Abstract
In order to develop the most effective T helper type-1 (Th1) immunity, naïve CD4+ T cells must acquire the capacity to express IFN-γ while silencing T helper type-2 (Th2) cytokine-producing potential. An Il4 gene silencer has been described. However, it is not completely understood how the silencer works. In this study, we examined whether IFN-γ can suppress permissive chromatin remodeling of regulatory region of the Il4 gene. We demonstrate that IFN-γ suppressed H3K4 dimethylation at the intronic enhancer region of the Il4 gene. The IFN-γ-mediated suppression of permissive chromatin remodeling was IFN-γ receptor-, STAT1-, and T-bet-dependent. Our study reveals a novel mechanism of how Th1 cells silence the Il4 gene.
Keywords: IFN-γ, Th1, Silencing Il4 gene expression, Chromatin remodeling
1. Introduction
In order to develop the most effective T helper type-1 (Th1) immunity, naïve CD4+ T cells must acquire the capacity to express IFN-γwhile silencing T helper type-2 (Th2) cytokine-producing potential. Ansel et al. have identified such a silencer region within the Il4 gene [1]. Deletion of the Il4 gene silencer results in a Th2 immune response following Leishmania major infection, which would normally trigger a strong Th1 immune response [1].
It has demonstrated that chromatin structure plays a pivotal role in Th2 cytokine gene expression [2]. As naïve CD4+ T cells differentiate into Th2 cells, the histone molecules that surround the regulatory regions of Th2 cytokine gene loci undergo covalent modifications. These modifications lead to conformational changes in chromatin structure, which allow transcription factors to gain access to cytokine gene loci. More generally, chromatin modifications can include both permissive and repressive modifications. Permissive modifications render the gene locus more accessible to transcription factors, whereas repressive modifications make the gene locus less accessible. Permissive modifications include demethylation of CpG islands in the regulatory regions of a gene, acetylation of the ninth and fourteenth lysine residues of histone 3 (H3K9/14ac), and dimethylation of the fourth lysine residue of H3 (H3K4me2) [3-7]. Repressive modifications include methylation of the ninth lysine residue of H3 (H3K9me) [6], methylation of the twenty-seventh lysine residue of H3 (H3K27me) [8] and methylation of the thirty-sixth lysine residue of H3 (H3K36me) [2].
Transcriptional active regions of the Il4 gene have also been mapped by DNase sensitivity assay. These DNase hypersensitive sites found in the Il4 locus includes HSS 0 in the intergenic region between the Il13 and the Il4 gene, HSS 1and HSS 2 in the conserved noncoding sequence 1 (CNS-1) region, HS I in the Il4 gene promoter region, HS II and HS III in the second intronic enhancer region (IE), and HSV in 3′ flanking region of exon 4 [9-11]. Rad50 hypersensitive site 7, the seventhIl4 hypersensitive site, was found to be a DNA element that supports high-levels of IL-4 expression in Th2 cells [12, 13]. This region is known as the locus control region (LCR). The Il4 gene silencer has been mapped to DNase hypersensitive site IV within the second consensus non-coding sequence (CNS-2), which is located in the 3′ untranslated region of the Il4 gene. T-bet and Runx3 have been shown to bind to the Il4 gene silencer, yet chromatin in the silencer region exhibits a permissive configuration rather than a repressive configuration [1]. Thus, it remains incompletely understood how the Il4gene silencer keeps the Il4genetranscriptionally inactive.
We previously showed that IFN-γ is essential in maintaining the Th1 phenotype by actively suppressingtheIl4 gene transcription potential [14]. We further demonstrated that both STAT4 and T-bet are required for silencing the IL-4-producing potential in committed Th1 cells [15, 16]. However, it has not been determined if IFN-γcan suppress permissive remodeling of regulatory regions in order to silence the Il4 gene. Here, we demonstrate that IFN-γ can directly suppress permissive chromatin remodeling in the regulatory region of the Il4 gene.
2. Materials and methods
2.1. Animals and cell cultures
C57BL/6 mice were purchased from The Jackson Laboratory (Bar Harbor, ME). T-bet-deficient (Tbx21−/−) mice [17], IFNGR−/− mice [18], and STAT1−/− mice [19] were also purchased from The Jackson Laboratory and maintained in the animal facility of National Jewish Health (Denver, CO). Naïve CD4+ T cells were purified from the spleen and lymph nodes of these various strains of mice using the CD4+CD62L+T Cell Isolation Kit (Miltenyi Biotec Inc, CA). Purified naïve CD4+T cells (0.2 × 106) were stimulated with irradiated T cell-depleted APC (0.8 × 106) in 2 ml of complete RPMI medium supplemented with IL-2 (30 U/ml), anti-CD3 (2C11, 3 μg/ml), and anti-CD28 antibody (3 μg/ml) for 2 to 7 days as described in more detail previously [16]. Briefly, for Th1-inducing conditions, IFN-γ (20 ng/ml, BD-PharMingen, San Diego, CA), and anti-IL-4 antibody (11B11, 10 μg/ml) were used; and for Th2-inducing conditions, IL-4 (5 ng/ml, BD-PharMingen), anti-IL-12 antibody (10 μg/ml), and anti-IFN-γ antibody (XMG, 10 μg/ml) were added; and for neutralized conditions anti-IL-4 antibody (10 μg/ml), anti-IL-12 antibody (10 μg/ml), and anti-IFN-γ antibody (10 μg/ml) were added (used as a control group). Anti-IL-4 antibody, anti-IL-12 antibodies, and anti-IFN-γ antibody were prepared by our lab from hybridoma culture supernatants using HiTrap Affinity Protein G column according to manufacturer’s instructions (GE Healthcare Life Science, Piscataway, NJ). The qualities of these purified antibodies were tested using ELISA as well as Th1 and Th2 priming [14-16, 20].
All animal protocols were approved by the Institutional Animal Care and Users Committee of National Jewish Health (IACUC#AS2703-03-14).
2.2. Intracellular staining of cytokine expression
Intracellular staining of IL-4 and IFN-γ was carried as described previously [15]. Briefly, the primed cells were washed and stimulated with PMA (50 ng/ml) and ionomycin (1 μM) in the presence of 2 μM of monensin (Calbiochem, La Jolla, CA). After 6 hours of stimulation, cells were fixed with 4% paraformaldehyde and stained with APC-labeled anti-CD4, FITC-labeled anti-IFN-γ and/or PE-labeled anti-IL-4 (11B11) monoclonal antibody (BD-PharMingen). Samples were collected using FACScan or FACScalibur (Becton-Dickinson) and analyzed using FlowJo software (Tree Star Inc, Ashland, OR). All FACS data were analyzed using a CD4+ gate and dead cells were excluded from FACS data using a low Forward Scatter.
2.3. Chromatin immunoprecipitation (ChIP) assay
Chromatin immunoprecipitation assay was performed using Chromatin Immunoprecipitation Assay Kit (Upstate Biotechnology Lake Placid, NY) according to manufacturer’s protocol. Briefly, cells (1×106) were treated with 1% formaldehyde for 10 minutes at 37°C and lysed in 200 μl of SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris, pH 8.1), freshly supplemented with 0.2 mM phenylmethylsulfonyl fluoride (PMSF), 10 μg/ml of pepstatin A, 10 μg/ml leupeptin, 10 μg/ml aprotinin. Cell lysates were sonicated and diluted 10 fold in 2 ml of dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.1, 167 mM NaCl). Twenty microliter (10% input) of lysates was set aside for input measurements. Lysates were pre-cleaned by incubating with 50 μ l of protein A agarose beads (50% slurry). To immunoprecipitate dimethyl-histone H3 lysine 4, anti-dimethyl-histone H3 lysine 4 antibody (Upstate Biotechnology) was added to 100 μl of lysate and incubated at 4°C for 1 hour. As a control, normal rabbit IgG was added to the other 100 μl of lysate. The immunocomplex was captured by incubating 50 μl of protein A agarose beads at 4°C overnight. The beads were subsequently washed for 3-5 minutes by low salt immune complex wash buffer (0.1% SDS, 1%Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, 150 mM NaCl) once, followed by high salt immune complex wash suffer ( 0.1% SDS, 1%Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, 500 mM NaCl) once. The washed beads were further washed with LiCl immune complex wash buffer (0.25 M LiCl, 1% IGEPAL-CA630, 1% deoxycholic acid (sodium salt), 1 mM EDTA, 10 mM Tris, pH 8.1) once and TE Buffer, (10 mM Tris-HCl, 1 mM EDTA pH 8.0) twice. After wash, immunocomplex was eluted in 500 μl of elution buffer (1% SDS, 0.1 M NaHCO3) at room temperature. Then crosslinking was reversed by adding 20 μl of 5 M NaCl and incubating at 65°C for 4 hour. Proteins were digested by Proteinase K (10 μl of 0.5M EDTA, 20 μl of 1 M Tris-HCl pH6.5, 2 μl of 10 mg/ml Proteinase K). The eluted DNA was purified by phenol/chloroform extraction and precipitated by ethanol with 20 μg of glycogen at −70°C for 30 minutes. The precipitated DNA was dissolved in 50 μl of TE buffer. The amount of DNA precipitated by the antibody was determined by Real-Time PCR. Ten percent of input was also used for Real-Time measurement of IE or silencer. Ct values of diluted input were adjusted to 100% of input by subtracting 3.322 cycles (i.e., log2 of 10) from the Ct value of diluted input. Real-time PCR primer sequences were as follows: IE, forward: 5′-GGGTGTGAATAAGCCATATTG-3′, reverse: 5′-CCCAGCGTTTACATGAGC-3′ [5]; Silencer, forward: 5′-TGCCCACATGAAATACCAGC-3′; reverse: 5′-GCATACCTTCCCTGATTGGC-3′ [1].The DNA amount precipitated by anti-H3K4me2 antibody was calculated as % input using the following formula: % of input = 2ΔCt ×100, where ΔCt=Ct input-Ct IP [21].
2.4. Statistical analysis
All of the error bars in this report represent the standard deviation. The difference between two samples was analyzed using Student’s t test.
3. Results
3.1. IFN-γ suppresses permissive chromatin remodeling in the intronic enhancer region
Our previous work has demonstrated that IFN-γ played critical role in stabilizing the Th1 phenotype. In the absence of IFN-γ signaling, Th1 cells retained the capacity to produce IL-4, indicating that the Il4 gene is not silenced in Th1 cells in the absence of IFN-γ signaling [14, 20]. Although an Il4 silencer has been found, it remains to be determined whether IFN-γ can actively suppress permissive chromatin remodeling in addition to target the Il4 silencer through T-bet and Runx3.
Purified naïve CD4+ T cells did not express IFN-γ or IL-4 protein as measured by intracellular staining (Fig. 1). Th1- or Th2-inducing conditions used throughout this study led to highly polarized Th1 and Th2 cells, while neutralized conditions (with addition of antibodies to Th1- and Th2-inducing cytokines) did not significantly enhance IL-4-producing or IFN-γ-producing capacity (Fig. 1), indicating that the Th1 and Th2 polarization protocol we used in this study, which is identical to that used in many of our previous studies [14, 16, 20, 22], is a highly reproducible process.
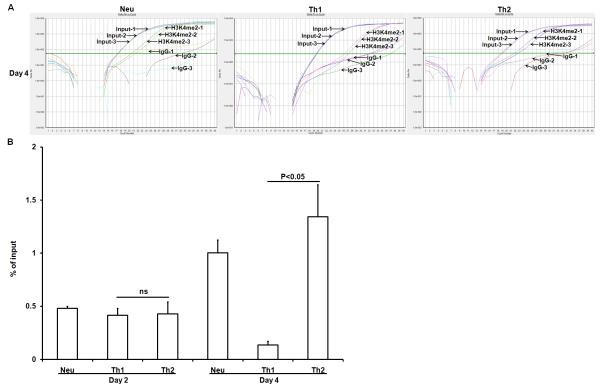
Fig. 1.
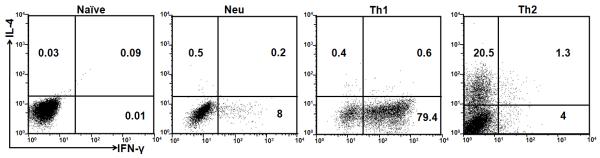
Th1- and Th2-inducing conditions lead to highly polarized IFN-γ expression and IL-4 expression, respectively. Naïve CD4+ T cells were not primed or primed under neutralized conditions [indicated as Neu, these conditions contained anti-IFN-γ antibody (10 μg/ml) and anti-IL-12 antibody (10 μg/ml)], Th1-inducing conditions [indicated as Th1), these conditions contained IFN-γ (20 ng/ml) plus anti-IL-4 antibody (10 μg/ml)] or Th2-inducing conditions [indicated as Th2, these conditions contained IL-4 (5 ng/ml) plus anti-IFN-γ antibody (10 μg/ml) and anti-IL-12 antibody (10 μg/ml)] for 7 days. IL-4 and IFN-γ protein expression was analyzed by intracellular staining. The numbers inside the FACS plots indicate the positive percentage within the gated CD4+ T cells.
We used a chromatin immunoprecipitation (ChIP) assay to examine the status of dimethylation modification of the fourth lysine residue of histone 3 (H3K4me2)in a regulatory region of the Il4 gene in CD4+ T cells that were primed under neutralized conditions, Th1-inducing or Th2-inducing conditions for either two or four days. Representative real-time PCR amplification plots of the immunoprecipitated IE region prepared from CD4+ T cells primed under neutralized conditions, Th1-inducing or Th2-inducing conditions were shown in Fig. 2A. We found that naïve CD4+ T cells showed very littleH3K4me2 modification in the intronic enhancer region (IE) of the Il4 gene. Two days after culturing under Th2-inducing conditions, the cultured CD4+ T cells displayed elevated levels of H3K4me2 modification in the IE region. Four days after Th2-priming, H3K4me2 modification was dramatically increased. CD4+ T cells that were primed under neutralized conditions also showed increased levels of H3K4me2 modification albeit to less degree than CD4+ T cells that were primed under Th2-inducing conditions. We interpret that the neutralized condition-induced H3K4me2 modification was mediated by TCR activation. We observed that in the presence of IFN-γ, H3K4me2 modification in the IE region was suppressed (Fig. 2B). We also observed a similar pattern for the acetylation of the H3K9/14 residues modification in the IE region of the Il4gene (data not shown). These data suggest that suppression of H3K4me2 modification in the IE region was actively mediated by IFN-γ.
Fig. 2.
IFN-γ suppresses H3K4me2 modification in the IE region of the Il4 gene. Naïve CD4+ T cells from C57BL/6 mice were primed under neutralized conditions, Th1-inducing conditions or Th2-inducing conditions for either 2 or 4 days. The resultant cells were harvested, cross-linked, and sonicated. H3K4me2 modification was measured by immunoprecipitation with anti-dimethylated lysine 4 residue antibody. The amount of immunoprecipitated IE fragment was measured by Real-Time PCR and shown as the percentage of input DNA. Values represent mean ± SD of triplicates. Ns= not significant. Data from one experiment represent two independent experiments with similar results.
3.2. IFN-γ depends on STAT1 and T-bet to suppress permissive chromatin remodeling in the intronic enhancer region
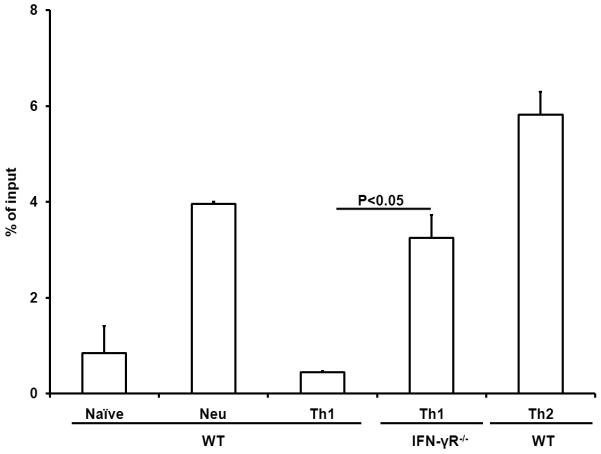
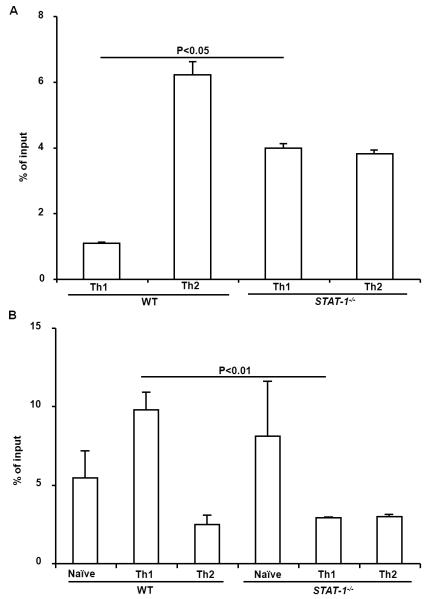
To analyze the pathway involved in IFN-γ-induced suppression of permissive chromatin remodeling in the IE region of the Il4 gene, we first tested the H3K4me2 modification in IFN-γ receptor deficient CD4+ T cells and found that the H3K4me2 modification in the IE region of the Il4 gene of IFNR−/− CD4+ that were primed under Th1 conditions for 7 days was greatly increased, demonstrating that IFN-γ directly suppresses H3K4me2 modification (Fig. 3). Representative real-time PCR amplification plots of the IE region were shown in Fig. S1. IFN-γ has been shown to exert its biological functions in either a STAT1-dependent or STAT1-independent manner. We found that STAT1−/− CD4+ T cells regardless they were under Th1 or Th2-inducing conditions showed markedly increased levels of H3K4me2 modification in the IE region (Fig. 4A). However, elevated levels of H3K4me2 modification were not found in the silencer region of STAT1−/− CD4+ T cells primed under Th1 or Th2-inducing conditions. On the contrary, the H3K4me2 modification was reduced significantly in the silencer region of STAT1−/− CD4+ T cells that were primed under Th1-inducing conditions compared to that of WT Th1 cells (Fig. 4B). Together, these data demonstrate that IFN-γ depended on STAT1 to suppress the H3K4me2 modification.
Fig. 3.
IFN-γ directly suppresses H3K4me2 modification. Naïve CD4+ T cells from C57BL/6 and IFNγR−/− mice were cultured for 7 days under the conditions indicated. The resultant cells were harvested, cross-linked, and sonicated. H3K4me2 modification was measured by immunoprecipitation with anti-dimethylated lysine 4 residue antibody. The amount of immunoprecipitated IE fragment was measured by Real-Time PCR and shown as the percentage of input DNA. Values represent mean ± SD of duplicates. Data from one experiment represent two independent experiments with similar finding.
Fig. 4.
IFN-γ depends on STAT1 to suppress H3K4me2 modification in the IE region but not the silencer region. (A) Naïve CD4+ T cells from C57BL/6 and STAT1−/− mice were cultured for 7 days under the conditions indicated. The resultant cells were harvested, cross-linked, and sonicated. H3K4me2 modification was measured by immunoprecipitation with anti-dimethylated lysine 4 residue antibody. The amount of immunoprecipitated IE fragment was measured by Real-Time PCR and shown as the percentage of input DNA. (B) The amount of immunoprecipitated silencer fragment was measured Real-Time PCR. Values represent mean ± SD of triplicates. Data from one experiment represent two independent experiments with similar results.
Our previous work showed that T-bet was required for maintaining the silencing state of the Il4 gene in committed Th1 cells; in the absence of T-bet, CD4+ T cells transcribed the Il4 gene even they were primed under Th1-inducing conditions [15]. However, it is not clear how T-bet maintains the silencing state of the Il4 gene. To determine whether T-bet mediates the IFN-γ-induced suppression of H3K4me2 modification, we examined H3K4me2 modification in the IE region of the Il4 gene in both WT and T-bet−/−CD4+ T cells. We showed that IFN-γ-induced suppression of H3K4me2 modification was abolished in the absence of T-bet (Fig. 5). The amounts of IE fragment immunoprecipitated by isotype control antibody ranged from 0.005% to 0.03% of input (data not shown because values were too low to be reflected in the figures). Thus, we concluded that IFN-γ depended on T-bet to suppress the H3K4me2 modification in the IE region of the Il4 gene.
Fig. 5.
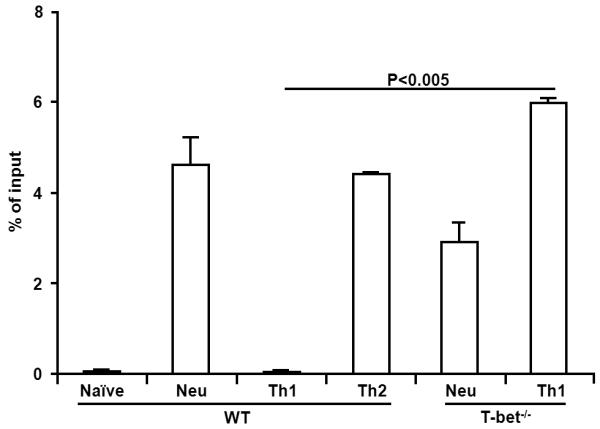
IFN-γ depends on T-bet to suppress H3K4me2 modification. Naïve CD4+ T cells from C57BL/6 and Tbx21−/− mice were cultured for 7 days under the conditions indicated. The amount of immunoprecipitated IE fragment was measured. Values represent mean ± SD of duplicates. Data from one experiment represent two independent experiments with similar results.
4. Discussion
In this study, we tested if IFN-γ can suppress permissive chromatin remodeling of regulatory regions other than the silencer region in order to silence the Il4 gene. Based on the experimental results obtained from primary cultures, we conclude that IFN-γ can actively suppress permissive chromatin remodeling in the IE region of the Il4 gene. Th1 and Th2 cell lines or clones, as the result of repeated stimulation under Th1 or Th2-inducing conditions, exhibit more stable phenotype [23, 24][25]. Measuring H3K4me2 modification in the IE region of Th1 cell lines or clones could further strengthen our conclusion. Furthermore, it would be interesting to test whether IFN-γ suppresses H3K4me2 modification in the regulatory regions of the Il13 genes in Th1 cells.
Th1-inducing conditions has been shown to induce elevated levels of H3K4me2 modification to histone molecules in the regulatory regions of the Ifng gene [26]. Authors of the paper showed that during the early phase of Th1/2 priming, levels of H3K4me2 modification in the regulatory regions of the Ifng gene were elevated in both Th1 and Th2 cells. After 28 days, H3K4me2 modification in the regulatory regions of the Ifng gene in Th2 cells disappeared, suggesting that Th2-inducing conditions act slowly to suppress H3K4me2 modification to histone molecules in the regulatory regions of the Ifng gene [26]. On the other hand, we observed a much faster action of IFN-γ on suppressing H3K4me2 modification in the IE region of the Il4 gene in Th1 cells. Consistently, we note that the fast IFN-γ action on H3K4me2 modification was IFNGR1-, STAT1- and T-bet-dependent. Our investigation reveals a novel mechanism by which IFN-γ can suppress Il4 gene transcription. These new findings might fill the gap in our understanding of how the silencers can interact with other regulatory regions of the Il4 gene to achieve silencing of the Il4 gene. For example, it is likely that a silencer is needed for recruiting enzymes that can induce repressive modifications into other distal regulatory regions of the Il4 gene, such as IE. Binding of T-bet and Runx3 to the silencer region could provide a contact point for long-distance interactions between the silencer region and other distal regulatory regions of the Il4 gene. This long-distance interaction is a prerequisite for recruiting repressive remodeling enzymes, such as HDAC1, to the IE region. Long-distance interaction has been documented for the Il4 gene. Spilianakis et al. showed that binding of GATA3 and STAT6 to the LCR resulted in a long-distance interaction of LCR with the Il4 promoter, which is located 30 kb away from the LCR [12]. Further studies using the Il4 silencer-deficient mice are needed to verify whether long-distance interaction between the silencer and the IE region can indeed induce reduction in permissive remodeling.
Supplementary Material
Highlights.
We demonstrate that IFN-γ suppressed H3K4 dimethylation at the intronic enhancer region of the Il4 gene.
The IFN-γ-mediated suppression of permissive chromatin remodeling was IFN-γ-R-, STAT1-, and T-bet-dependent.
Our study reveals a novel mechanism of how Th1 cells silence the Il4 gene.
Acknowledgements
This work was supported by the National Institutes of Health (RO1 AI048568 and RO1 AI059170).
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: J.N. and H.H. are involved in experimental designs, performing experiment, data analysis and manuscript preparation; Y. L., Y.Z. and Z. H. are involved in experimental designs, performing experiments, revising the manuscript.
Financial Disclosure
Authors have no financial conflict of interest.
References
- [1].Ansel KM, Greenwald RJ, Agarwal S, Bassing CH, Monticelli S, Interlandi J, et al. Deletion of a conserved Il4 silencer impairs T helper type 1-mediated immunity. Nat Immunol. 2004;5:1251–9. doi: 10.1038/ni1135. [DOI] [PubMed] [Google Scholar]
- [2].Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–56. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- [3].Avni O, Lee D, Macian F, Szabo SJ, Glimcher LH, Rao A. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat Immunol. 2002;3:643–51. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- [4].Yamashita M, Ukai-Tadenuma M, Kimura M, Omori M, Inami M, Taniguchi M, et al. Identification of a conserved GATA3 response element upstream proximal from the interleukin-13 gene locus. J Biol Chem. 2002;277:42399–408. doi: 10.1074/jbc.M205876200. [DOI] [PubMed] [Google Scholar]
- [5].Baguet A, Bix M. Chromatin landscape dynamics of the Il4-Il13 locus during T helper 1 and 2 development. Proc Natl Acad Sci U S A. 2004;101:11410–5. doi: 10.1073/pnas.0403334101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Grogan JL, Wang ZE, Stanley S, Harmon B, Loots GG, Rubin EM, et al. Basal chromatin modification at the IL-4 gene in helper T cells. J Immunol. 2003;171:6672–9. doi: 10.4049/jimmunol.171.12.6672. [DOI] [PubMed] [Google Scholar]
- [7].Fields PE, Kim ST, Flavell RA. Cutting edge: changes in histone acetylation at the IL-4 and IFN-gamma loci accompany Th1/Th2 differentiation. J Immunol. 2002;169:647–50. doi: 10.4049/jimmunol.169.2.647. [DOI] [PubMed] [Google Scholar]
- [8].Koyanagi M, Baguet A, Martens J, Margueron R, Jenuwein T, Bix M. EZH2 and histone 3 trimethyl lysine 27 associated with Il4 and Il13 gene silencing in Th1 cells. J Biol Chem. 2005;280:31470–7. doi: 10.1074/jbc.M504766200. [DOI] [PubMed] [Google Scholar]
- [9].Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–75. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- [10].Takemoto N, Koyano-Nakagawa N, Yokota T, Arai N, Miyatake S, Arai K. Th2-specific DNase I-hypersensitive sites in the murine IL-13 and IL-4 intergenic region. Int Immunol. 1998;10:1981–5. doi: 10.1093/intimm/10.12.1981. [DOI] [PubMed] [Google Scholar]
- [11].Lee DU, Agarwal S, Rao A. Th2 lineage commitment and efficient IL-4 production involves extended demethylation of the IL-4 gene. Immunity. 2002;16:649–60. doi: 10.1016/s1074-7613(02)00314-x. [DOI] [PubMed] [Google Scholar]
- [12].Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol. 2004;5:1017–27. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- [13].Lee GR, Spilianakis CG, Flavell RA. Hypersensitive site 7 of the TH2 locus control region is essential for expressing TH2 cytokine genes and for long-range intrachromosomal interactions. Nat Immunol. 2005;6:42–8. doi: 10.1038/ni1148. [DOI] [PubMed] [Google Scholar]
- [14].Zhang Y, Apilado R, Coleman J, Ben-Sasson S, Tsang S, Hu-Li J, et al. Interferon gamma stabilizes the T helper cell type 1 phenotype. The Journal of experimental medicine. 2001;194:165–72. doi: 10.1084/jem.194.2.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhuang Y, Huang Z, Nishida J, Brown M, Zhang L, Huang H. A continuous T-bet expression is required to silence the interleukin-4-producing potential in T helper type 1 cells. Immunology. 2009;128:34–42. doi: 10.1111/j.1365-2567.2009.03049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhuang Y, Huang Z, Nishida J, Zhang L, Huang H. Signaling pathways that lead to the silencing of the interleukin-4-producing potential in Th1 cells. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2009;29:399–406. doi: 10.1089/jir.2008.0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science. 2002;295:338–42. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- [18].Huang S, Hendriks W, Althage A, Hemmi S, Bluethmann H, Kamijo R, et al. Immune response in mice that lack the interferon-gamma receptor. Science. 1993;259:1742–5. doi: 10.1126/science.8456301. [DOI] [PubMed] [Google Scholar]
- [19].Meraz MA, White JM, Sheehan KC, Bach EA, Rodig SJ, Dighe AS, et al. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–42. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- [20].Huang Z, Xin J, Coleman J, Huang H. IFN-gamma suppresses STAT6 phosphorylation by inhibiting its recruitment to the IL-4 receptor. J Immunol. 2005;174:1332–7. doi: 10.4049/jimmunol.174.3.1332. [DOI] [PubMed] [Google Scholar]
- [21].Allan RS, Zueva E, Cammas F, Schreiber HA, Masson V, Belz GT, et al. An epigenetic silencing pathway controlling T helper 2 cell lineage commitment. Nature. 2012;487:249–53. doi: 10.1038/nature11173. [DOI] [PubMed] [Google Scholar]
- [22].Huang H, Paul WE. Impaired interleukin 4 signaling in T helper type 1 cells. The Journal of experimental medicine. 1998;187:1305–13. doi: 10.1084/jem.187.8.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hu-Li J, Huang H, Ryan J, Paul WE. In differentiated CD4+ T cells, interleukin 4 production is cytokine-autonomous, whereas interferon gamma production is cytokine-dependent. Proc Natl Acad Sci U S A. 1997;94:3189–94. doi: 10.1073/pnas.94.7.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Messi M, Giacchetto I, Nagata K, Lanzavecchia A, Natoli G, Sallusto F. Memory and flexibility of cytokine gene expression as separable properties of human T(H)1 and T(H)2 lymphocytes. Nat Immunol. 2003;4:78–86. doi: 10.1038/ni872. [DOI] [PubMed] [Google Scholar]
- [25].Murphy E, Shibuya K, Hosken N, Openshaw P, Maino V, Davis K, et al. Reversibility of T helper 1 and 2 populations is lost after long-term stimulation. The Journal of experimental medicine. 1996;183:901–13. doi: 10.1084/jem.183.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hamalainen-Laanaya HK, Kobie JJ, Chang C, Zeng WP. Temporal and spatial changes of histone 3 K4 dimethylation at the IFN-gamma gene during Th1 and Th2 cell differentiation. J Immunol. 2007;179:6410–5. doi: 10.4049/jimmunol.179.10.6410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.