Abstract
Our intestine is host to a large microbial community (microbiota) that educates the immune system and confers niche protection. Profiling of the gut-associated microbial community reveals a dominance of obligate anaerobic bacteria in healthy individuals. However, intestinal inflammation is associated with a disturbance of the microbiota—known as dysbiosis—that often includes an increased prevalence of facultative anaerobic bacteria. This group contains potentially harmful bacterial species, the bloom of which can further exacerbate inflammation. Here, we review the mechanisms that generate changes in the microbial community structure during inflammation. One emerging concept is that electron acceptors generated as by-products of the host inflammatory response feed facultative anaerobic bacteria selectively, thereby increasing their prevalence within the community. This new paradigm has broad implications for understanding dysbiosis during gut inflammation and identifies potential targets for intervention strategies.
Keywords: intestinal inflammation, dysbiosis, anaerobic respiration, Enterobacteriaceae
See Glossary for abbreviations used in this article.
Glossary.
- ABC
ATP-binding cassette
- DMSO
dimethyl S-oxide
- DSS
dextran sulphate sodium
- DUOX2
dual function NAD(P)H oxidase 2
- ExbB/D
excretion of enterobactin gene B/D
- IFN-γ
interferon gamma
- IL-22
interleukin 22
- iNOS
inducible nitric oxide synthase
- LamB
outer membrane receptor for phage λ
- MaLE/F/G/K/S
maltose utilization genes E/F/G/K/S
- MoaA
molybdopterin synthesis protein A
- MPO
myeloperoxidase
- NADPH
nicotinamide adenine dinucleotide phosphate
- NF-κB
nuclear factor kappa B
- SOD
superoxide dismutase
- SopE
Salmonella outer protein E
- Sus
starch utilization system
- TonB
phage T1 resistance locus B
Introduction
Over 90% of the cells in the human body are microbes, most of which reside in bacterial communities—known collectively as microbiota—that inhabit the large intestine. Advances in high-throughput microbiota sequencing provide a powerful tool for profiling the previously hidden microbial diversity in the gut. For example, such metagenomic analysis shows that the large intestine is host to a diverse bacterial community the structure of which, at the phylum level, is maintained through unknown mechanisms. The bacterial species dominating the microbiota in the large bowel are strict anaerobes, which lack the ability to respire oxygen and rely on fermentation of complex polysaccharides for growth [1]. Towering above all others are obligate anaerobic bacteria belonging to the phyla Bacteroidetes (class Bacteroidia) and Firmicutes (class Clostridia; [2]). This dominance of Bacteroidia and Clostridia is a conserved feature of the large intestine microbiota of both humans and mice [2,3].
However, intestinal inflammation can lead to a microbial imbalance—known as dysbiosis—which is characterized by a marked decrease in the representation of obligate anaerobic bacteria and an increased relative abundance of facultative anaerobic bacteria. For example, acute intestinal inflammation triggered by pathogenic Enterobacteriaceae (class Gammaproteobacteria, phylum Proteobacteria)—such as Salmonella enterica or Citrobacter rodentium—is accompanied by changes in the bacterial community structure that are marked by an outgrowth of the respective facultative anaerobic pathogen [4,5,6]. Similarly, a reduction of strictly anaerobic members of the classes Bacteroidia and Clostridia, and a concomitant increase in facultative anaerobic commensal bacteria belonging to the class Gammaproteobacteria (most commonly members of Enterobacteriaceae) or to the class Bacilli (phylum Firmicutes), is seen in individuals with inflammatory bowel disease [7,8,9,10,11,12,13]. Likewise, a marked decrease in Bacteroidia and Clostridia and an increased relative abundance of Enterobacteriaceae is also observed in mice when colitis is induced chemically [6] or through genetically engineered immune defects [14]. Although metagenomics provides a powerful lens for viewing these changes in the microbial community structure during conditions of intestinal inflammation, the mechanisms responsible have remained an enigma. Here, we review advances in the understanding of the fundamental principles that govern the phylum-level changes in the structure of host-associated microbial communities in the inflamed gut.
Enterobacteriaceae in the healthy gut
The lumen of the distal gut is a fairly anaerobic environment. The traces of oxygen present in this habitat are readily consumed by facultative anaerobic bacteria—such as Enterobacteriaceae—that constitute a small fraction (approximately 0.1%) of the microbiota [2]. The amount of available oxygen seems to limit the growth of Enterobacteriaceae in this environment, because elevated oxygen levels increase their relative abundance. For example, the ileostomy of small bowel transplant patients provides a portal that allows oxygen to reach the otherwise anaerobic distal ileum. An increase in the relative abundance of Enterobacteriaceae is observed in close proximity to the ileostomy and the microbial community returns to its normal composition after its surgical closure [15]. Thus, once the available oxygen is consumed, Enterobacteriaceae are apparently poorly equipped to compete with obligate anaerobic bacteria for high-energy nutrients to support their growth by fermentation.
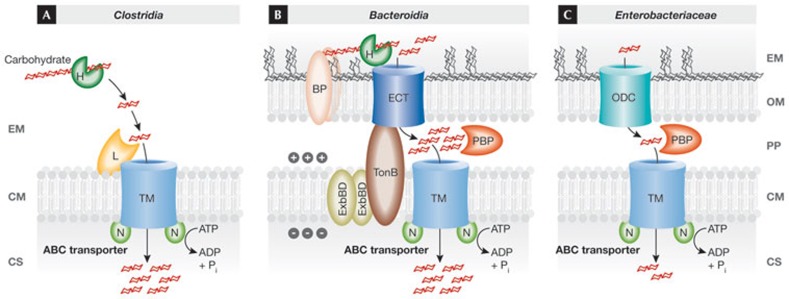
The dense bacterial communities that inhabit the distal gut compete fiercely for a limited quantity of diet-derived or host mucus-derived carbohydrate available for fermentation [16,17]. Changes in the diet can alter the microbial community structure at the species level, but the dominance persists of obligate anaerobic Clostridia and Bacteroidia over Enterobacteriaceae [18,19,20,21,22]. A comparison of the strategies by which Clostridia, Bacteroidia and Enterobacteriaceae acquire fermentable nutrients illustrates why the latter might be at a disadvantage during anaerobic growth on a limited quantity of carbohydrate (Fig 1).
Figure 1.
Carbohydrate acquisition strategies of Clostridia, Bacteroidia and Enterobacteriaceae. The transport of carbohydrate across the cytoplasmic membrane (CM) is typically mediated by ATP-binding cassette (ABC) transporters. These contain either periplasmic binding proteins (PBPs), present in Gram-negative bacteria (shown in panels B and C), or a membrane-anchored lipoprotein involved in binding (L), present in Gram-positive bacteria (shown in panel A), a transmembrane transporter (TM) and membrane-associated nucleotide-binding proteins (Ns) involved in energizing transport by mediating hydrolysis of ATP. Bacteroidia use outer membrane proteins (BPs) to bind carbohydrate, which is degraded into oligosaccharides using glycoside hydrolases (Hs). The resulting oligosaccharides are transported by an energy-coupled outer membrane transporter (ECT) in a process energized through the proton motive force generated by the TonB ExbBD proteins. Enterobacteriaceae rely on outer membrane diffusion channels (ODCs) for passive transport of oligosaccharides across the outer membrane (OM) along a concentration gradient. CS, cytosol; EM, extracellular milieu; ExbBD, excretion of enterobactin gene B/D; PP, periplasmic space; TonB, phage T1 resistance locus B.
The Gram-positive Clostridia lack an outer membrane and use glycoside hydrolases to degrade complex carbohydrates. The oligosaccharides generated through this process are transported actively against a concentration gradient across the cytoplasmic membrane by using ABC transporters (Fig 1A; [16]).
The starch utilization system encoded by the sus gene cluster of Bacteroides thetaiotaomicron is a well-studied glycan acquisition strategy conserved among the Bacteroidia. The outer membrane proteins SusD, SusE and SusF bind starch to the bacterial surface [23]. The outer membrane protein SusG degrades starch into malto-oligosaccharides [24], which are subsequently actively transported against the concentration gradient by the energy-coupled outer membrane import protein SusC [25]. The energy required for transport through this class of outer membrane import proteins is provided by the proton motive force of the cytoplasmic membrane, which is transmitted to the outer membrane through the TonB, ExbB and ExbD proteins (reviewed in [26]). Finally, periplasmic malto-oligosaccharides are further degraded by SusA to glucose, which is actively transported into the cytosol to support growth by fermentation [27]. Sequencing of the B. thetaiotaomicron genome revealed the presence of 88 gene clusters related to the sus system, suggesting that this general strategy is used for degradation of a multitude of carbohydrate sources in the intestinal lumen (Fig 1B; [28]).
Escherichia coli is a well-studied member of the Enterobacteriaceae commonly present in the large bowel. Malto-oligosaccharides and maltose cross the bacterial outer membrane passively along a concentration gradient through an outer membrane diffusion channel (LamB; [29]). In the periplasm, malto-oligosaccharides are degraded to maltose by the α-amylase MalS [30,31]. Maltose is bound by the periplasmic binding protein MalE [32] and actively transported into the cytosol by an ABC transporter formed by the MalF, MalG and MalK proteins [33,34,35,36,37,38]. Passive transport through outer membrane diffusion channels is a conserved feature of carbohydrate acquisition within the Enterobacteriaceae (Fig 1C). The only energy-coupled outer membrane import proteins present in Enterobacteriaceae transport low-molecular-weight iron chelators—known as siderophores—and vitamin B12 [26].
The above examples illustrate the main difference between the carbohydrate acquisition strategies of Clostridia, Bacteroidia and Enterobacteriaceae. Both Clostridia and Bacteroidia use glycoside hydrolases to degrade complex carbohydrates, make use of binding proteins to concentrate carbohydrate at their surface and then use an active transport system—that is, an ABC transporter or an energy-coupled outer membrane import protein—to import substrates against a concentration gradient across the first diffusion barrier, be it across the cytoplasmic membrane in Clostridia or across the outer membrane in Bacteroidia (Fig 1A,B). By contrast, a paucity of secreted glycoside hydrolases make Enterobacteriaceae ill-equipped to degrade complex carbohydrate. Instead, Enterobacteriaceae rely on the presence of oligosaccharides that are transported passively across the first diffusion barrier—the outer membrane—through diffusion channels (Fig 1C). Using more effective nutrient-uptake mechanisms is predicted to confer a competitive growth advantage on Clostridia and Bacteroidia over Enterobacteriaceae during anaerobic growth on a limited quantity of carbohydrate. This competitive growth advantage might at least in part explain the dominance of obligate anaerobic Clostridia and Bacteroidia over the facultative anaerobic Enterobacteriaceae in the healthy gut.
From metagenomics to mechanisms
One important consequence of intestinal inflammation is diarrhoea, the flushing action of which limits the availability of fermentable high-energy carbon sources to host mucus-derived carbohydrate. The more effective nutrient-uptake mechanisms used by Clostridia and Bacteroidia are predicted to confer an advantage in this environment. However, intestinal inflammation is associated with phylum-level changes in the microbiota composition, characterized by an increase in facultative anaerobic bacteria [39]. How can inflammation diminish the competitive anaerobic growth advantage of obligate anaerobic bacteria over Enterobacteriaceae?
To answer this question, it is important to understand how inflammation alters the nutritional environment in the distal gut. Intestinal inflammation is accompanied by the release of antimicrobials, a host defence mechanism designed to eradicate microbes from tissue or from close vicinity to the epithelium. Some antimicrobials withhold nutrients by interfering with the microbial acquisition of trace elements, such as iron or zinc [40,41]. For example, on stimulation with IL-22, epithelial cells release the antimicrobial lipocalin 2 into the intestinal lumen [41]. Lipocalin 2 binds to enterobactin, thereby preventing bacteria from using this siderophore for iron acquisition [42,43,44]. The release of lipocalin 2 can provide a selective advantage for enteric pathogens that have specific lipocalin 2 resistance mechanisms [41]. However, many commensal Enterobacteriaceae are susceptible to lipocalin 2, suggesting that its release is probably not responsible for the phylum-level changes in microbial communities associated with gut inflammation.
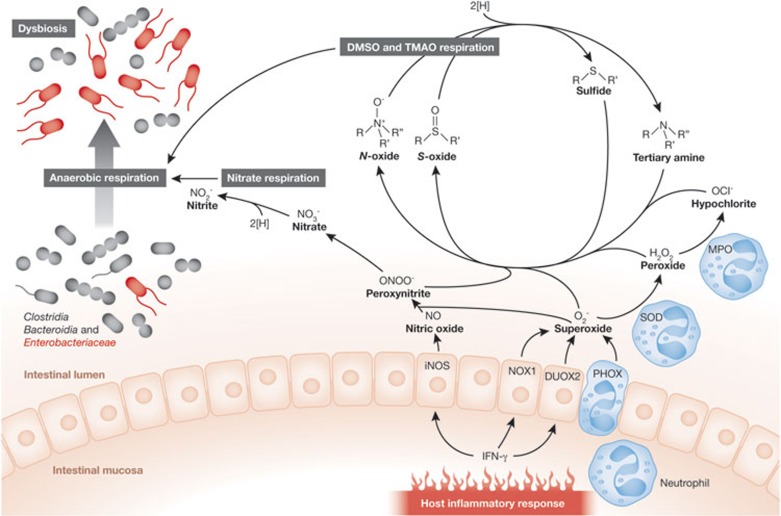
A second group of antimicrobials produced during inflammation are reactive oxygen species (ROS) and reactive nitrogen species (RNS). On stimulation with pro-inflammatory cytokines, such as IFN-γ, the intestinal epithelium can produce hydrogen peroxide (H2O2) by activating DUOX2 (Fig 2; [45]). In addition, IFN-γ induces expression of the NADPH oxidase 1 (Nox1) gene, encoding a second NADPH oxidase of epithelial cells that generates superoxide radicals (O2−; [46]). Severe intestinal inflammation can be accompanied by transmigration of neutrophils into the intestinal lumen and subsequent generation of superoxide radicals by the phagocyte NADPH oxidase (PHOX). The generation of superoxide radicals by phagocytes is essential for host defence, as illustrated by the existence of recurrent bacterial infections in individuals with chronic granulomatous disease, an illness caused by PHOX-deficiency [47,48,49]. Neutrophils also express SOD and MPO, which convert superoxide radicals to hydrogen peroxide and hypochlorite (OCl−). Furthermore, stimulation with IFN-γ can induce expression of the Nos2 gene in the intestinal epithelium [50]. The enzyme encoded by Nos2, iNOS, catalyses the production of nitric oxide from L-arginine [51]. Phagocytes recruited to the gut mucosa during inflammation are another cellular source of iNOS [52]. Elevated levels of iNOS during inflammation can alter the luminal environment of the large bowel, as indicated by raised nitric oxide concentrations in colonic luminal gas of individuals with inflammatory bowel disease [53,54,55]. Finally, nitric oxide can react with a superoxide radical, giving rise to peroxynitrite (ONOO−), a potent bactericidal RNS [56,57].
Figure 2.
Phylum-level changes in the microbiota composition on intestinal inflammation. Inflammatory infiltrates—such as neutrophils, and cytokines—such as IFN-γ, are a source of or induce the expression of enzymes (iNOS, NOX1, DUOX2, PHOX, SOD and MPO) that generate antimicrobial radicals, such as superoxide, peroxide, hypochloride, nitric oxide and peroxynitrite. In the intestinal lumen, these radicals react to form harmless oxidation products—such as nitrate, N-oxides or S-oxides—that serve as electron acceptors. These support the growth of facultative anaerobic bacteria by anaerobic respiration. The resulting outgrowth of facultative anaerobic bacteria gives rise to phylum-level changes in the microbiota composition, including an increased relative abundance of Enterobacteriaceae and a marked decrease in obligate anaerobic Clostridia and Bacteroidia. DMSO, dimethyl S-oxide; DUOX2, dual function NAD(P)H oxidase 2; IFN-γ, interferon gamma; iNOS, inducible nitric oxide synthase; MPO, myeloperoxidase; NOX1, NADPH oxidase 1; PHOX, phagocyte NADPH oxidase; SOD, superoxide dismutase; TMAO, trimethylamine N-oxide.
Although the production of RNS and ROS creates a hostile environment in close proximity to the mucosal surface, the generation of these radicals has important side effects. As peroxynitrite, superoxide, hydrogen peroxide and hypochlorite diffuse away from the epithelium, they quickly react with organic sulphides and tertiary amines present in the intestinal lumen to form the respective S-oxides (R2-SO) and N-oxides (R3-N+-O−; Fig 2; [58,59]). For example, when dietary contents have been flushed out by diarrhoea, enterocytes released from the tips of villi are the main source of membrane lipids, such as phosphatidylcholine and sphingomyelin, in the intestinal lumen. A nutrient derived from phosphatidylcholine or sphingomyelin is choline. Choline is degraded by the gut microbiota to trimethylamine (TMA; [60])—a compound that can be oxidized by peroxynitrite, superoxide, hydrogen peroxide and hypochlorite to trimethylamine N-oxide (TMAO; [58,59]). Alternatively, peroxynitrite can be converted to nitrate (NO3−) in a reaction catalysed by carbon dioxide [61]. As a result, nitrate production in the gut lumen is a by-product of chemically induced colitis [62]. Feeding mice the iNOS-inhibitor aminoguanidine hydrochloride prevents nitrate production during colitis [63]. iNOS is responsible for the production of nitrate during inflammation, suggesting that nitrate is host-derived rather than originating from the diet. Ultimately, these processes convert bactericidal RNS and ROS into non-toxic products—that is, S-oxides, N-oxides and nitrate—the presence of which causes a marked change in the growth conditions encountered in the distal gut.
The lumen of the large bowel is largely devoid of exogenous electron acceptors that would support the growth of bacteria by anaerobic respiration. As a result, fermentation of carbohydrates is the main strategy by which microbial communities in the healthy large intestine support their anaerobic growth. However, the generation of S-oxides, N-oxides and nitrate as by-products of the host inflammatory response opens a new alternative for facultative anaerobic microbes to grow in this environment (Fig 2). Enterobacteriaceae can use S-oxides, N-oxides and nitrate as terminal electron acceptors for anaerobic respiration by expressing DMSO, TMAO and nitrate reductases, respectively [64]. By contrast, Clostridia and Bacteroidia have a primitive electron transport chain and lack the terminal oxidoreductases needed to use the exogenous electron acceptors generated during inflammation [17].
E. coli encodes three nitrate reductases in the narGHJI, narZYWV and napFDAGHBC operons, two DMSO reductases in the dmsABC and ynfFGH operons, and three TMAO reductases in the torCAD, torYZ and yedYZ operons [65]. Nitrate, DMSO and TMAO reductases, as well as two formate dehydrogenases (FdnG and FdoG), contain molybdenum as a crucial catalyst for electron transfer reactions. The functions of FdnG and FdoG are linked to respiration, because they couple respiratory electron acceptors to the electron donor formate, a fermentation end product present in the large intestine [66,67]. Formate dehydrogenases and respiratory reductases contain molybdenum within a molybdopterin cofactor [68]. MoaA catalyses the first reaction in the biosynthesis of this molybdopterin cofactor [69]. Therefore, an E. coli moaA mutant is deficient for several respiratory pathways, including nitrate, DMSO and TMAO respiration. When mice with DSS-induced colitis are challenged with various E. coli strains, the E. coli wild-type is recovered from the large intestine at a 100-fold higher level than an E. coli moaA mutant. By contrast, wild-type and moaA mutants are recovered in similar numbers from the non-inflamed intestine of mock-treated control mice [63]. These results suggest that the presence of exogenous electron acceptors in the inflamed gut confers a substantial fitness advantage to E. coli—and probably to other commensal Enterobacteriaceae—by supporting their growth through anaerobic respiration. This fitness advantage contributes to a bloom of commensal Enterobacteriaceae, thereby giving rise to the phylum-level changes in the microbiota composition that accompany intestinal inflammation (Fig 2). In other words, one of the mechanisms responsible for dysbiosis in the inflamed gut is that the host response selectively feeds facultative anaerobic bacteria.
Virulence factors change what is on the menu
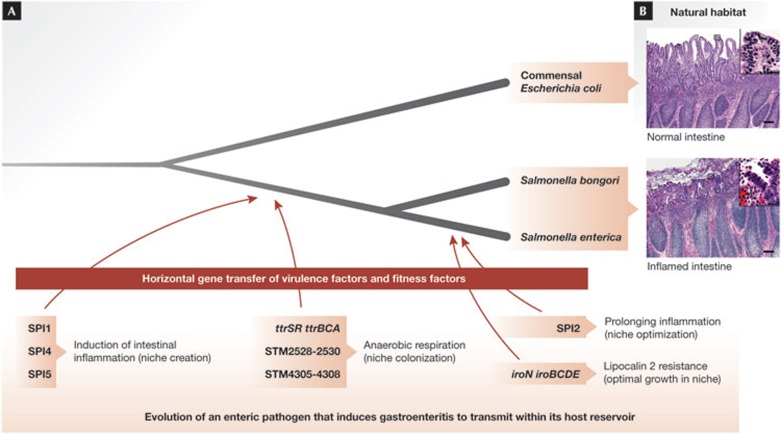
The enhanced growth of commensal Enterobacteriaceae in the inflamed gut of individuals with inflammatory bowel disease could be viewed as an accident in which, as Louis Pasteur would put it, “chance favours the prepared microbe”. However, the fitness advantage conferred by host-derived electron acceptors also represents a potent selective force that is probably responsible for the evolution of pathogenic species within the Enterobacteriaceae (Sidebar A; Fig 3). For example, the genus Salmonella comprises a group of pathogens that are closely related to E. coli. Since diverging from a common ancestor with E. coli, the Salmonella lineage acquired several virulence factors through plasmid or phage-mediated horizontal gene transfer [70]. The genes encoding these virulence factors are located on Salmonella pathogenicity islands (SPIs), defined as horizontally acquired DNA regions that are absent from the otherwise colinear E. coli genome [71]. SPIs that are present in all members of the genus Salmonella include SPI1, which encodes the invasion-associated type III secretion system (T3SS-1; [72,73]), SPI4, which encodes a large non-fimbrial adhesin required for epithelial invasion [74] and SPI5, which encodes proteins (known as effectors) that are injected into host cells by the T3SS-1 (Fig 3A; [75]). The genus Salmonella can be divided into two species, S. bongori (containing 23 serovars) and S. enterica (containing 2,587 serovars; [76]). All members of S. enterica encode a second type III secretion system (T3SS-2) encoded by SPI2—which is absent from S. bongori and E. coli [77,78]—and serves to prolong intestinal inflammation [79]. The presence of these pathogenicity islands enables a fraction of the Salmonella population to invade the intestinal epithelium (SPI1, SPI4 and SPI5) and survive in macrophages (SPI2), thereby triggering acute intestinal inflammation (gastroenteritis) [75,80,81,82,83,84].
Sidebar A | In need of answers.
Although it seems clear that intestinal inflammation can result in changes in the microbial community structure, relatively little is known about possible consequences of the resulting dysbiosis. Does inflammation select for more harmful bacterial species that can further exacerbate host responses? If so, what are the properties that make them more potent irritants? This area needs to be explored further by using both metagenomic and mechanistic approaches.
Although an increased relative abundance of facultative anaerobic bacteria is common in individuals with inflammatory bowel disease, not all patients show these changes in their microbiota. Thus, it is tempting to speculate that other mechanisms—in addition to anaerobic respiration—can influence the microbial community structure during intestinal inflammation. These mechanisms remain to be identified.
How does the quality of the host response alter the nutritional environment in the inflamed gut? This seems to be particularly relevant to understand the evolution of enteric pathogens that use their virulence factors to manipulate host responses. For example, acquisition of the sopE gene by horizontal gene transfer enhances the ability of Salmonella enterica sv Typhimurium to elicit production of host-derived nitrate, thereby altering the nutritional environment in the intestinal lumen [87]. Deeper insights into how virulence factors change the nutrient availability in the gut are necessary to appreciate how the host response has shaped the evolution of enteric pathogens.
Figure 3.
Selective forces driving the evolution of pathogenic Enterobacteriaceae species. (A) The genus Salmonella comprises enteric pathogens that are closely related to commensal Escherichia coli, with whom they have a common ancestor as indicated by the schematic drawing of their phylogenetic tree (not to scale). The timing of horizontal gene transfer events introducing virulence factors or fitness factors is indicated. Acquisition of the indicated DNA regions conferred the ability to induce inflammation, benefit from the resulting host response and enhance their transmission. (B) The images show the natural habitat of commensal E. coli (normal intestine, top panel) and of pathogenic Salmonella species (inflamed intestine, bottom panel). The images of calf intestine were reproduced from [112] with permission. SPI1/2/4/5; Salmonella pathogenicity island 1/2/4/5.
Interestingly, by using their virulence factors to actively induce intestinal inflammation, Salmonella serovars can tip the balance in their favour in competition with the intestinal microbiota [4,5]. For instance, when S. enterica serovar Typhimurium (S.e. sv typhimurium) induces colitis in a mouse model, the pathogen edges out competing microbes in the gut lumen to become a prominent member of the microbiota. Inactivation of both type III secretion systems renders S.e. sv typhimurium unable to trigger intestinal inflammation, thereby markedly reducing its ability to colonize the lumen of the large intestine. However, when mice that develop colitis spontaneously—such as IL-10-deficient mice—are infected with a S.e. sv typhimurium mutant lacking both type III secretion systems, an enrichment for the pathogen and a concomitant depletion of Clostridia and Bacteroidia is observed [4]. Although virulence factors are necessary for inducing intestinal inflammation, these data suggest that T3SS-1 and T3SS-2 are not required for securing the growth advantage S.e. sv typhimurium gains in the lumen of the inflamed gut.
Which factors confer a growth advantage on Salmonella serovars during gastroenteritis? Optimal growth in the environment of the inflamed gut requires resistance to antimicrobial proteins—such as lipocalin 2—which is conferred by the iroN iroBCDE gene cluster [41], a DNA region present in all members of S. enterica but absent from S. bongori [85,86]. Lipocalin 2 resistance might confer an advantage on S. enterica during its competition with commensal Enterobacteriaceae, provided that the latter rely on enterobactin for iron acquisition. Rivalry with commensal Enterobacteriaceae probably arises because exogenous electron acceptors—such as nitrate—enhance their growth in the inflamed gut [63]. As S.e. sv typhimurium can also use host-derived nitrate to boost its luminal growth [87], the pathogen and commensal Enterobacteriaceae have to compete for this limited resource. However, Salmonella serovars have improved their ability to outgrow other gut microbes during inflammation by acquiring additional fitness factors that are absent from E. coli. Among these are gene clusters encoding putative DMSO reductases (STM2528-STM2530 and STM4305-STM4308; [88]) and the ttrSR ttrBCA gene cluster, which confers the ability to use tetrathionate (S4O62−) as an electron acceptor for anaerobic respiration (Fig 3A; [89,90]). The ability to respire tetrathionate has been used since 1923 to enrich for Salmonella serovars in biological samples containing competing microbes [91]. However, the fact that tetrathionate is a by-product of the host inflammatory response in the gut was only recently discovered [92]. Hydrogen sulphide (H2S) and methanethiol (CH3SH) are fermentation end-products of gut microbes that are converted to thiosulphate (S2O32−) by the colonic epithelium to avoid toxicity [93,94]. ROS generated during intestinal inflammation oxidize thiosulphate (S2O32−) to tetrathionate (S4O62−), thereby boosting luminal growth of S.e. sv typhimurium through tetrathionate respiration whilst reducing the relative abundance of Clostridia and Bacteroidia [92]. Importantly, enhanced growth in the intestinal lumen promotes transmission of S.e. sv typhimurium by the faecal–oral route [95]. Ultimately, the necessity to spread from an infected to a naive host places virulence factors and fitness factors under selection. Although taxonomists identified tetrathionate respiration empirically as a characteristic that helps distinguish Salmonella serovars from close relatives [91], the picture emerging from research is that this function is part of a ‘business plan’ that defines the genus Salmonella.
These findings suggest that an important initial event in the evolution of the Salmonella lineage was the acquisition of virulence factors the deployment of which induces intestinal inflammation, thereby creating a new niche in the host (Fig 3B). At the same time, the Salmonella lineage acquired fitness factors that enabled these pathogens to occupy this new niche to ensure transmission (Fig 3A). Evolution is still at work to fine-tune this ‘business plan’ by incorporating new virulence factors to instigate subtle alterations in host responses that generate exogenous electron acceptors. These processes probably contribute to the rise of new epidemic clones, as illustrated by an analysis of the prophage-encoded T3SS-1 effector protein SopE [87].
The T3SS-1 induces host responses by injecting effector proteins into host cells [81]. SPI1 and SPI5 encode effector proteins that are conserved among Salmonella serotypes, whereas other effector proteins are encoded by prophages and have a limited distribution. One of these prophage-encoded T3SS-1 effector proteins is SopE [96]. On injection into host cells, SopE activates small Rho GTPases to induce NF-κB-dependent gene expression [97], which leads to a modest increase in the severity of intestinal inflammation in animal models [98]. Remarkably, deployment of SopE significantly enhances the production of iNOS in the intestinal mucosa, thereby increasing luminal growth of S.e. sv typhimurium by nitrate respiration [87]. SopE is encoded by a prophage present in only a few S.e. sv typhimurium clones, which caused an epidemic among cattle and humans in Europe during the 1970s and ‘80s [99]. Collectively, these data suggest that phage-mediated horizontal transfer of the sopE gene confers a nitrate respiration-dependent fitness advantage that might have contributed to the emergence of an epidemic S.e. sv typhimurium clone.
Side-stepping the competition
The findings reviewed above point to anaerobic respiration as one of the fundamental principles that governs the phylum-level changes in the composition of gut-associated microbial communities during inflammation. But how does anaerobic respiration enable Enterobacteriaceae to outcompete Clostridia and Bacteroidia, which can thrive on a limited quantity of carbohydrate? One possibility is that anaerobic respiration is more efficient for energy production than fermentation. Although this might be true for electron acceptors with a high standard redox potential, such as the nitrate–nitrite redox couple (E° = 433 mV; [100]), this explanation seems less convincing for the tetrathionate–thiosulphate redox couple, which has a relatively low standard redox potential (E° = 170 mV; [101]). A second possibility is that exogenous electron acceptors enable Enterobacteriaceae to use carbon sources that cannot be fermented. The removal of gut contents during diarrhoea limits nutrients to mucus-derived carbohydrate and nutrients derived from the release of enterocytes from the tips of villi. Membranes of enterocytes contain phosphatidylethanolamine as their most abundant phospholipid [102]. Ethanolamine—a non-fermentable substrate present in the intestinal contents of calves at a concentration of approximately 2 mM [103]—is a nutrient derived from phosphatidylethanolamine. S.e. sv typhimurium requires tetrathionate for anaerobic growth on ethanolamine as a sole carbon source in vitro [104]. In the lumen of the inflamed intestine, the ability to consume ethanolamine bestows a marked growth advantage on S.e. sv typhimurium, which depends on the ability of the pathogen to perform tetrathionate respiration [105]. These data suggest that anaerobic respiration enables S.e. sv typhimurium to consume an abundant simple substrate, ethanolamine, which is provided by the host but cannot be readily fermented by competing obligate anaerobic bacteria. Thus, a significant benefit of anaerobic respiration is the ability of S.e. sv typhimurium to side-step nutritional competition with Clostridia and Bacteroidia, thereby fostering its own growth in the gut.
Mechanistic insights guide future research efforts
The mechanistic insights discussed above suggest that the phylum-level changes in gut-associated microbial communities are a consequence rather than a cause of intestinal inflammation. However, one possible consequence of this dysbiosis is an exacerbation of pre-existing inflammatory conditions (Sidebar A). The presence of gut microbes is a prerequisite for the development of chronic intestinal inflammation in genetically predisposed mice [106,107], and studies suggest that a bloom of Enterobacteriaceae can be associated with enhanced intestinal inflammation. For example, in a mouse model of inflammatory bowel disease, changes in the microbial community structure characterized by an increased luminal abundance of Enterobacteriaceae can be transferred to other animals, resulting in an exacerbation of intestinal inflammation [14,108]. Adherent-invasive E. coli (AIEC) are isolated more commonly from the intestinal mucosa of individuals with Crohn's disease than from healthy controls [109,110]. AIEC colonize and exacerbate gut inflammation in mice with DSS-injured colon [111]. Thus, the mechanisms leading to dysbiosis might also select for intestinal colonization with more harmful members of the Enterobacteriaceae—such as AIEC—thereby exacerbating inflammation and interfering with its resolution.
Anaerobic respiration emerges as a potential target for new intervention strategies aimed at restoring a normal microbial community structure. A formal proof of principle for this intervention strategy is the fact that the iNOS-inhibitor aminoguanidine hydrochloride can blunt nitrate respiration-dependent growth of E. coli in mice with DSS-induced colitis [63]. This approach would have to be broadened to block the use of multiple respiratory electron acceptors, thereby ending the bloom of Enterobacteriaceae in the distal gut by essentially suffocating these facultative anaerobic bacteria. Exploring this approach for the treatment of intestinal inflammatory disorders represents an exciting direction for future research.

Sebastian E Winter, Andreas J Bäumler & Christopher A Lopez
Acknowledgments
Work in the laboratory of A.J.B. is supported by Public Health Service Grants AI044170, AI076246 and AI096528. C.A.L. was supported by Public Health Service Grant AI060555.
Footnotes
The authors declare that they have no conflict of interest.
References
- Mahowald MA et al. (2009) Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci USA 106: 5859–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA (2005) Diversity of the human intestinal microbial flora. Science 308: 1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JL (2005) Obesity alters gut microbial ecology. Proc Natl Acad Sci USA 102: 11070–11075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B et al. (2007) Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol 5: 2177–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barman M, Unold D, Shifley K, Amir E, Hung K, Bos N, Salzman N (2008) Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun 76: 907–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB (2007) Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2: 119–129 [DOI] [PubMed] [Google Scholar]
- Krook A, Lindström B, Kjellander J, Järnerot G, Bodin L (1981) Relation between concentrations of metronidazole and Bacteroides spp in faeces of patients with Crohn's disease and healthy individuals. J Clin Pathol 34: 645–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaffer MH, Holdsworth CD, Duerden BI (1991) The assessment of faecal flora in patients with inflammatory bowel disease by a simplified bacteriological technique. J Med Microbiol 35: 238–243 [DOI] [PubMed] [Google Scholar]
- Seksik P, Rigottier-Gois L, Gramet G, Sutren M, Pochart P, Marteau P, Jian R, Doré J (2003) Alterations of the dominant faecal bacterial groups in patients with Crohn's disease of the colon. Gut 52: 237–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR (2007) Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA 104: 13780–13785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart M et al. (2007) Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum. ISME J 1: 403–418 [DOI] [PubMed] [Google Scholar]
- Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJ (2006) Differences between tissue-associated intestinal microfloras of patients with Crohn's disease and ulcerative colitis. J Clin Microbiol 44: 4136–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AW et al. (2011) High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol 11: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett WS et al. (2010) Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe 8: 292–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman AL, Lough DM, Barupal DK, Fiehn O, Fishbein T, Zasloff M, Eisen JA (2009) Human gut microbiome adopts an alternative state following small bowel transplantation. Proc Natl Acad Sci USA 106: 17187–17192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatkin NM, Cameron EA, Martens EC (2012) How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol 10: 323–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach MA, Sonnenburg JL (2011) Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe 10: 336–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, McNulty NP, Rey FE, Gordon JI (2011) Predicting a human gut microbiota's response to diet in gnotobiotic mice. Science 333: 101–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez I, Kim J, Duffy PR, Schlegel VL, Walter J (2010) Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects. PLoS ONE 5: e15046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL (2010) Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell 141: 1241–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AW (2011) Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J 5: 220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GD et al. (2011) Linking long-term dietary patterns with gut microbial enterotypes. Science 334: 105–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves AR, Wang GR, Salyers AA (1997) Characterization of four outer membrane proteins that play a role in utilization of starch by Bacteroides thetaiotaomicron. J Bacteriol 179: 643–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipman JA, Cho KH, Siegel HA, Salyers AA (1999) Physiological characterization of SusG, an outer membrane protein essential for starch utilization by Bacteroides thetaiotaomicron. J Bacteriol 181: 7206–7211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves AR, D'Elia JN, Frias J, Salyers AA (1996) A Bacteroides thetaiotaomicron outer membrane protein that is essential for utilization of maltooligosaccharides and starch. J Bacteriol 178: 823–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V (2003) Iron uptake by Escherichia coli. Front Biosci 8: s1409–s1421 [DOI] [PubMed] [Google Scholar]
- D'Elia JN, Salyers AA (1996) Contribution of a neopullulanase, a pullulanase, and an alpha-glucosidase to growth of Bacteroides thetaiotaomicron on starch. J Bacteriol 178: 7173–7179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI (2003) A genomic view of the human–Bacteroides thetaiotaomicron symbiosis. Science 299: 2074–2076 [DOI] [PubMed] [Google Scholar]
- Ishii JN, Okajima Y, Nakae T (1981) Characterization of lamB protein from the outer membrane of Escherichia coli that forms diffusion pores selective for maltose-maltodextrins. FEBS Lett 134: 217–220 [DOI] [PubMed] [Google Scholar]
- Freundlieb S, Boos W (1986) Alpha-amylase of Escherichia coli, mapping and cloning of the structural gene, malS, and identification of its product as a periplasmic protein. J Biol Chem 261: 2946–2953 [PubMed] [Google Scholar]
- Schneider E, Freundlieb S, Tapio S, Boos W (1992) Molecular characterization of the MalT-dependent periplasmic alpha-amylase of Escherichia coli encoded by malS. J Biol Chem 267: 5148–5154 [PubMed] [Google Scholar]
- Duplay P, Bedouelle H, Fowler A, Zabin I, Saurin W, Hofnung M (1984) Sequences of the malE gene and of its product, the maltose-binding protein of Escherichia coli K12. J Biol Chem 259: 10606–10613 [PubMed] [Google Scholar]
- Shuman HA, Silhavy TJ (1981) Identification of the malK gene product. A peripheral membrane component of the Escherichia coli maltose transport system. J Biol Chem 256: 560–562 [PubMed] [Google Scholar]
- Froshauer S, Beckwith J (1984) The nucleotide sequence of the gene for malF protein, an inner membrane component of the maltose transport system of Escherichia coli. Repeated DNA sequences are found in the malE-malF intercistronic region. J Biol Chem 259: 10896–10903 [PubMed] [Google Scholar]
- Dassa E, Hofnung M (1985) Sequence of gene malG in E. coli K12: homologies between integral membrane components from binding protein-dependent transport systems. EMBO J 4: 2287–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassa E (1993) Sequence–function relationships in MalG, an inner membrane protein from the maltose transport system in Escherichia coli. Mol Microbiol 7: 39–47 [DOI] [PubMed] [Google Scholar]
- Schneider E, Linde M, Tebbe S (1995) Functional purification of a bacterial ATP-binding cassette transporter protein (MalK) from the cytoplasmic fraction of an overproducing strain. Protein Expr Purif 6: 10–14 [DOI] [PubMed] [Google Scholar]
- Bohm A, Diez J, Diederichs K, Welte W, Boos W (2002) Structural model of MalK, the ABC subunit of the maltose transporter of Escherichia coli: implications for mal gene regulation, inducer exclusion, and subunit assembly. J Biol Chem 277: 3708–3717 [DOI] [PubMed] [Google Scholar]
- Peterson DA, Frank DN, Pace NR, Gordon JI (2008) Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe 3: 417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ et al. (2012) Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe 11: 227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffatellu M et al. (2009) Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe 5: 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger T et al. (2006) Lipocalin 2-deficient mice exhibit increased sensitivity to Escherichia coli infection but not to ischemia-reperfusion injury. Proc Natl Acad Sci USA 103: 1834–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A (2004) Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 432: 917–921 [DOI] [PubMed] [Google Scholar]
- Goetz DH, Holmes MA, Borregaard N, Bluhm ME, Raymond KN, Strong RK (2002) The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol Cell 10: 1033–1043 [DOI] [PubMed] [Google Scholar]
- Harper RW, Xu C, Eiserich JP, Chen Y, Kao CY, Thai P, Setiadi H, Wu R (2005) Differential regulation of dual NADPH oxidases/peroxidases, Duox1 and Duox2, by Th1 and Th2 cytokines in respiratory tract epithelium. FEBS Lett 579: 4911–4917 [DOI] [PubMed] [Google Scholar]
- Kuwano Y, Kawahara T, Yamamoto H, Teshima-Kondo S, Tominaga K, Masuda K, Kishi K, Morita K, Rokutan K (2006) Interferon-gamma activates transcription of NADPH oxidase 1 gene and upregulates production of superoxide anion by human large intestinal epithelial cells. Am J Physiol Cell Physiol 290: C433–C443 [DOI] [PubMed] [Google Scholar]
- Hohn DC, Lehrer RI (1975) NADPH oxidase deficiency in X-linked chronic granulomatous disease. J Clin Invest 55: 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhail LC, DeChatelet LR, Shirley PS, Wilfert C, Johnston RB Jr, McCall CE (1977) Deficiency of NADPH oxidase activity in chronic granulomatous disease. J Pediatr 90: 213–217 [DOI] [PubMed] [Google Scholar]
- Seger RA, Tiefenauer L, Matsunaga T, Wildfeuer A, Newburger PE (1983) Chronic granulomatous disease due to granulocytes with abnormal NADPH oxidase activity and deficient cytochrome-b. Blood 61: 423–428 [PubMed] [Google Scholar]
- Salzman A, Denenberg AG, Ueta I, O'Connor M, Linn SC, Szabó C (1996) Induction and activity of nitric oxide synthase in cultured human intestinal epithelial monolayers. Am J Physiol 270: G565–G573 [DOI] [PubMed] [Google Scholar]
- Palmer RM, Ashton DS, Moncada S (1988) Vascular endothelial cells synthesize nitric oxide from L-arginine. Nature 333: 664–666 [DOI] [PubMed] [Google Scholar]
- Jagannath C, Actor JK, Hunter RL Jr (1998) Induction of nitric oxide in human monocytes and monocyte cell lines by Mycobacterium tuberculosis. Nitric Oxide 2: 174–186 [DOI] [PubMed] [Google Scholar]
- Lundberg JO, Hellström PM, Lundberg JM, Alving K (1994) Greatly increased luminal nitric oxide in ulcerative colitis. Lancet 344: 1673–1674 [DOI] [PubMed] [Google Scholar]
- Singer II, Kawka DW, Scott S, Weidner JR, Mumford RA, Riehl TE, Stenson WF (1996) Expression of inducible nitric oxide synthase and nitrotyrosine in colonic epithelium in inflammatory bowel disease. Gastroenterology 111: 871–885 [DOI] [PubMed] [Google Scholar]
- Enocksson A, Lundberg J, Weitzberg E, Norrby-Teglund A, Svenungsson B (2004) Rectal nitric oxide gas and stool cytokine levels during the course of infectious gastroenteritis. Clin Diagn Lab Immunol 11: 250–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Gunn C, Beckman JS (1992) Bactericidal activity of peroxynitrite. Arch Biochem Biophys 298: 452–457 [DOI] [PubMed] [Google Scholar]
- De Groote MA, Granger D, Xu Y, Campbell G, Prince R, Fang FC (1995) Genetic and redox determinants of nitric oxide cytotoxicity in a Salmonella typhimurium model. Proc Natl Acad Sci USA 92: 6399–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagam B, Richardson DE (2008) The mechanism of carbon dioxide catalysis in the hydrogen peroxide N-oxidation of amines. Inorg Chem 47: 1173–1178 [DOI] [PubMed] [Google Scholar]
- Schoneich C (2005) Methionine oxidation by reactive oxygen species: reaction mechanisms and relevance to Alzheimer's disease. Biochim Biophys Acta 1703: 111–119 [DOI] [PubMed] [Google Scholar]
- de la Huerga J, Popper H (1951) Urinary excretion of choline metabolites following choline administration in normals and patients with hepatobiliary diseases. J Clin Invest 30: 463–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo C, Ischiropoulos H, Radi R (2007) Peroxynitrite: biochemistry, pathophysiology and development of therapeutics. Nat Rev Drug Discov 6: 662–680 [DOI] [PubMed] [Google Scholar]
- Dudhgaonkar SP, Tandan SK, Kumar D, Raviprakash V, Kataria M (2007) Influence of simultaneous inhibition of cyclooxygenase-2 and inducible nitric oxide synthase in experimental colitis in rats. Inflammopharmacology 15: 188–195 [DOI] [PubMed] [Google Scholar]
- Winter SE et al. (2013) Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339: 708–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennis RB, Stewart V (1996) Respiration. In Escherichia coli and Salmonella. Cellular and Molecular Biology 2nd edn, vol 1 (eds Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE), pp. 217–261. Washington DC, USA: ASM [Google Scholar]
- Blattner FR et al. (1997). The complete genome sequence of Escherichia coli K-12. Science 277: 1453–1462 [DOI] [PubMed] [Google Scholar]
- Benoit S, Abaibou H, Mandrand-Berthelot MA (1998) Topological analysis of the aerobic membrane-bound formate dehydrogenase of Escherichia coli. J Bacteriol 180: 6625–6634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg BL, Stewart V (1990) Structural genes for nitrate-inducible formate dehydrogenase in Escherichia coli K-12. Genetics 125: 691–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan KV, Johnson JL (1992) The pterin molybdenum cofactors. J Biol Chem 267: 10199–10202 [PubMed] [Google Scholar]
- Rivers SL, McNairn E, Blasco F, Giordano G, Boxer DH (1993) Molecular genetic analysis of the moa operon of Escherichia coli K-12 required for molybdenum cofactor biosynthesis. Mol Microbiol 8: 1071–1081 [DOI] [PubMed] [Google Scholar]
- Bäumler AJ (1997) The record of horizontal gene transfer in Salmonella. Trends Microbiol 5: 318–322 [DOI] [PubMed] [Google Scholar]
- Kingsley RA, Bäumler AJ (2002) Pathogenicity islands and host adaptation of Salmonella serovars. Curr Top Microbiol Immunol 264: 67–87 [PubMed] [Google Scholar]
- Mills DM, Bajaj V, Lee CA (1995) A 40kb chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 chromosome. Mol Microbiol 15: 749–759 [DOI] [PubMed] [Google Scholar]
- Galán JE, Curtiss R 3rd (1989) Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci USA 86: 6383–6387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach RG, Claudio N, Rohde M, Jackel D, Wagner C, Hensel M (2008) Cooperation of Salmonella pathogenicity islands 1 and 4 is required to breach epithelial barriers. Cell Microbiol 10: 2364–2376 [DOI] [PubMed] [Google Scholar]
- Wood MW, Jones MA, Watson PR, Hedges S, Wallis TS, Galyov EE (1998) Identification of a pathogenicity island required for Salmonella enteropathogenicity. Mol Microbiol 29: 883–891 [DOI] [PubMed] [Google Scholar]
- Guibourdenche M, Roggentin P, Mikoleit M, Fields PI, Bockemuhl J, Grimont PA, Weill FX (2010) Supplement 2003–2007 (No. 47) to the White-Kauffmann-Le Minor scheme. Res Microbiol 161: 26–29 [DOI] [PubMed] [Google Scholar]
- Hensel M, Shea JE, Bäumler AJ, Gleeson C, Blattner F, Holden DW (1997) Analysis of the boundaries of Salmonella pathogenicity island 2 and the corresponding chromosomal region of Escherichia coli K-12. J Bacteriol 179: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Soncini FC, Solomon F, Groisman EA (1996) Identification of a pathogenicity island required for Salmonella survival in host cells. Proc Natl Acad Sci USA 93: 7800–7804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapfelmeier S et al. (2005) The Salmonella pathogenicity island (SPI)-2 and SPI-1 type III secretion systems allow Salmonella serovar typhimurium to trigger colitis via MyD88-dependent and MyD88-independent mechanisms. J Immunol 174: 1675–1685 [DOI] [PubMed] [Google Scholar]
- Tsolis RM, Adams LG, Ficht TA, Bäumler AJ (1999) Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect Immun 67: 4879–4885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Santos RL, Tsolis RM, Stender S, Hardt WD, Bäumler AJ, Adams LG (2002) The Salmonella enterica serotype typhimurium effector proteins SipA, SopA, SopB, SopD, and SopE2 act in concert to induce diarrhea in calves. Infect Immun 70: 3843–3855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel M, Hapfelmeier S, Quintanilla-Martinez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Russmann H, Hardt WD (2003) Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 71: 2839–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn B, Li Y, Owen D, Vallence BA, Finlay BB (2005) Salmonella enterica serovar Typhimurium pathogenicity island 2 is necessary for complete virulence in a mouse model of infectious enterocolitis. Infect Immun 73: 3219–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach RG, Jäckel D, Stecher B, Wagner C, Lupas A, Hardt WD, Hensel M (2007) Salmonella pathogenicity island 4 encodes a giant non-fimbrial adhesin and the cognate type 1 secretion system. Cell Microbiol 9: 1834–1850 [DOI] [PubMed] [Google Scholar]
- Bäumler AJ, Norris TL, Lasco T, Voight W, Reissbrodt R, Rabsch W, Heffron F (1998) IroN, a novel outer membrane siderophore receptor characteristic of Salmonella enterica. J Bacteriol 180: 1446–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäumler AJ, Heffron F, Reissbrodt R (1997) Rapid detection of Salmonella enterica with primers specific for iroB. J Clin Microbiol 35: 1224–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez CA, Winter SE, Rivera-Chavez FM, Xavier N, Poon V, Nuccio SP, Tsolis RM, Bäumler AJ (2012) Phage-mediated acquisition of a type III secreted effector protein boosts growth of salmonella by nitrate respiration. MBio 3: e00143–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernikos GS, Thomson NR, Parkhill J (2007) Genetic flux over time in the Salmonella lineage. Genome Biol 8: R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel M, Nikolaus T, Egelseer C (1999) Molecular and functional analysis indicates a mosaic structure of Salmonella pathogenicity island 2. Mol Microbiol 31: 489–498 [DOI] [PubMed] [Google Scholar]
- Hensel M, Hinsley AP, Nikolaus T, Sawers G, Berks BC (1999) The genetic basis of tetrathionate respiration in Salmonella typhimurium. Mol Microbiol 32: 275–287 [DOI] [PubMed] [Google Scholar]
- Muller L (1923) A new medium of enrichment for the research of typhic and paratyphic bacillus. Comptes Rendus des Seances de la Societe de Biologie et de ses Filiales 89: 434–437 [Google Scholar]
- Winter SE et al. (2010) Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467: 426–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furne J, Springfield J, Koenig T, DeMaster E, Levitt MD (2001) Oxidation of hydrogen sulfide and methanethiol to thiosulfate by rat tissues: a specialized function of the colonic mucosa. Biochem Pharmacol 62: 255–259 [DOI] [PubMed] [Google Scholar]
- Levitt MD, Furne J, Springfield J, Suarez F, DeMaster E (1999) Detoxification of hydrogen sulfide and methanethiol in the cecal mucosa. J Clin Invest 104: 1107–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Bouley DM, Hoy YE, Gerke C, Relman DA, Monack DM (2008) Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infect Immun 76: 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardt WD, Urlaub H, Galán JE (1998) A substrate of the centisome 63 type III protein secretion system of Salmonella typhimurium is encoded by a cryptic bacteriophage. Proc Natl Acad Sci USA 95: 2574–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardt WD, Chen LM, Schuebel KE, Bustelo XR, Galan JE (1998) S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell 93: 815–826 [DOI] [PubMed] [Google Scholar]
- Zhang S, Santos RL, Tsolis RM, Mirold S, Hardt WD, Adams LG, Bäumler AJ (2002) Phage mediated horizontal transfer of the sopE1 gene increases enteropathogenicity of Salmonella enterica serotype Typhimurium for calves. FEMS Microbiol Lett 217: 243–247 [DOI] [PubMed] [Google Scholar]
- Mirold S, Rabsch W, Rohde M, Stender S, Tschape H, Russmann H, Igwe E, Hardt WD (1999) Isolation of a temperate bacteriophage encoding the type III effector protein SopE from an epidemic Salmonella typhimurium strain. Proc Natl Acad Sci USA 96: 9845–9850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauer RK, Jungermann K, Decker K (1977) Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev 41: 100–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaprálek F (1972) The physiological role of tetrathionate respiration in growing citrobacter. J Gen Microbiol 71: 133–139 [DOI] [PubMed] [Google Scholar]
- Kawai K, Fujita M, Nakao M (1974) Lipid components of two different regions of an intestinal epithelial cell membrane of mouse. Biochim Biophys Acta 369: 222–233 [PubMed] [Google Scholar]
- Bertin Y, Girardeau JP, Chaucheyras-Durand F, Lyan B, Pujos-Guillot E, Harel J, Martin C (2010) Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ Microbiol 13: 365–377 [DOI] [PubMed] [Google Scholar]
- Price-Carter M, Tingey J, Bobik TA, Roth JR (2001) The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar typhimurium on ethanolamine or 1,2-propanediol. J Bacteriol 183: 2463–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiennimitr P et al. (2011) Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci USA 108: 17480–17485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadlack B, Merz H, Schorle H, Schimpl A, Feller AC, Horak I (1993) Ulcerative colitis-like disease in mice with a disrupted interleukin-2 gene. Cell 75: 253–261 [DOI] [PubMed] [Google Scholar]
- Sellon RK, Tonkonogy S, Schultz M, Dieleman LA, Grenther W, Balish E, Rennick DM, Sartor RB (1998) Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun 66: 5224–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett WS, Lord GM, Punit S, Lugo-Villarino GS Mazmanian K, Ito S, Glickman JN, Glimcher LH (2007) Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell 131: 33–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darfeuille-Michaud A et al. (2004) High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology 127: 412–421 [DOI] [PubMed] [Google Scholar]
- Darfeuille-Michaud A et al. (1998) Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn's disease. Gastroenterology 115: 1405–1413 [DOI] [PubMed] [Google Scholar]
- Carvalho FA, Barnich N, Sauvanet P, Darcha C, Gelot A, Darfeuille-Michaud A (2008) Crohn's disease-associated Escherichia coli LF82 aggravates colitis in injured mouse colon via signaling by flagellin. Inflamm Bowel Dis 14: 1051–1060 [DOI] [PubMed] [Google Scholar]
- Zhang S, Kingsley RA, Santos RL, Andrews-Polymenis H, Raffatellu M, Figueiredo J, Nunes J, Tsolis RM, Adams LG, Bäumler AJ (2003) Molecular pathogenesis of Salmonella enterica serotype typhimurium-induced diarrhea. Infect Immun 71: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]