynamic light scattering
- EGFR
Epidermal growth factor receptor
- Eps8
EGFR kinase substrate 8
- Grb2
Growth factor receptor binder 2
- GuHCl
Guanidine hydrochloride
- HF
Histone fold
- LIC
Ligation-independent cloning
- MAP
Mitogen-activated protein
- MD
Molecular dynamics
- PH
Pleckstrin homology
- PPII
Polyproline II (helix)
- PR
Proline-rich
- REM
Ras exchange motif
- RTK
Receptor tyrosine kinase
- SAXS
Small-angle x-ray scattering
- SEC
Size-exclusion chromatography
- SH3
Src homology 3
- Sos1
Son of sevenless 1
- TMAO
Trimethylamine N-oxide
REFERENCES
- 1.Chardin P, Camonis JH, Gale NW, van Aelst L, Schlessinger J, Wigler MH, Bar-Sagi D. Human Sos1: a guanine nucleotide exchange factor for Ras that binds to GRB2. Science. 1993;260:1338–1343. doi: 10.1126/science.8493579. [DOI] [PubMed] [Google Scholar]
- 2.Li N, Batzer A, Daly R, Yajnik V, Skolnik E, Chardin P, Bar-Sagi D, Margolis B, Schlessinger J. Guanine-nucleotide-releasing factor hSos1 binds to Grb2 and links receptor tyrosine kinases to Ras signalling. Nature. 1993;363:85–88. doi: 10.1038/363085a0. [DOI] [PubMed] [Google Scholar]
- 3.Rozakis-Adcock M, Fernley R, Wade J, Pawson T, Bowtell D. The SH2 and SH3 domains of mammalian Grb2 couple the EGF receptor to the Ras activator mSos1. Nature. 1993;363:83–85. doi: 10.1038/363083a0. [DOI] [PubMed] [Google Scholar]
- 4.Nimnual AS, Yatsula BA, Bar-Sagi D. Coupling of Ras and Rac guanosine triphosphatases through the Ras exchanger Sos. Science. 1998;279:560–563. doi: 10.1126/science.279.5350.560. [DOI] [PubMed] [Google Scholar]
- 5.Innocenti M, Tenca P, Frittoli E, Faretta M, Tocchetti A, Di Fiore PP, Scita G. Mechanisms through which Sos-1 coordinates the activation of Ras and Rac. J Cell Biol. 2002;156:125–136. doi: 10.1083/jcb.200108035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nimnual A, Bar-Sagi D. The two hats of SOS. Sci STKE. 2002;2002:PE36. doi: 10.1126/stke.2002.145.pe36. [DOI] [PubMed] [Google Scholar]
- 7.Pierre S, Bats AS, Coumoul X. Understanding SOS (Son of Sevenless) Biochem Pharmacol. 2011;82:1049–1056. doi: 10.1016/j.bcp.2011.07.072. [DOI] [PubMed] [Google Scholar]
- 8.Robinson MJ, Cobb MH. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 9.Reuther GW, Der CJ. The Ras branch of small GTPases: Ras family members don't fall far from the tree. Curr Opin Cell Biol. 2000;12:157–165. doi: 10.1016/s0955-0674(99)00071-x. [DOI] [PubMed] [Google Scholar]
- 10.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 11.Ridley AJ. Rho GTPases and cell migration. J Cell Sci. 2001;114:2713–2722. doi: 10.1242/jcs.114.15.2713. [DOI] [PubMed] [Google Scholar]
- 12.Zhao C, Du G, Skowronek K, Frohman MA, Bar-Sagi D. Phospholipase D2-generated phosphatidic acid couples EGFR stimulation to Ras activation by Sos. Nat Cell Biol. 2007;9:706–712. doi: 10.1038/ncb1594. [DOI] [PubMed] [Google Scholar]
- 13.Jorge R, Zarich N, Oliva JL, Azanedo M, Martinez N, de la Cruz X, Rojas JM. HSos1 contains a new amino-terminal regulatory motif with specific binding affinity for its pleckstrin homology domain. J Biol Chem. 2002;277:44171–44179. doi: 10.1074/jbc.M204423200. [DOI] [PubMed] [Google Scholar]
- 14.Sondermann H, Soisson SM, Boykevisch S, Yang SS, Bar-Sagi D, Kuriyan J. Structural analysis of autoinhibition in the Ras activator Son of sevenless. Cell. 2004;119:393–405. doi: 10.1016/j.cell.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 15.Gureasko J, Kuchment O, Makino DL, Sondermann H, Bar-Sagi D, Kuriyan J. Role of the histone domain in the autoinhibition and activation of the Ras activator Son of Sevenless. Proc Natl Acad Sci U S A. 2010;107:3430–3435. doi: 10.1073/pnas.0913915107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kriwacki RW, Hengst L, Tennant L, Reed SI, Wright PE. Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: conformational disorder mediates binding diversity. Proc Natl Acad Sci U S A. 1996;93:11504–11509. doi: 10.1073/pnas.93.21.11504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daughdrill GW, Chadsey MS, Karlinsey JE, Hughes KT, Dahlquist FW. The C-terminal half of the anti-sigma factor, FlgM, becomes structured when bound to its target, sigma 28. Nat Struct Biol. 1997;4:285–291. doi: 10.1038/nsb0497-285. [DOI] [PubMed] [Google Scholar]
- 18.Penkett CJ, Redfield C, Dodd I, Hubbard J, McBay DL, Mossakowska DE, Smith RA, Dobson CM, Smith LJ. NMR analysis of main-chain conformational preferences in an unfolded fibronectin-binding protein. J Mol Biol. 1997;274:152–159. doi: 10.1006/jmbi.1997.1369. [DOI] [PubMed] [Google Scholar]
- 19.Liu D, Ishima R, Tong KI, Bagby S, Kokubo T, Muhandiram DR, Kay LE, Nakatani Y, Ikura M. Solution structure of a TBP-TAF(II)230 complex: protein mimicry of the minor groove surface of the TATA box unwound by TBP. Cell. 1998;94:573–583. doi: 10.1016/s0092-8674(00)81599-8. [DOI] [PubMed] [Google Scholar]
- 20.Kim AS, Kakalis LT, Abdul-Manan N, Liu GA, Rosen MK. Autoinhibition and Activation Mechanisms of the Wiskott-Aldrich Syndrome Protein. Nature. 2000;404:151–158. doi: 10.1038/35004513. [DOI] [PubMed] [Google Scholar]
- 21.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr. Opin. Struct. Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 22.Farooq A, Zeng L, Yan KS, Ravichandran KS, Zhou M-M. Coupling of Folding and Binding in the PTB Domain of the Signaling Protein Shc. Structure. 2003;11:905–913. doi: 10.1016/s0969-2126(03)00134-5. [DOI] [PubMed] [Google Scholar]
- 23.McDonald CB, Balke JE, Bhat V, Mikles DC, Deegan BJ, Seldeen KL, Farooq A. Multivalent binding and facilitated diffusion account for the formation of the Grb2-Sos1 signaling complex in a cooperative manner. Biochemistry. 2012;51:2122–2135. doi: 10.1021/bi3000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein Identification and Analysis Tools on the ExPASy Server. In: Walker JM, editor. The Proteomics Protocols Handbook. Totowa, New Jersey, USA: Humana Press; 2005. pp. 571–607. [Google Scholar]
- 25.Flory PJ. Principles of Polymer Chemistry. Ithaca: Cornell University Press; 1953. [Google Scholar]
- 26.De Gennes PG. Scaling Concepts in Polymer Physics. Ithaca: Cornell University Press; 1979. [Google Scholar]
- 27.Wilkins DK, Grimshaw SB, Receveur V, Dobson CM, Jones JA, Smith LJ. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry. 1999;38:16424–16431. doi: 10.1021/bi991765q. [DOI] [PubMed] [Google Scholar]
- 28.Kohn JE, Millett IS, Jacob J, Zagrovic B, Dillon TM, Cingel N, Dothager RS, Seifert S, Thiyagarajan P, Sosnick TR, Hasan MZ, Pande VS, Ruczinski I, Doniach S, Plaxco KW. Random-coil behavior and the dimensions of chemically unfolded proteins. Proc Natl Acad Sci U S A. 2004;101:12491–12496. doi: 10.1073/pnas.0403643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allaire M, Yang L. Biomolecular solution X-ray scattering at the National Synchrotron Light Source. J Synchrotron Radiat. 2011;18:41–44. doi: 10.1107/S0909049510036022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 31.Hess B. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 32.Oostenbrink C, Villa A, Mark AE, van Gunsteren WF. A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J Comp Chem. 2004;25:1656–1676. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- 33.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J Comp Chem. 1983;4:187–217. [Google Scholar]
- 36.Brooks BR, Brooks CLr, Mackerell ADJ, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, JZ P, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: The biomolecular simulation program. J Comp Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jorgensen WL, Tirado-Rives J. The OPLS Force Field for Proteins: Energy Minimizations for Crystals of Cyclic Peptides and Crambin. J Am Chem Soc. 1988;110:1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 38.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. Evaluation and Reparametrization of the OPLS-AA Force Field for Proteins via Comparison with Accurate Quantum Chemical Calculations on Peptides. J Phys Chem B. 2001;105:6474–6487. [Google Scholar]
- 39.Toukan K, Rahman A. Molecular-dynamics study of atomic motions in water. Physical Review B. 1985;31:2643–2648. doi: 10.1103/physrevb.31.2643. [DOI] [PubMed] [Google Scholar]
- 40.Berendsen HJC, Grigera JR, Straatsma TP. The Missing Term in Effective Pair Potentials. J Phys Chem. 1987;91:6269–6271. [Google Scholar]
- 41.Darden TA, York D, Pedersen L. Particle mesh Ewald: An N.log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 42.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: A linear constraint solver for molecular simulations. J Comput Chem. 1997;18:1463–1472. [Google Scholar]
- 43.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 44.Uversky VN. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mao AH, Crick SL, Vitalis A, Chicoine CL, Pappu RV. Net charge per residue modulates conformational ensembles of intrinsically disordered proteins. Proc Natl Acad Sci U S A. 2010;107:8183–8188. doi: 10.1073/pnas.0911107107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marsh JA, Forman-Kay JD. Sequence determinants of compaction in intrinsically disordered proteins. Biophys J. 2010;98:2383–2390. doi: 10.1016/j.bpj.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rabanal F, Ludevid MD, Pons M, Giralt E. CD of proline-rich polypeptides: application to the study of the repetitive domain of maize glutelin-2. Biopolymers. 1993;33:1019–1028. doi: 10.1002/bip.360330704. [DOI] [PubMed] [Google Scholar]
- 48.Woody RW. Circular dichroism spectrum of peptides in the poly(Pro)II conformation. J Am Chem Soc. 2009;131:8234–8245. doi: 10.1021/ja901218m. [DOI] [PubMed] [Google Scholar]
- 49.Hamburger JB, Ferreon JC, Whitten ST, Hilser VJ. Thermodynamic mechanism and consequences of the polyproline II (PII) structural bias in the denatured states of proteins. Biochemistry. 2004;43:9790–9799. doi: 10.1021/bi049352z. [DOI] [PubMed] [Google Scholar]
- 50.Cortajarena AL, Lois G, Sherman E, O'Hern CS, Regan L, Haran G. Non-random-coil behavior as a consequence of extensive PPII structure in the denatured state. J Mol Biol. 2008;382:203–212. doi: 10.1016/j.jmb.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brandts JF, Halvorson HR, Brennan M. Consideration of the Possibility that the slow step in protein denaturation reactions is due to cis-trans isomerism of proline residues. Biochemistry. 1975;14:4953–4963. doi: 10.1021/bi00693a026. [DOI] [PubMed] [Google Scholar]
- 52.Zimmerman SS, Scheraga HA. Stability of cis, trans, and nonplanar peptide groups. Macromolecules. 1976;9:408–416. doi: 10.1021/ma60051a005. [DOI] [PubMed] [Google Scholar]
- 53.Wedemeyer WJ, Welker E, Scheraga HA. Proline cis-trans isomerization and protein folding. Biochemistry. 2002;41:14637–14644. doi: 10.1021/bi020574b. [DOI] [PubMed] [Google Scholar]
- 54.Tanford C. Protein denaturation. Adv Protein Chem. 1968;23:121–282. doi: 10.1016/s0065-3233(08)60401-5. [DOI] [PubMed] [Google Scholar]
- 55.Makhatadze GI, Privalov PL. Protein interactions with urea and guanidinium chloride. A calorimetric study. J Mol Biol. 1992;226:491–505. doi: 10.1016/0022-2836(92)90963-k. [DOI] [PubMed] [Google Scholar]
- 56.Bennion BJ, Daggett V. The molecular basis for the chemical denaturation of proteins by urea. Proc Natl Acad Sci U S A. 2003;100:5142–5147. doi: 10.1073/pnas.0930122100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Almarza J, Rincon L, Bahsas A, Brito F. Molecular mechanism for the denaturation of proteins by urea. Biochemistry. 2009;48:7608–7613. doi: 10.1021/bi9007116. [DOI] [PubMed] [Google Scholar]
- 58.England JL, Haran G. Role of solvation effects in protein denaturation: from thermodynamics to single molecules and back. Annu Rev Phys Chem. 2011;62:257–277. doi: 10.1146/annurev-physchem-032210-103531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 60.Zagrovic B, Lipfert J, Sorin EJ, Millett IS, van Gunsteren WF, Doniach S, Pande VS. Unusual compactness of a polyproline type II structure. Proc Natl Acad Sci U S A. 2005;102:11698–11703. doi: 10.1073/pnas.0409693102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Makowska J, Rodziewicz-Motowidlo S, Baginska K, Vila JA, Liwo A, Chmurzynski L, Scheraga HA. Polyproline II conformation is one of many local conformational states and is not an overall conformation of unfolded peptides and proteins. Proc Natl Acad Sci U S A. 2006;103:1744–1749. doi: 10.1073/pnas.0510549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shi Z, Chen K, Liu Z, Kallenbach NR. Conformation of the backbone in unfolded proteins. Chem Rev. 2006;106:1877–1897. doi: 10.1021/cr040433a. [DOI] [PubMed] [Google Scholar]
- 63.Choi UB, McCann JJ, Weninger KR, Bowen ME. Beyond the random coil: stochastic conformational switching in intrinsically disordered proteins. Structure. 2011;19:566–576. doi: 10.1016/j.str.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 65.Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. Intrinsic disorder in transcription factors. Biochemistry. 2006;45:6873–6888. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Intrinsic disorder and functional proteomics. Biophys J. 2007;92:1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Galea CA, Wang Y, Sivakolundu SG, Kriwacki RW. Regulation of cell division by intrinsically unstructured proteins: intrinsic flexibility, modularity, and signaling conduits. Biochemistry. 2008;47:7598–7609. doi: 10.1021/bi8006803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Uversky VN, Oldfield CJ, Midic U, Xie H, Xue B, Vucetic S, Iakoucheva LM, Obradovic Z, Dunker AK. Unfoldomics of human diseases: linking protein intrinsic disorder with diseases. BMC Genomics. 2009;10(Suppl 1):S7. doi: 10.1186/1471-2164-10-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Uversky VN. Intrinsically disordered proteins from A to Z. Int J Biochem Cell Biol. 2011;43:1090–1103. doi: 10.1016/j.biocel.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 70.Uversky VN. Intrinsically disordered proteins may escape unwanted interactions via functional misfolding. Biochim Biophys Acta. 2011;1814:693–712. doi: 10.1016/j.bbapap.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 71.Uversky VN. Multitude of binding modes attainable by intrinsically disordered proteins: a portrait gallery of disorder-based complexes. Chem Soc Rev. 2011;40:1623–1634. doi: 10.1039/c0cs00057d. [DOI] [PubMed] [Google Scholar]
- 72.Follis AV, Galea CA, Kriwacki RW. Intrinsic protein flexibility in regulation of cell proliferation: advantages for signaling and opportunities for novel therapeutics. Adv Exp Med Biol. 2012;725:27–49. doi: 10.1007/978-1-4614-0659-4_3. [DOI] [PubMed] [Google Scholar]
- 73.Hsu WL, Oldfield C, Meng J, Huang F, Xue B, Uversky VN, Romero P, Dunker AK. Intrinsic protein disorder and protein-protein interactions. Pac Symp Biocomput. 2012:116–127. [PubMed] [Google Scholar]
- 74.Shoemaker BA, Portman JJ, Wolynes PG. Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc Natl Acad Sci U S A. 2000;97:8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN. Living with water stress: evolution of osmolyte systems. Science. 1982;217:1214–1222. doi: 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]
- 76.Baskakov I, Bolen DW. Forcing thermodynamically unfolded proteins to fold. J Biol Chem. 1998;273:4831–4834. doi: 10.1074/jbc.273.9.4831. [DOI] [PubMed] [Google Scholar]
- 77.Gursky O. Probing the conformation of a human apolipoprotein C-1 by amino acid substitutions and trimethylamine-N-oxide. Protein Sci. 1999;8:2055–2064. doi: 10.1110/ps.8.10.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Henkels CH, Kurz JC, Fierke CA, Oas TG. Linked folding and anion binding of the Bacillus subtilis ribonuclease P protein. Biochemistry. 2001;40:2777–2789. doi: 10.1021/bi002078y. [DOI] [PubMed] [Google Scholar]
- 79.Wu P, Bolen DW. Osmolyte-induced protein folding free energy changes. Proteins. 2006;63:290–296. doi: 10.1002/prot.20868. [DOI] [PubMed] [Google Scholar]
- 80.Lin TY, Timasheff SN. Why do some organisms use a urea-methylamine mixture as osmolyte? Thermodynamic compensation of urea and trimethylamine N-oxide interactions with protein. Biochemistry. 1994;33:12695–12701. doi: 10.1021/bi00208a021. [DOI] [PubMed] [Google Scholar]
- 81.Wang A, Bolen DW. A naturally occurring protective system in urea-rich cells: mechanism of osmolyte protection of proteins against urea denaturation. Biochemistry. 1997;36:9101–9108. doi: 10.1021/bi970247h. [DOI] [PubMed] [Google Scholar]
- 82.Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 83.Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 84.Yang ZR, Thomson R, McNeil P, Esnouf RM. RONN: the bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics. 2005;21:3369–3376. doi: 10.1093/bioinformatics/bti534. [DOI] [PubMed] [Google Scholar]