Abstract
Myotonic dystrophy type 1 (DM1) is caused by DM protein kinase (DMPK) transcripts containing an expanded (CUG)n repeat. Antisense oligonucleotide (AON)-mediated suppression of these mutant RNAs is considered a promising therapeutic strategy for this severe disorder. Earlier, we identified a 2′-O-methyl (2′-OMe) phosphorothioate (PT)–modified (CAG)7 oligo (PS58), which selectively silences mutant DMPK transcripts through recognition of the abnormally long (CUG)n tract. We present here a comprehensive collection of triplet repeat AONs and found that oligo length and nucleotide chemistry are important determinants for activity. For significant reduction of expanded DMPK mRNAs, a minimal length of five triplets was required. 2′-O,4′-C-ethylene-bridged nucleic acid (ENA)–modified AONs appeared not effective, probably due to lack of nuclear internalization. Selectivity for products from the expanded DMPK allele in patient myoblasts, an important requirement to minimize unwanted side effects, appeared also dependent on AON chemistry. In particular, RNase-H–dependent (CAG)n AONs did not show (CUG)n length specificity. We provide evidence that degradation of long DMPK transcripts induced by PS58-type AONs is an RNase-H independent process, does not involve oligo-intrinsic RNase activity nor does it interfere with splicing of DMPK transcripts. Our collection of triplet repeat AONs forms an important resource for further development of a safe therapy for DM1 and other unstable microsatellite diseases.
Keywords: antisense oligonucleotides, myotonic dystrophy, RNA processing, RNA silencing, triplet repeat expansion
Introduction
Myotonic dystrophy type 1 (DM1) is an autosomal dominant, trinucleotide repeat expansion disease and the most common form of adult onset muscular dystrophy.1,2,3 The multisystemic symptoms are highly variable and involve different tissues and organs, including skeletal muscle, heart, brain, eyes, reproductive systems, and the gastrointestinal tract. No cure to slow or stop the disease process is currently available.
The genetic basis of DM1 is an unstable (CTG)n expansion in the 3′-untranslated region of the DM protein kinase (DMPK) gene. Length of the (CTG)n segment is correlated with age of onset and severity of disease.1 In the healthy population, the CTG repeat is polymorphic in length, with a triplet number that ranges from 5 to 37 (mostly below 15),4 whereas disease alleles may contain between 50 and >2,000 triplets.1
The DM1 pathogenic mechanism involves an RNA gain-of-function. Repeat-containing transcripts from the expanded allele accumulate in cell nuclei and alter the function of RNA-binding proteins, whose association with disease has been most intensively studied for muscleblind (MBNL) and CUG-binding protein 1 (CUGBP1).2 In adult DM1 tissue, the resulting imbalance in availability of these antagonistic splicing regulators causes misregulation of events in which embryonic patterns of splicing occur. For example, myotonia and insulin resistance, two key features of DM1, may be caused by altered splicing of muscle chloride channel and insulin receptor transcripts, respectively.2 Also effects on microRNA processing and function have been reported.5
Results obtained with DM1 mouse models that inducibly express (CUG)n RNA show that features of RNA toxicity are reversible,6 strongly suggesting that reducing the level of expanded DMPK transcripts will be beneficial to patients. In the past decade, different antisense strategies have been developed with the intention to eliminate expanded DMPK transcripts via either antisense RNA expression, ribozyme activity, oligo-mediated RNase-H activity or RNA interference (RNAi) technology.7 Reduction of DMPK transcript levels was achieved by several of these methods, but selectivity between toxic and normal-sized transcripts was generally low.
To be functional and safe for therapeutic use in DM1 patients, an antisense approach should ideally silence transcripts from the expanded allele only and leave normal-sized DMPK transcripts or unrelated transcripts containing (CUG)n segments intact. Since length of the (CUG)n repeat is the only consistent polymorphic difference between mutant and normal DMPK (pre)mRNAs, the repeat tract itself forms an attractive therapeutic target. Besides, despite the fact that normal and mutant DMPK transcripts are both made in the nucleus, (CUG)n length selectivity may be enhanced by directing active compounds to this compartment where mutant transcripts accumulate after synthesis.
Earlier, we published on the identification and characterization of PS58, an antisense oligonucleotide (AON) complementary to the (CUG)n repeat.8 PS58 is a (CAG)7 fully 2′-O-methyl (2′-OMe) phosphorothioate (PT)-modified AON, which silences expanded (CUG)n transcripts in DM1 cell and animal models. Other in vivo AON studies directed at the expansion mutation made use of a (CAG)n morpholino to interfere with the (CUG)n repeat–MBNL interaction9 or LNA (locked nucleic acid)/MOE (2′-O-methoxyethyl)-PT–type (CAG)n gapmers to recruit RNase-H activity for degradation of (CUG)n-containing transcripts.10 François et al. engineered a hU7-(CAG)15-small nuclear RNA which preferentially reduced pathogenic DMPK mRNA in DM1 cells, via a yet unknown mechanism.11
Here, we report on specific properties that (CUG)n-directed AONs require for effective knockdown of expanded DMPK RNA. Triplet repeat AONs with different chemistries and lengths were tested in an unbiased screen for silencing capacity in a transgenic DM1 cell culture model expressing human DMPK (CUG)500 transcripts. AON length and nucleotide chemistry appeared to be major parameters for therapeutic efficacy. A selection of effective AONs was tested in DM1 patient myoblasts. Findings of this study are important for transition towards in vivo studies with AONs and ultimately for development of a specific AON-mediated therapy for DM1. As a first step in support of this transition, we report on effects on in vitro complement activation by some of these AONs.
Results
AONs complementary to DMPK's (CUG)n repeat
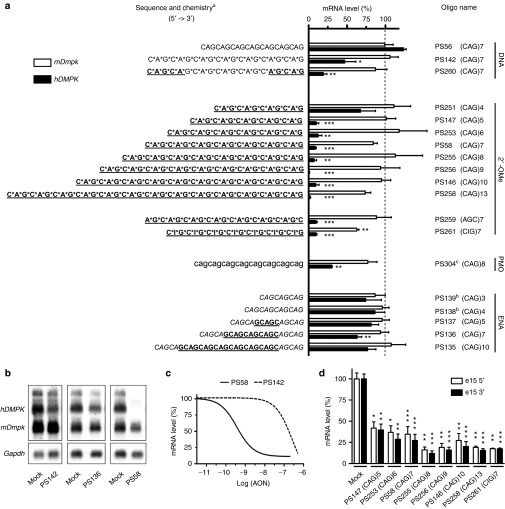
A series of triplet repeat AONs that differed in length, chemistry, and nucleotide composition was designed and analyzed for their ability to silence expanded DMPK transcripts in cultured cells (see AON description in Figure 1a). AONs were tested via transfection in the DM500 myoblast–myotube cell model.8 The human DMPK (hDMPK) transgene in this model is under near-normal myogenic transcriptional control and produces hDMPK transcripts bearing a (CUG)500 repeat at levels comparable to that of endogenous mouse Dmpk (mDmpk) transcripts. It should be noted that mDmpk transcripts contain a CCG(CUG)2(CAG)2CUG sequence instead of the generally believed to be uninterrupted (CUG)n segment found in hDMPK transcripts. Monitoring AON-silencing effects was done by northern blotting to follow steady-state levels of full-length transgenic hDMPK transcripts and possible stable breakdown products. Analysis of endogenous mDmpk transcripts was used as a control. Reverse transcription-quantitative PCR (RT-qPCR) was included for quantitative assessment of expression of defined transcript segments.
Figure 1.
Collection of triplet repeat AONs tested for their ability to silence expanded hDMPK mRNA in DM500 myotubes. (a) Summary of silencing efficiency. Endogenous mDmpk RNA was included as a negative control, since it lacks a pure (CUG)n repeat. AONs are grouped according to their main chemical modification: aDNA (capital); RNA (capital, underlined); PT, phosphorothioate linkage (*); 2′-OMe, 2′-O-methyl sugar modification (capital, underlined, bold); ENA, 2′-O,4′-C-ethylene-bridged nucleic acid (capital, italics); PMO, morpholino phosphorodiamidate (lower case); bCy3-labeled; cCoupled to octaguanidine dendrimer. The means of at least three independent experiments for each AON are shown. Dashed line indicates 100% levels (mock samples). (b) Representative northern blots with RNA isolated from AON-treated DM500 myotubes probed with a hDMPK and a Gapdh probe. Results of treatment with three AONs (200 nmol/l) or mock treatment is shown. (c) Concentration–response curves of PS58 and PS142. DM500 myotubes were treated with a concentration series of 0.01–500 nmol/l AON (see also Supplementary Figure S2). (d) RT-qPCR analysis of hDMPK RNA levels in DM500 myotubes after treatment with a selection of 2′-OMe AONs (200 nmol/l, n ≥ 3). PCR amplicons were located in exon 15, either 5′ or 3′ to the (CUG)n tract. *P < 0.05; **P < 0.01; ***P < 0.001. AON, antisense oligonucleotide; Gapdh, glyceraldehyde 3-phosphate dehydrogenase; hDMPK, human DM protein kinase; mDmpk, mouse Dmpk; RT-qPCR, reverse transcription-quantitative PCR.
DNA-based (CAG)n AONs
DNA-type oligonucleotides are known to allow specific RNase-H–mediated cleavage of RNA in a DNA–RNA heteroduplex.12 For these AONs, several chemical modifications have been developed that improve their metabolic stability, while maintaining hybridization affinity to target RNA and ability to recruit RNase H.13 We tested three DNA-type (CAG)7 AONs in DM500 myotubes: PS56, a pure DNA AON; PS142, a PT-modified DNA AON; and PS260, a PT-modified chimeric AON, so called gapmer, comprised of 2′-OMe wings on the 5′ and 3′ sides of a DNA gap. DNA AON PS56 did not show silencing activity (Figure 1a), probably because it was quickly degraded inside cells (Supplementary Figure S1). In contrast, PS142 and PS260–both stabilized against nuclease breakdown–were able to silence expanded hDMPK mRNA for ~50 and 80%, respectively (Figure 1a,b). Endogenous mDmpk mRNA was left intact. A concentration–response analysis yielded an IC50 (half maximal inhibitory concentration) of 0.3 ± 0.4 µmol/l for DNA/PT oligo PS142, which is about 1,000-fold lower than that of 2′-OMe/PT lead compound PS58 (Figure 1c, Supplementary Figure S2 and ref. 8).
Oligo length and base variations of 2′-OMe AONs
To investigate length as a potential parameter in the performance of PS58-type oligos, we tested a series of fully 2′-OMe–modified (CAG)n AONs consisting of 4–13 triplets in the DM500 myotube culture system. All, except PS251 (CAG)4, effectively and selectively silenced (CUG)500 transcripts (>80% efficiency) (Figure 1a,b). A (CUG)7 2′-OMe sense oligo or 2′-OMe oligos directed against target sequences in the hDMPK transcript outside the repeat did not have a silencing effect (data not shown).8 The lack of activity of the (CAG)4 AON is best explained by assuming that a minimum oligo length is necessary for effective hybridization to the (CUG)n target needed for AON activity.
We wondered whether the nature of the nucleotide residue at the 5′ or 3′ end of the AON might be crucial for silencing and tested a repeat-shifted version, 2′-OMe–modified (AGC)7 (PS259). PS259 treatment resulted in a significant reduction of 85% of (CUG)500 transcripts without silencing mDmpk mRNA (Figure 1a).
Finally, a 2′-OMe–modified (CIG)7 AON was tested. The inosine residues should enable this AON, PS261, to hybridize to (CUG)n, (CAG)n, (CCG)n repeats and any combination of these triplets. Similar to PS58, PS261 induced 85% reduction of expanded mRNA (Figure 1a). Notably, endogenous mDmpk mRNA was also significantly reduced, albeit with only ~35% efficiency. This effect may be best explained by binding of (CIG)7 to the CCG(CUG)2(CAG)2CUG sequence in mDmpk mRNA. Besides, three low-affinity–binding sites were identified that could have a cooperative, additive effect (11–13 nucleotides match; not present in hDMPK transcripts).
Silencing of expanded hDMPK transcripts by triplet repeat 2′-OMe AONs in DM500 myotubes was independently confirmed by RT-qPCR analysis (Figure 1d). The lower silencing efficiency measured by RT-qPCR (60–90 versus 85–95% detected by northern blotting) is attributed to technical differences in sensitivity intrinsic to the methods.
Morpholino phosphorodiamidate (CAG)n AONs
Morpholino phosphorodiamidate oligos are uncharged AONs widely used as blocking oligos for selective inhibition of gene expression.14 A (CAG)8 morpholino (PS304) was tested for its ability to silence expanded hDMPK RNA in DM500 myotubes. The morpholino was coupled to an octaguanidine dendrimer (so called Vivo-Porter) to facilitate cellular uptake. PS304 treatment caused a 70% reduction of (CUG)500 transcripts, leaving mDmpk mRNA intact (Figure 1a).
2′-O,4′-C-ethylene-bridged nucleic acid-modified (CAG)n AONs
The bicyclic modification in 2′-O,4′-C-ethylene-bridged nucleic acid (ENA) confers strong nuclease resistance and high-binding affinity to complementary single-stranded RNA (average δTm (δmelting temperature)/modification = 5.5 versus 1.3 °C for 2′-OMe)15 (Supplementary Figure S1). We tested two fully ENA-modified AONs consisting of three and four CAG triplets, and three ENA/2′-OMe gapmers of 5, 7, and 10 CAGs (PS139-135, Figure 1a). Only treatment with PS136 resulted in a small, yet significant reduction (30%) of hDMPK RNA expression.
The discrepancy in efficacy between ENA- and 2′-OMe–modified AONs may be caused by differences in subcellular targeting. To verify this possibility, we followed cellular uptake and routing of two fluorescently labeled AONs with these modifications. For these analyses, we switched to the use of DM500 myoblasts, which display a rather flat morphology, rendering them more suitable for live cell imaging of AON localization than myotubes. Forty minutes after transfection, fluorescent PS58 oligo was present in the cell nucleus and signal intensity further increased up to 60 minutes (Supplementary Figure S3). This behavior was independent of the type of fluorophore used and fluorophore coupling did not affect PS58 silencing activity (data not shown). In contrast, labeled PS138, as a representative of the ENA AON collection, was concentrated in vesicle-like structures and rarely present in the nucleus itself. This pattern remained unchanged up to 48 hours after transfection (data not shown). We conclude that lower silencing efficiency of ENA-modified (CAG)n AONs observed in this study is due to their inability to reach muscle cell nuclei, where most expanded (CUG)n transcripts reside.
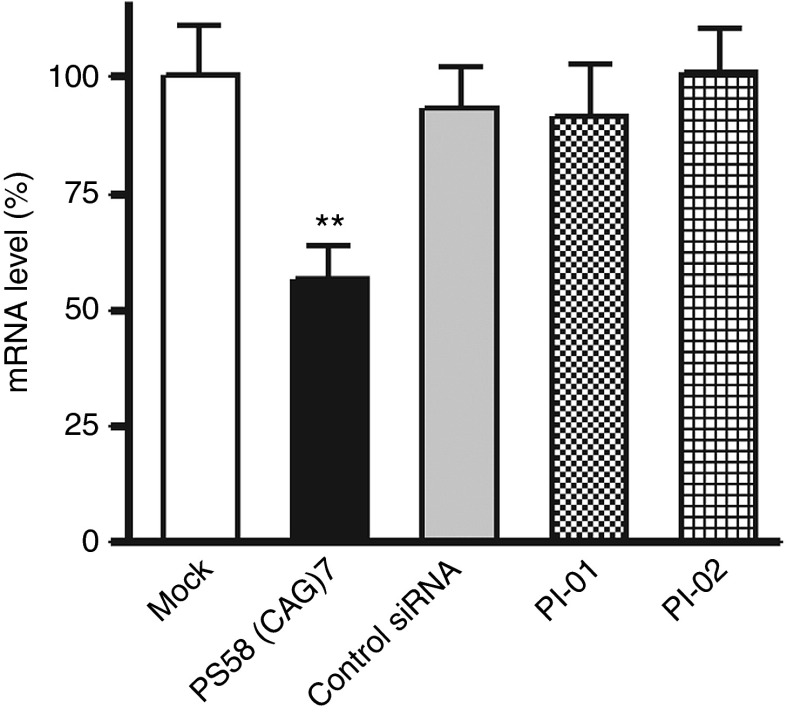
Triplet repeat siRNAs
Krol et al.16 reported that (CXG)n triplet repeat hairpins in mRNAs are substrates for Dicer and produce short repeat RNAs that in turn may act as short-interfering RNAs (siRNAs) against complementary repeats. We wanted to investigate whether in our cell model expanded hDMPK mRNAs could be silenced by introducing synthetic (CAG)n/(CUG)n siRNAs. Two different RNA duplexes were transfected in DM500 myotubes, PI-01 ((CAG)7/(GCU)7) and PI-02 ((GCA)7/(CUG)7) (Supplementary Table S1). An unrelated siRNA and PS58 were included as controls. RT-qPCR analysis demonstrated that none of the repeat siRNAs silenced hDMPK mRNA expression (Figure 2). From our AON stability assay, we conclude that it is unlikely that the inability to reduce expanded hDMPK transcripts is due to rapid intracellular degradation of the chemically unprotected siRNAs (Supplementary Figure S1).
Figure 2.
RT-qPCR analysis of hDMPK (CUG)500 RNA levels in DM500 myotubes after treatment with siRNAs (50 nmol/l) targeting the (CUG)n repeat (n = 6). PS58 (50 nmol/l) was used as a control. Sequences of PI-01 ((CAG)7/(GCU)7 duplex), PI-02 ((GCA)7/(CUG)7 duplex), and control siRNA are listed in Supplementary Table S1. **P < 0.01. hDMPK, human DM protein kinase; RT-qPCR, reverse transcription-quantitative PCR; siRNA, short-interfering RNA.
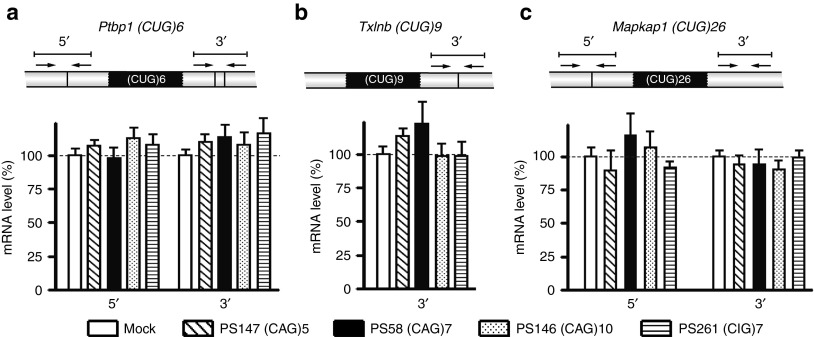
Mouse transcripts containing a small (CUG)n segment
Transcripts that contain a (CUG)n segment, other than encoded by the hDMPK transgene, may also serve as targets for (CAG)n AONs in DM500 cells. Genes carrying a (CTG)n segment are rare, but at least nine transcripts in the mouse transcriptome contain a repeat of more than six triplets.8 We investigated whether 2′-OMe/PT AONs of variable length and sequence ((CAG)5, (CAG)7, (CAG)10, and (CIG)7) acted on a subset of these transcripts expressed in DM500 myotubes. We analyzed transcript levels by RT-qPCR based on amplicons next to the repeat tracts (Figure 3; primer locations are depicted in more detail in Supplementary Figure S4). Ptbp1 (CUG)6, Txlnb (CUG)9, and Mapkap1 (CUG)26–the longest (CUG)n tract in the mouse transcriptome–showed no significant changes in expression after AON treatment (Figure 3). Note that due to the location of the (CUG)9 tract in the very 5′ end of the Txlnb gene, no suitable 5′ amplicon could be designed to measure Txlnb transcripts.
Figure 3.
Expression of three mouse transcripts, carrying a small (CUG)n repeat, after transfecting DM500 myotubes with a selection of AONs. (a) Ptbp1 (CUG)6, (b) Txlnb (CUG)9, and (c) Mapkap1 (CUG)26. RNA levels were analyzed by RT-qPCR amplifying segments flanking the triplet repeat (n = 3). Schemes illustrate exon–exon junctions, locations of PCR primers, and amplicons. Dashed lines indicate 100% levels (mock samples). AON, antisense oligonucletide; RT-qPCR, reverse transcription-quantitative PCR.
We also wanted to analyze RNA expression via amplification across the (CUG)n triplet repeat. Semi-quantitative RT-PCR for Ptbp1 and Txlnb transcripts confirmed the RT-qPCR findings and showed no significant change in expression (Supplementary Figure S5a,b). Remarkably, quantification across the Mapkap1 (CUG)26 repeat did record significant silencing after AON treatment, especially by PS146 (CAG)10 (Supplementary Figure S5c). This effect was not seen, however, when segments flanking the (CUG)26 tract were amplified in the same assay. Also northern blotting, visualizing two types of Mapkap1 transcripts as a result of alternative polyadenylation17 (Supplementary Figure S4), revealed only a minor reduction in RNA expression, whereas western blotting revealed no significant changes in Mapkap1 protein expression (Supplementary Figure S5d,e). We therefore decided that PCR-based quantification should not rely on amplification across a medium-sized (CUG)n tract.
Given these observations, we wanted to exclude the possibility that traces of AONs that might still be present in purified RNA samples would interfere with RT-PCR analysis. Using a hybridization–ligation method, we determined that free AON concentration in our RNA preparations was <0.2 nmol/l. To rule out interference of (CAG)n AONs with RT or PCR amplification, untreated DM500 RNA samples were spiked with 0.1–10 nmol/l PS147, PS58 or PS146 and correspondingly used in RT-PCR analysis. Presence of AONs during RT-PCR amplification did not have an effect on Mapkap1 (CUG)26 levels nor on quantification of hDMPK (CUG)500 RNA (Supplementary Figure S6). In sum, we conclude that transcripts containing small (CUG)n tracts were largely unaffected by the (CAG)n and (CIG)n repeat oligos tested.
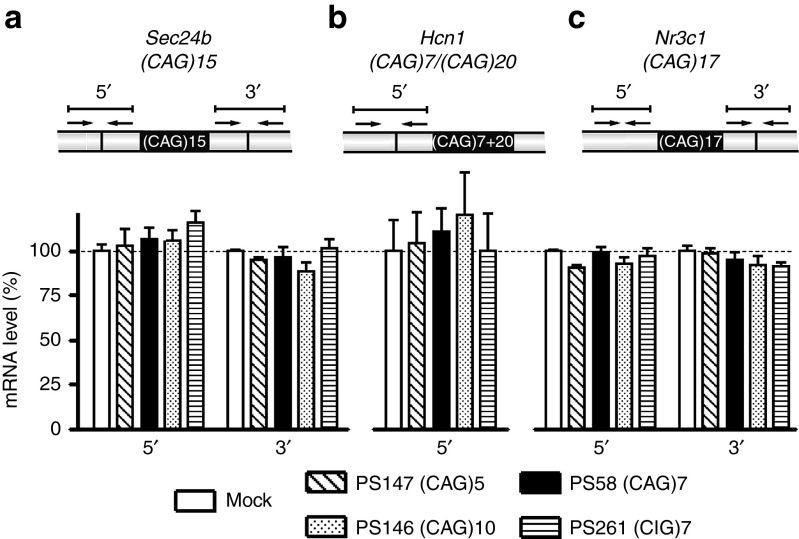
Mouse transcripts carrying a small (CAG)n segment
To examine the possibility that oligo-mediated hDMPK RNA breakdown would generate short (CUG)n RNAs that in turn might act as siRNAs on complementary (CAG)n transcripts (compare, ref. 16), we measured levels of mouse transcripts with a small (CAG)n segment: Sec24b (CAG)15, Hcn1 (CAG)7+(CAG)20, and Nr3c1 (CAG)17. After treatment with PS147, PS58, PS146, and PS261; no significant change in expression of any of these transcripts was detectable by RT-qPCR analysis (Figure 4) ruling out an indirect effect of (CAG)n AONs on transcripts bearing a (CAG)n segment in the transcriptome.
Figure 4.
Analysis of expression levels of three mouse transcripts, containing a (CAG)n repeat, in DM500 myotubes after transfection with a selection of AONs. (a) Sec24b (CAG)15, (b) Hcn1 carrying a (CAG)7 and a (CAG)20 repeat located close together (see Supplementary Figure S4), and (c) Nr3c1 (CAG)17. Expression was analyzed by RT-qPCR amplifying segments flanking the triplet repeat (n = 3). Schemes illustrate exon–exon junctions, locations of PCR primers, and amplicons. Dashed lines indicate 100% levels (mock samples). AON, antisense oligonucletide; RT-qPCR, reverse transcription-quantitative PCR.
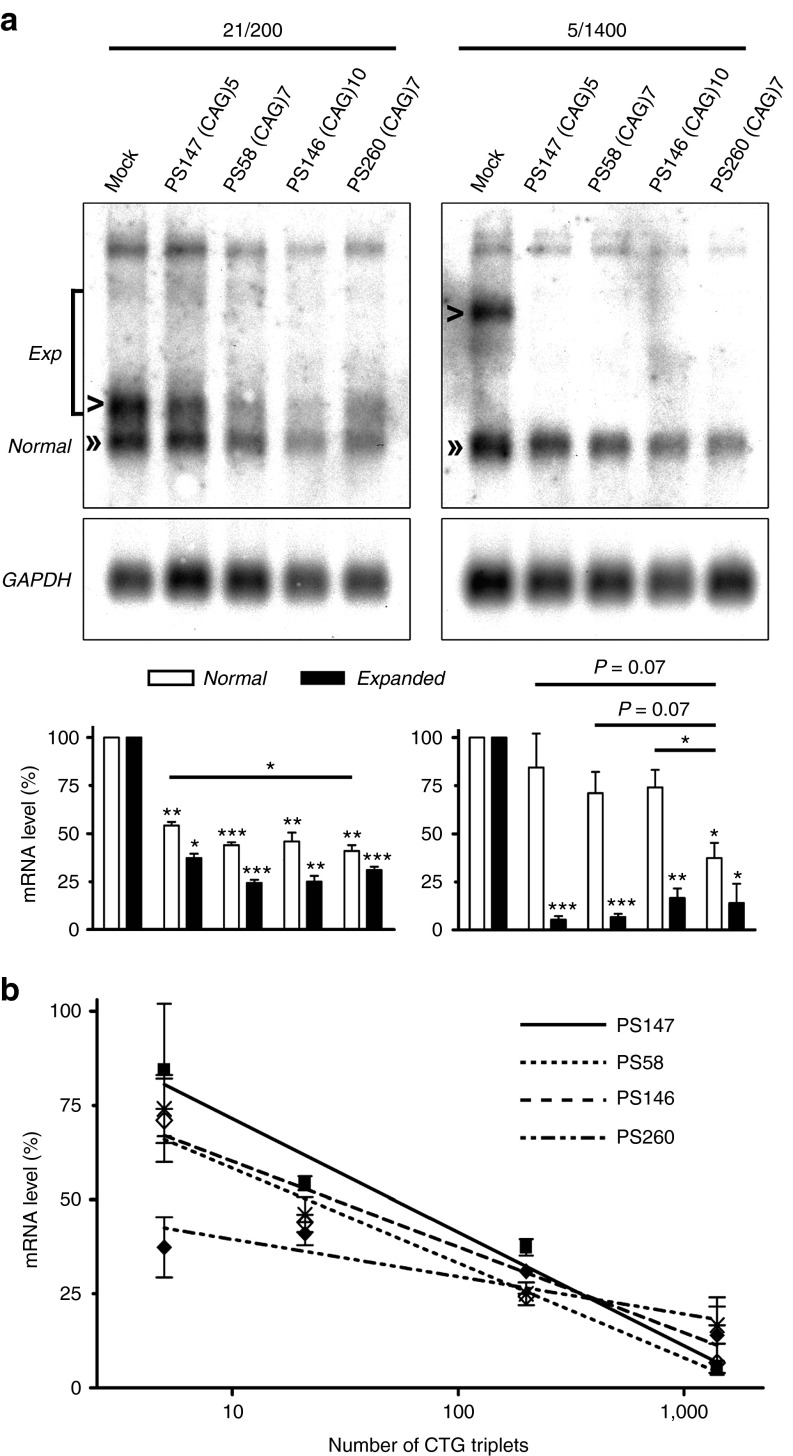
(CUG)n length selectivity in DM1 patient myoblasts
A safe, antisense strategy for DM1 should differentiate between expanded and normal-sized hDMPK transcripts. To test this requirement, we used two DM1 myoblast cultures: one expressing hDMPK transcripts with 21 (normal) and 200 (mutant) triplets (abbreviated 21/200) and another with 5 and 1,400 CUG triplets (abbreviated 5/1,400).18 Activities of three 2′-OMe AONs, PS147, PS58, and PS146 were examined in these cultures, with use of northern blotting to discriminate between normal and mutant hDMPK RNA. The three pure 2′-OMe/PT AONs behaved equally (Figure 5a): levels of (CUG)1400 and (CUG)200 transcripts were strongly decreased (65–95%). (CUG)21 transcripts appeared less susceptible to breakdown (45–55%) and as expected, (CUG)5 hDMPK mRNA was left intact. Silencing efficacy significantly correlated with the number of CUG triplets in the transcripts (Figure 5b).
Figure 5.
(CUG)n length selectivity in DM1 myoblasts. (a) Human myoblasts expressing either hDMPK (CUG)21 and (CUG)200 transcripts (21/200) or hDMPK (CUG)5 and (CUG)1400 transcripts (5/1400) were treated with a selection of CAG AONs. Northern blot analysis indicated that expanded hDMPK transcripts (“exp”, arrow heads) were efficiently silenced by all AONs. (CUG)21 transcripts (“normal”, double arrow heads) were moderately silenced. Only RNase-H recruiting PS260 substantially degraded (CUG)5 transcripts (“normal”, double arrow head). One representative blot of three independent experiments is shown for each cell culture. Quantification of signals is summarized in the graphs at the bottom. *P < 0.05; **P < 0.01; ***P < 0.001. (b) A significant correlation was demonstrated between the number of CTG triplets and silencing activity of PS147, PS58, and PS146 (P < 0.05, Pearson's correlation, r = −0.98), but not PS260. AON, antisense oligonucleotide; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; hDMPK, human DM protein kinase.
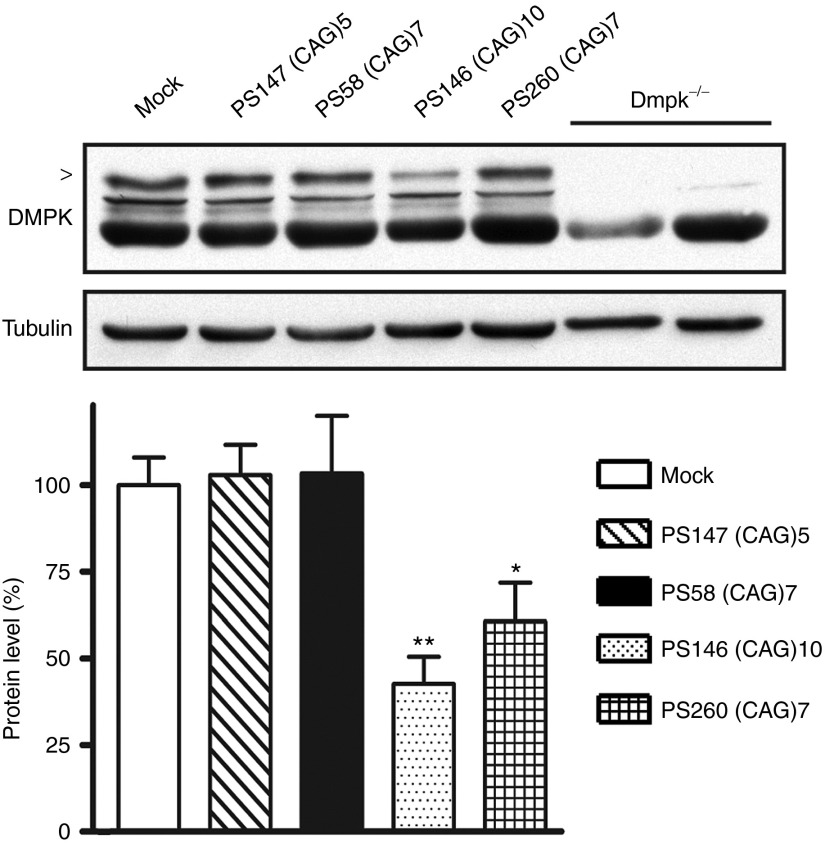
Western blot analysis showed that silencing of hDMPK transcripts by PS146 (CAG)10 resulted in around 50% loss of DMPK protein after 72 hours in 21/200 myoblasts (Figure 6). No effect of PS147 (CAG)5 or PS58 (CAG)7 treatment was observed at this time point.
Figure 6.
Analysis of DMPK protein expression after AON treatment. Human myoblasts (21/200) expressing hDMPK (CUG)21 and (CUG)200 transcripts were treated with a selection of (CAG)n AONs. After 72 hours, protein lysates were made and DMPK protein (arrow head) levels were analyzed by western blotting (n = 3). Tubulin protein was used as a loading control. Lysate from Dmpk knockout myoblasts served as reference (right two lanes). *P < 0.05; **P < 0.01. AON, antisense oligonucleotide; hDMPK, human DM protein kinase.
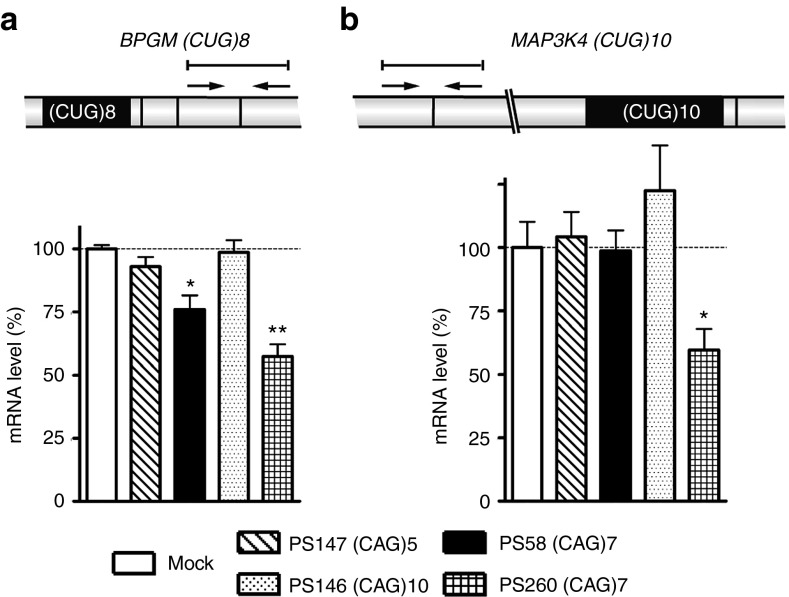
Human transcripts containing a small (CUG)n segment
Also in the human transcriptome (CUG)n tracts are relatively rare.8,19 Effects of a subset of 2′-OMe/PT AONs on transcripts bearing a (CUG)n repeat expressed in 5/1,400 myoblasts were examined for two transcripts: BPGM (CUG)8 and MAP3K4 (CUG)10 by RT-qPCR. No reduction of BPGM transcripts was detected after treatment with PS147 and PS146, whereas only a small effect was seen with PS58 (25% reduction) (Figure 7a). Similar effects for BPGM (CUG)8 were detected in 21/200 myoblast cultures (data not shown).
Figure 7.
Analysis of expression of two transcripts, each containing a small (CUG)n repeat, in DM1 patient myoblasts (5/1400) after transfection with a selection of AONs. RNA levels of (a) BPGM (CUG)8 and (b) MAP3K4 (CUG)10 were measured by RT-qPCR (n = 3). Schemes illustrate exon–exon junctions and locations of PCR primers and amplicons. Dashed lines indicate 100% levels (mock samples). *P < 0.05; **P < 0.01. AON, antisense oligonucleotide; RT-qPCR, reverse transcription-quantitative PCR.
Total MAP3K4 mRNA levels were not affected by 2′-OMe AON treatment (Figure 7b). A conspicuous observation was made with regard to an effect on MAP3K4 pre-mRNA processing. Analysis of the fate of alternative exon 17,20 which contains the short (CUG)10 repeat that might serve as target, revealed that PS146 (CAG)10 was able to enhance exon 17 skipping (Supplementary Figure S7). Effects on alternative splicing by shorter (CAG)n oligos were only minor.
Investigating potential mechanisms of expanded hDMPK transcript silencing
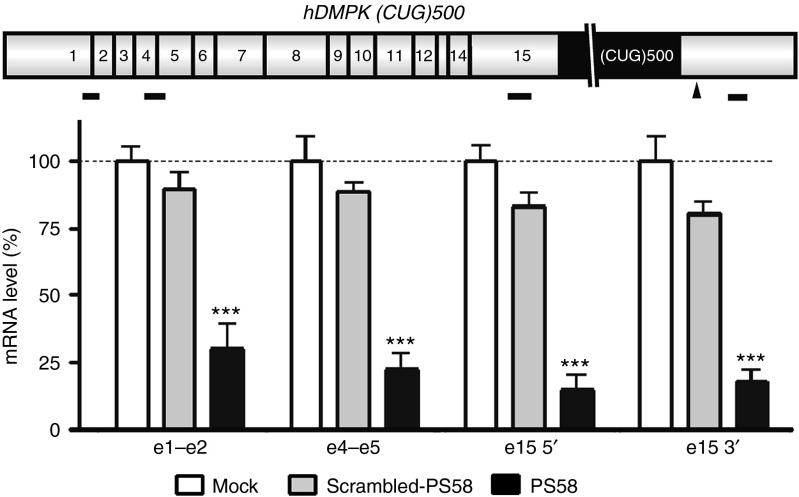
We wondered whether exon skipping could also be the underlying or initiating mechanism of PS58-induced silencing of expanded hDMPK transcripts. hDMPK pre-mRNAs are subject to extensive alternative splicing and events involving final exon 15, in which the (CUG)n tract is located, have been reported.21,22 For analysis of fate of different hDMPK RNA segments in presence of AONs, we used a transcript scanning RT-qPCR approach for amplicons along the expanded hDMPK transcript in DM500 myotubes. Amplicons were designed in the 5′ end of the gene (exon 1–exon 2, exon 4– exon 5) and on either side of the (CUG)n tract in exon 15 (Figure 8). No significant differences were found in signal strength for any of these hDMPK segments (70–85% reduction for all segments across the transcript, compared with mock-treated samples). This finding indicates that PS58-induced skipping of the (CUG)n tract during hDMPK pre-mRNA splicing is unlikely to be mechanistically involved.
Figure 8.
Human DMPK (CUG)500 RNA expression in DM500 myoblasts after transfection with PS58 or scrambled-PS58. Expression of four different transcript segments (little black bars) was analyzed by RT-qPCR (n = 3): two in the 5′ end of the transcript and two flanking the triplet repeat in the 3′ end of the transcript. Scheme illustrates exon composition (numbered boxes) of hDMPK transcript, arrowhead indicates reported cryptic splicing site 3′ to the triplet repeat.22 Dashed lines indicate 100% levels (mock samples). No significant differences in response to the oligos were observed between the four segments. ***P < 0.001. hDMPK, human DM protein kinase; RT-qPCR, reverse transcription-quantitative PCR.
Test tube incubation of radioactively labeled, in vitro-transcribed (CUG)90 RNA with a concentration series of PS58 (CAG)7 caused a mobility shift illustrating direct oligo–RNA association (Supplementary Figure S8). Similar shifts were seen after incubation of different (CAG)n and (CUG)n AONs with (CUG)90 or (CAG)90 RNAs (Supplementary Figure S8). To exclude that triplet repeat 2′-OMe AONs had an intrinsic RNase activity, similar to ribozymes or DNAzymes,23 (CUG)n RNA was incubated with a series of different AONs in the presence of various concentrations of Mg2+, Mn2+ or Zn2+ (Supplementary Figure S8; data not shown). No (CUG)n breakdown products could be detected, indicating that under the conditions tested, none of these AONs displayed RNase-like activity when complexed to (CUG)n RNA.
Using the same (CUG)90 RNA template, we could confirm that PS58, a fully modified 2′-OMe AON, was unable to induce RNase-H–mediated degradation of expanded (CUG)n transcripts in vitro (Supplementary Figure S8). As positive controls, we used DNA-type AON PS142 and gapmer PS260, both of which were able to recruit RNase-H to cleave (CUG)90 RNA. Activity of gapmer PS260 was also examined in human myoblasts, but with an apparent lack of specificity (Figures 5 and 6). More than 60% of both expanded and normal-sized hDMPK transcripts were degraded. Similar lack of specificity was observed for BPGM (CUG)8 and MAP3K4 (CUG)10 RNAs, for which a 40% reduction in steady-state copy number was observed (Figure 7). Since the MAP3K4 exon 17+ variant and the exon 17− variant (which lacks the AON target site) were equally reduced by PS260, we conclude that RNase-H–dependent degradation must have occurred in the nucleus, before exon 17 splicing (Supplementary Figure S7).
Complement activation by triplet repeat AONs
All triplet repeat oligos were well tolerated by cells in culture and essentially no toxic effects on growth or behavior were observed (data not shown). A well-known toxic side effect of AONs in vivo, especially PT-modified versions, is activation of the complement pathway.24 As a prelude to later preclinical studies in animals and humans, we tested a selection of our PT-modified AONs with different chemistries and sequences for their ability to activate complement in plasma in vitro: full 2′-OMe (PS58, PS146, PS147, PS261), chimeric 2′-OMe/DNA (PS260), and pure DNA (PS142). A first generation oligonucleotide known to activate the complement pathway was included as a positive control.25 Formation of complement split products C3a (marker for the classical and alternative pathway) and Bb (marker for the alternative complement pathway) was measured in plasma from cynomolgus monkeys and humans after addition of AONs (Supplementary Figure S9). In particular, PS261 (CIG)7, and to a lesser extent PS146, PS147, and PS260, caused a significant dose-dependent increase in the concentration of Bb in cynomolgus plasma. Only PS261 and PS260 activated C3a formation in human plasma. Other AONs showed no significant or minor effects.
Discussion
This study demonstrates that a range of triplet repeat AONs is capable of silencing expanded hDMPK transcripts. The long (CUG)n target that characterizes toxic hDMPK transcripts is a special RNA sequence in the sense that it can form a stem-loop structure that folds almost like an antiparallel double-stranded helix, despite the periodic U•U mismatches.26 We conclude that the thermodynamically stable hairpin structure, known to sequester members of the MBNL protein family, does not hamper accessibility of antisense repeat AONs, when in the cellular context. The (CUG)n repeat, whose length differs between normal-short and mutant-expanded hDMPK transcripts, is therefore an attractive therapeutic target.
Chemical modifications of the sugar phosphate backbone of AONs serve to confer resistance against nucleases to extend cellular half-life.13 The silencing activity of modified AONs PS142 and PS260 versus the inability of pure DNA AON PS56 to induce breakdown clearly illustrates the relationship between stability and efficacy. Oligo chemistry may also determine nucleocytoplasmic distribution of AONs. As target transcripts in DM1 are concentrated in the nucleus, an AON must be capable to enter this compartment to become useful. Microscopic evidence is provided that ENA-modified AONs remain in vesicular structures and only inefficiently reach the nucleus, rendering them a poor choice for repeat-directed antisense DM1 strategies. We have no simple explanation for this finding, as similarly modified 2′-OMe RNA/ENA chimeras have shown activity in the nucleus during exon skipping for Duchenne muscular dystrophy.27 Effects of nucleotide sequence on subcellular oligo localization cannot be excluded at this point. Also the type of cell model and transfection method used may have an effect.
Not all types of AONs were equally efficient and specific, but as we compared only triplet repeat oligos here, some conspicuous observations are worth mentioning. First, as an explanation for the failure of (CUG)n/(CAG)n triplet repeat siRNAs to induce an RNAi response, we assume that a combination of chemistry and oligo sequence must be responsible. The poor performance of the two duplexes tested can be the result of unfavorable subcellular distribution of siRNAs, although the RNAi machinery is known to be active in the nucleus.28 Alternatively, the (CUG)n RNA hairpin may be insufficiently accessible to certain components of the RNAi pathway in our myogenic cell model. Very recently, a comparable triplet repeat siRNA was successfully tested in vivo in skeletal muscle in a mouse model for DM1. (CUG)220 expanded RNA and also Txlnb (CUG)9 transcripts were reduced, but not Mapkap1 (CUG)25 RNA.29
Second, oligo PS261 (CIG)7 was initially considered a promising new lead compound, because of its predicted ability to bind to (CUG)n as well as (CAG)n transcripts, with potential for treatment of DM1 and polyQ diseases like Huntington's disease and several spinocerebellar ataxias. From the complement activation assay we learned, however, that this AON activated the complement cascade, rendering it a less favorable candidate for further development. Virtually none of the other AONs showed major adverse effects in vitro, with the best performing oligo being PS58. Screening for toxicity profiles in animals in vivo will be a logical next step.
Third, RNase-H recruiting AON PS260 appeared to have prominent silencing capacity, but also suffered of lack of target specificity. Further characteristics of these DNA-type AONs will be discussed below in the context of mechanistic considerations.
The main focus of our study had to do with AONs based on 2′-OMe/PT chemistry. For these AONs, we demonstrated that oligo length may provide a means to introduce selectivity between expanded transcripts from a mutated hDMPK allele and RNA containing a normal, short (CUG)n tract. The shortest variant in our series of 4–13 CAG triplets was unable to knockdown expanded transcripts in DM500 myotubes. The hybridization strength of the four CAG triplet AON may have been insufficient for a stable interaction with a (CUG)n segment in the living cell, even though the theoretical Tm for this interaction is around 39 °C.
AONs longer than four triplets efficiently silenced transgenic (CUG)500 transcripts. Significant (CUG)n repeat length-dependent activity in patient cells was displayed by (CAG)5, (CAG)7, and (CAG)10 2′-OMe/PT AONs. Between them they showed no significant difference in activity towards normal-sized hDMPK transcripts: (CUG)5 transcripts, expressed by ~40% of the Caucasian population,4 were left intact and (CUG)21 transcripts were ~50% reduced by all three AONs. In contrast, differential effects were observed at the DMPK protein level, which might indicate a correlation with translation efficiency of remaining hDMPK transcripts. It should be noted that we cannot predict at the moment whether partial reduction of normal-sized transcripts will have any adverse consequences. We know that Dmpk knockout mice have relatively mild phenotypes,30 hence a low level of remaining Dmpk protein might be sufficient to protect functional and structural integrity of cells in which it is expressed. Another still unresolved issue is the minimum efficacy needed for a beneficial effect. Partial suppression of toxic (CUG)n RNA may already be of clinical relevance in DM1 patients.
Expression of genes with a small (CTG)n repeat in myogenic cells was essentially unchanged upon treatment with 2′-OMe/PT AONs, except for MAP3K4. PS146 (CAG)10 enhanced skipping of MAP3K4 exon 17 in which the (CUG)10 repeat is located. This (CUG)10 repeat is unusual as it is located in an alternative exon, where it might function as an exon-splicing enhancer.31 To what extent exon 17+ and exon 17− transcript variants contribute to protein production is unknown. More importantly, it is unclear whether a shift in MAP3K4 isoform ratio would have a biological effect. Nothing is known about functional differentiation generated by inclusion of the 50 amino acid stretch encoded by in-frame exon 17 (including 10 alanines encoded by the repeat tract). The minor shift in MAP3K4 variant ratio induced by smaller (CAG)n AONs is essentially within the normal variation observed in mock samples. Note that since splicing is a nuclear process, we can conclude that PS146 (CAG)10 must have been active inside the nucleus.
It is difficult to reconcile all observations regarding Mapkap1 (CUG)26 transcripts. First, as also seen with MAP3K4 transcripts, oligo length is an important parameter for AON activity and side effects seem to intensify with increased oligo length. Second, we provide ample evidence that our oligos did not interfere with RT-PCR quantification. Third, we propose that special care should be taken with RNA quantification based on amplicons covering a triplet repeat of >10 units. These amplicons will never meet requirements for RT-qPCR analysis, but they may also be useless in semi-quantitative RT-PCR. For that reason, we generally used segments outside the repeat region for PCR quantification. Similar warnings regarding RT-PCR and use of AONs have been issued by others.32 Given the location of the (CTG)26 repeat in between two alternative polyadenylation sites of the Mapkap1 gene,17 we cannot exclude length-dependent AON effects on splicing, transcription termination or polyadenylation. Clearly, more work is needed to be able to unequivocally interpret these data.
What do we know about the molecular mechanism underlying silencing activity of hDMPK transcripts by triplet repeat AONs examined here? Although the goal of this study was not to identify distinct pathways or describe novel interference principles, our findings provide important new insight in potential activities of triplet repeat AONs.
In a series of simple biochemical test tube experiments, we first investigated whether triplet repeat 2′-OMe/PT AONs might carry intrinsic endoribonuclease activity, similar to ribozymes or DNAzymes.23 In none of the experiments RNase activity was detected. Although we also tested several cell-free or protein-free conditions (data not shown), it cannot be excluded that the right cellular reaction conditions were not fully reproduced.
We provide evidence that PS260, a (CAG)7 gapmer, efficiently reached various (CUG)n RNA targets, irrespective of repeat length, and recruited endogenous RNase-H for a first endonucleolytic cleavage. We expect that unprotected 5′ and 3′ RNA segments will be degraded after initial cleavage by conventional exonucleases as part of the quality control system in the cell.33 Surprisingly, PS260 showed only little selectivity between expanded and normal-sized hDMPK transcripts in patient cells, suggesting that either RNase-H activity took place in both the nucleus and the cytosol, or that degradation already occurred swiftly, during or shortly after RNA synthesis in the nucleus. Our observations contrast with a report that an LNA-based repeat gapmer did cleave hDMPK (CUG)40, but not (CUG)12 transcripts;10 and also with a study in which a (CAG)6 LNA gapmer did not reduce CASK (CUG)16 RNA.34 The reason for these discrepancies should probably be found in oligo chemistry, concentration or model system used.
Nucleotide analogs like morpholinos or modifications like 2′-OMe do not support RNase-H–dependent degradation. These AONs must therefore use distinct pathways, explaining why PS58, a 2′-OMe/PT (CAG)7 AON, and PS260, displayed differential (CUG)n length-dependent silencing.
The RNAi pathway is unlikely to be involved in activity of fully 2′-OMe/PT AONs or (CAG)n morpholinos. Imperfect hybrids of 2′-OMe/PT or morpholino (CAG)n AONs, or perfect hybrids that these AONs may form with (CUG)n transcripts, will not be recognized by the RNAi machinery.35,36,37 Furthermore, the size of the shortest effective AON (15 nucleotides) and tolerated 5′ modifications like a fluorescent group (as in FAM-PS58) or a peptide (data not shown) render involvement of the RNAi pathway highly unlikely.38,39 Also, we did not find evidence for AON-induced processing of expanded hDMPK transcripts into (CUG)n siRNAs that could trigger degradation of (CAG)n transcripts.
Fully 2′-OMe/PT-modified AONs, like PS58, and morpholinos are usually designed and employed to serve as blocking oligos to modulate splicing or prevent translation.13,40 As discussed above, PS146, a 30 nucleotide long 2′-OMe/PT AON, exhibits splicing modulating activity. We could not detect a similar activity of (CAG)n AONs towards hDMPK (CUG)500 transcripts. The most logical explanation for this observation is the location of the (CUG)n repeat in the final exon of the hDMPK gene. The only reported splice acceptor, located around 35 nucleotides 3′ of the (CUG)n tract, is only rarely used22,41 and probably too weak to be amenable to manipulation.
A (AGC)8A morpholino reportedly blocked Mbnl1 protein binding to transgenic (CUG)250 transcripts in a mouse model for DM1-related RNA toxicity.9 In that study, cytoplasmic levels of expanded transcripts raised upon treatment, but the total amount of (CUG)250 transcripts was ~50% reduced. A comparable reduction was reported for expanded Huntingtin (CAG)n RNA using a (GCT)6G LNA-type blocking oligonucleotide.42 The molecular mechanism underlying the activity of AGC morpholino is unknown, but it was suggested that the displacement of expanded transcripts to the cytoplasm may have induced hDMPK mRNA decay. We propose that a similar mechanism may be at play for our (CAG)n morpholino and the 2′-OMe/PT AONs examined here. Alternatively, this class of AONs may activate a yet to be identified RNase or RNA decay program in nucleus or cytoplasm. Our current working hypotheses are that the interaction between (CAG)n AONs and the (CUG)n repeat attracts proteins involved in RNA degradation or that AON binding displaces RNA-protecting proteins resulting in naked RNA, which is extremely sensitive to RNases. Further detailed studies into these interesting possibilities are warranted.
In sum, we provide further evidence that the expanded (CUG)n segment in hDMPK mRNA, being the product from the only known mutation in DM1, forms an attractive target for AON therapy. For development of future treatment modalities, chemical structure and oligo length in combination with the rather special sequence content needed—i.e., the complementary (CAG)n repeat character—have now been identified as crucial determinants. By extrapolation of findings, our results also open up interesting possibilities for therapy of other triplet repeat diseases.
Materials and Methods
Cell culture. Immortalized DM500 myoblasts were derived from DM300-328 mice43 and cultured and differentiated to myotubes as described.8 Human DM1 myoblast lines were derived and cultured as described.18
Oligonucleotides. AONs and siRNAs were complementary to the (CUG)n repeat. Their sequence and chemistry are listed in Figure 1a and Supplementary Table S1, except for control oligos scrambled-PS58: 5′-CAGAGGACCACC AGACCAAGG-3′ and PS57: 5′-CUGCUGCUGCUGCUGCU GCUG-3′ (both fully 2′-OMe/PT-modified). All AONs and siRNAs were provided by Prosensa (Leiden, The Netherlands), except morpholino PS304, which was purchased coupled to an octaguanidine dendrimer from Gene Tools (Philomath, OR) and control siRNA, purchased from Thermo Scientific Dharmacon (Lafayette, CO).
Oligotransfection. AONs were transfected using polyethyleneimine (ExGen 500; Fermentas, Glen Burnie, MD) according to the manufacturer's instructions. Typically, 5 µl polyethyleneimine/µg AON was added at a final AON concentration of 200 nmol/l in differentiation medium to myotubes on day 5 of myogenesis or in Opti-MEM (Invitrogen, Carlsbad, CA) to myoblasts. For concentration–response experiments, oligo concentrations between 0.01 and 500 nmol/l were applied. Four hours later, fresh medium was supplemented to a maximum volume of 2 ml medium/well. Medium was changed 20 hours later. RNA was isolated 48 hours after start of transfection.
PS304 was tested following the protocol above with the exception that no transfection reagent was used.
siRNAs were administered twice (on subsequent days: day 4 and 5 of myogenesis) using a Lipofectamine 2000-based transfection protocol (Invitrogen) according to the manufacturer's instructions, at a final concentration of 50 nmol/l. Four hours following addition of the transfection mix and 24 hours after start of the second transfection, medium was changed. RNA was isolated 48 hours after the second transfection.
Oligo stability. AONs (7.5 pmol) were radioactively labeled by T4 polynucleotide kinase in the presence of [γ-32P]-ATP and then purified on G50 columns followed by ethanol precipitation.
DM500 myoblast whole cell extract was prepared by scraping ~1 × 106 cells per well in phosphate-buffered saline (PBS) containing 1 mmol/l EDTA. Cells were collected via centrifugation (1,000 rpm, 5 minutes), packed cell volume was estimated and cells were resuspended in 1× packed cell volume of 10 mmol/l Tris-HCl; pH 8.0, 1.5 mmol/l MgCl2, 10 mmol/l KCl, 1 mmol/l DTT and 1× Protease Inhibitory Cocktail (Roche, Almere, The Netherlands). Cell suspensions were placed on ice for 15 minutes and homogenized using a pestle. Whole cell lysate was collected after centrifugation (14,000g, 10 minutes).
AONs (~2 pmol) were incubated with 5 µl whole cell lysate at 37 °C for 0, 3 or 24 hours, reaction was stopped by adding 140 mmol/l sodium acetate; pH 5.0, phenol/CHCI3 and water phase was collected (14,000g, 10 minutes, 4 °C). Oligo was precipitated by the addition of three volumes of ethanol. The oligo precipitate was washed with 70% (vol/vol) ethanol, air dried and dissolved in sample buffer (75% deionized formamide, 25% glycerol). Samples were loaded on a 12% (wt/vol) acrylamide/bisacrylamide (37.5:1) denaturing urea (8 mol/l) PAGE gel in 1× MOPS buffer. After running, gels were dried at 70 °C for 2 hours and exposed to X-ray film (X-Omat AR; Kodak, Rochester, NY).
RNA isolation. RNA was isolated using the Aurum Total RNA Mini Kit (Bio-Rad, Hercules, CA), according to the manufacturer's protocol.
Northern blotting. Northern blotting was done as described.8 Random-primed 32P-labeled human DMPK (2.6 kb cDNA), rat Gapdh (1.1 kb cDNA), and mouse Mapkap1 (1.1 kb cDNA) probes were used. Signals were quantified by phospho-imager analysis (GS-505 or Molecular Imager FX; Bio-Rad) and analyzed with Quantity One (Bio-Rad) or ImageJ software. Gapdh levels were used for normalization. RNA levels of control samples were set at 100%.
RT-PCR. Primer sets for PCR were designed using Primer-BLAST44 in the National Center for Biotechnology Information database and validated in silico using OligoAnalyzer 3.145 to prevent formation of hairpins and dimers during amplification. Resulting products were visualized on agarose gels and sequenced to verify identity and triplet repeat length. Primer sequences are listed in Supplementary Table S2.
For RT, typically 0.5 µg RNA was subjected to cDNA synthesis using the SuperScript first-strand synthesis system with random hexamer primers in a total volume of 20 µl. For RT-PCR experiments in presence of AONs, AONs were added to the RNA to reach the indicated final concentration and incubated for 15 minutes on ice before starting the RT reaction.
For semi-quantitative RT-PCR, 1 µl of cDNA preparation was used in a PCR according to standard procedures. In RT− control experiments, reverse transcriptase was omitted. The signal for β-actin was used for normalization. Cycle number was 18 for β-actin and 22-31 for other primer sets. PCR products were analyzed on 1.5–2.5% agarose gels stained by ethidium bromide. Quantification of signals was done using Labworks 4.0 software (UVP BioImaging Systems, Cambridge, UK). Values for control samples were set at 100%.
For RT-qPCR, all procedures were performed following the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines.46 Amplification efficiency of primer sets was determined using standard curves (serial dilutions; tenfold to 5,000-fold range). Primers sets displaying <90% efficiency in product amplification were ruled out and redesigned. cDNA preparations (see RT reaction above) were diluted 200–500-fold to avoid PCR inhibition by RT components; 3 µl of this dilution was mixed in a final volume of 10 µl containing 5 µl 2X Sybr Green mix (Roche Applied Science, Almere, The Netherlands) and 400 nmol/l of each primer using the CAS-1200 automated pipetting system (Corbett Life Science/Qiagen, Venlo, The Netherlands). No template control and no reverse transcriptase control (RT−) were included in each qPCR run to detect possible contaminations. Samples were analyzed using the Rotor-Gene 6000 (Corbett Life Science) or CFX96 Real-time System (Bio-Rad). A melting curve was obtained for each sample to confirm single product amplification. Relative mRNA levels were calculated using the δδCt method.47 Both Actb and Gapdh were used for normalization. We validated that these reference genes were not affected by the treatment and had an optimal M stability value48 under all experimental conditions. Values of control samples were set at 100%.
AON hybridization–ligation assay. PS58 concentrations were determined by a hybridization–ligation assay as published by Yu et al.49 with a few modifications: Template DNA probe (5′-GAATAGACGCTGCTGCTGCTGCTGCTGCTG-biotin-3′) and ligation DNA probe (5′-P-CGTCTATTC-digoxigenin (DIG)-3′) were used. Samples were incubated with the template probe (50 nmol/l) at 37 °C for 1 hour and hybridized samples were transferred to streptavidin-coated plates. Subsequently, plates were washed four times and the DIG-labeled ligation probe (2 nmol/l) was added and incubated for 30 minutes at ambient temperature. DIG label was detected using an anti-DIG-POD (1:7,500; Roche Diagnostics, Almere, The Netherlands), visualized with a 3,3′,5,5′-tetramethylbenzidine substrate (Sigma Aldrich, Zwijndrecht, The Netherlands). The reaction was stopped using maleic acid (345 mmol/l; Sigma Aldrich). Absorption was measured at 450 nm using a BioTek Synergy HT plate reader (Beun de Ronde, Abcoude, The Netherlands). All samples and calibration curves were diluted (fit to criteria of the assay) in PBS. Absorption was read against a calibration curve of PS58 in PBS. A sample containing only PBS was used as correction for background.
Live cell imaging. DM500 myoblasts were cultured on gelatin-coated glass-bottom Willco dishes (Willco Wells, Amsterdam, The Netherlands). Cells were analyzed directly after addition of the transfection mix containing FAM-PS58 or Cy3-PS138 in RPMI medium without phenol red and including 10 mmol/l HEPES and 10% fetal calf serum (Invitrogen). A Zeiss LSM510 meta confocal laser-scanning microscope was used, equipped with a temperature controlled CO2 incubator (type S) and sample stage. For recording of oligo uptake, we used a PlanApochromatic 63×, 1.4 NA oil immersion DIC lens (Carl Zeiss, Jena, Germany).
Western blotting. DM1 patient 21/200 myoblasts or DM500 mouse myotubes were transfected with PS147, PS58, PS146, PS260 or PS261 as indicated (200 nmol/l) or mock transfected and 72 hours later lysed in lysis buffer (TBS, 1% Nonidet P-40, 1 mmol/l PMSF, and Protease Inhibitory Mixture (Roche)) on ice. Cleared lysates were separated on an 8% SDS/PAGE gel and transferred to PVDF (Amersham Pharmacia Biotech, Piscataway, NJ). Blots were incubated with DMPK-specific antibody B7921 or anti-SIN1/Mapkap1 antibody (ab71152; Abcam, Cambridge, UK) and β-tubulin monoclonal antibody E7 (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA), followed by electrochemiluminescence detection. Quantification of signals was done using ImageJ software. Values for mock-treated samples were set at 100%.
Experiments using in vitro-transcribed triplet repeat RNA. (CUG)n and (CAG)n RNA was generated by in vitro transcription of a hDMPK (CTG)90 DNA template carrying flanking T7 and SP6 promoters. The hDMPK (CTG)90 template was generated by PCR using a hDMPK cDNA construct bearing a (CTG)90 repeat,50 T7-hDMPK forward primer: 5′-GAATTTAATACGACTCACTATAGGGAGAACGGGGCTCGAAGGGT-3′ and SP6-hDMPK reverse primer: 5′-ATTTAGGTGACACTATAGAAGGGCGTCATGCACAAGAAA-3′ and sequenced to verify triplet repeat number. (CUG)90 and (CAG)90 RNAs were synthesized in presence of [α-32P]GTP, using the MEGAscript in vitro transcription kit (Applied Biosystems/Ambion, Austin, TX) according to the manufacturer's protocol.
To confirm AON binding to expanded triplet repeat RNA, (CUG)90 RNA was incubated with a concentration series (1, 10, and 100 µmol/l) of PS58 in 50 mmol/l Tris-HCl; pH 7.5 and 25 mmol/l MgCl2 for 2 hours at 37 °C. Sample buffer was added to a final concentration of 50% deionized formamide and 30% glycerol and samples were run at room temperature on a denaturing polyacrylamide gel (5% acryl/bisacryl, 8 mol/l urea) in 1× MOPS. Gels were dried and exposed to X-ray film (X-Omat AR; Kodak).
To reveal binding specificity, (CUG)90 and (CAG)90 RNAs were incubated with 100 µmol/l AONs in the presence of 50 mmol/l Tris-HCl; pH 7.5, 50 mmol/l NaCl, 25 mmol/l MgCl2, and 1 mmol/l EDTA for 2 hours at 37 °C. Samples were next analyzed by electrophoresis as described above.
To examine RNase-H–dependent activity, (CUG)n RNA was dissolved in 50 mmol/l Tris-HCl; pH 7.5, 50 mmol/l KCl, 5 mmol/l MgCl2, and 1 mmol/l DTT and incubated with 60–100 µmol/l AON in the presence of 20 U/ml RNase-H for 1 hour at 37 °C. Samples were analyzed by electrophoresis as described above with the exception that the 1× MOPS running buffer was heated to 70 °C before start of electrophoresis to induce dissociation between AONs and (CUG)n RNA.
To examine intrinsic oligo catalytic activity, (CUG)n RNA was dissolved in 100 mmol/l Tris-HCl; pH 7.5, 100 mmol/l NaCl, and 1 mmol/l EDTA and incubated with 100 µmol/l AON for 2 hours at 37 °C in the presence of 3.5 or 25 mmol/l MgCl2. Samples were analyzed by electrophoresis as described above.
Complement activation assay. The potency of AONs to activate the complement pathway was demonstrated using Li+ heparin plasma from cynomolgus monkeys (kindly provided by CIT, France) and Li+ heparin plasma from healthy human donors (Sanquin, Amsterdam, The Netherlands). AONs were added to plasma in various concentrations (25, 50, 150, and 300 µg/ml in a dilution of ~1:20 (volume AON/volume plasma)) and the samples were incubated at 37 °C for 30 minutes. The reaction was terminated by putting the samples on ice and adding ice-cold diluents according to the manufacturer's protocol (Quidel, San Diego, CA). Bb and C3a concentrations were determined by ELISA (Quidel). First generation oligonucleotide PS455 (ISIS 5132) was included as a positive control, as this oligo has been shown to activate the complement pathway.25
Statistical analysis. Northern blot and PCR signals were analyzed using an unpaired Student's t-test or a one-way analysis of variance. All values in graphs are presented as the mean ± SEM. Pearson's correlation was applied to evaluate whether a correlation exists between silencing efficacy and number of CTG triplets in hDMPK alleles in human myoblasts. Differences between groups were considered significant when P < 0.05: *P < 0.05; **P < 0.01; ***P < 0.001. Statistical analyses and sigmoidal concentration curve fitting were performed with GraphPad Prism 4 software (GraphPad Software, La Jolla, CA).
SUPPLEMENTARY MATERIAL Figure S1. AON stability in cell extract. Figure S2. Concentration–response curve of PS142 including raw data. Figure S3. Differential uptake and localization of AONs in DM500 myoblasts. Figure S4. Schematic representation of triplet repeat transcripts and segments thereof, analyzed by RT-(q)PCR. Figure S5. Expression of mouse transcripts, carrying a small (CUG)n repeat, after transfecting DM500 myotubes with a selection of AONs. Figure S6. Control experiments to exclude perturbing effects of AONs during RT-(q)PCR quantification. Figure S7. (CAG)n oligos can modulate alternative splicing of the (CUG)n-containing exon in the MAP3K4 primary transcript. Figure S8. Test tube experiments using in vitro-synthesized RNA bearing a (CUG)90 or (CAG)90 tract. Figure S9. Complement activation assay. Table S1. siRNAs (RNA duplexes) used in this study. Table S2. PCR primers used in this study (5′ -> 3′).
Acknowledgments
This work was supported by grants from SenterNovem (a Dutch agency in the Ministry of Economic Affairs), the Prinses Beatrix Spierfonds (in particular, the Stichting Spieren voor Spieren (War07-34)), the Association Française contre les Myopathies, and the Muscular Dystrophy Association–USA. We thank members of the Department of Cell Biology for discussions and continued support and Sjef de Kimpe and Gerard Platenburg (Prosensa Therapeutics, Leiden, The Netherlands) for help during the initial period of this project. We are grateful to Geneviève Gourdon (Hôpital Necker-Enfants Malades, Paris, France) for kindly providing mice from the DM300-328 line to establish the immorto DM500 cell model and to Denis Furling (Institut de Myologie, Paris, France) for providing human DM1 myoblast lines. We thank Makoto Koizumi (Sankyo Co., Ltd., Tokyo, Japan) for providing ethylene-bridged nucleic acid nucleosides as building blocks for the synthesis of ethylene-bridged nucleic acid-modified oligonucleotides. Data described in this paper are the subject of a patent application (inventors D.G.W. et al.). A.G.-B., S.A.M.M., J.van de G., S.B., and J.C.T.van D. report being employed by or having an equity interest in Prosensa Therapeutics B.V. The other authors declared no conflict of interest.
Supplementary material
AON stability in cell extract.
Concentration–response curve of PS142 including raw data.
Differential uptake and localization of AONs in DM500 myoblasts.
Schematic representation of triplet repeat transcripts and segments thereof, analyzed by RT-(q)PCR.
Expression of mouse transcripts, carrying a small (CUG)n repeat, after transfecting DM500 myotubes with a selection of AONs.
Control experiments to exclude perturbing effects of AONs during RT-(q)PCR quantification.
(CAG)n oligos can modulate alternative splicing of the (CUG)n-containing exon in the MAP3K4 primary transcript.
Test tube experiments using in vitro-synthesized RNA bearing a (CUG)90 or (CAG)90 tract.
Complement activation assay.
siRNAs (RNA duplexes) used in this study.
PCR primers used in this study (5′ -> 3′).
References
- Groenen, P and Wieringa, B (1998). Expanding complexity in myotonic dystrophy. Bioessays 20: 901–912. [DOI] [PubMed] [Google Scholar]
- Wheeler, TM and Thornton, CA (2007). Myotonic dystrophy: RNA-mediated muscle disease. Curr Opin Neurol 20: 572–576. [DOI] [PubMed] [Google Scholar]
- Harper, PS (2001). Myotonic Dystrophy. WB Saunders: London, UK. [Google Scholar]
- Zerylnick, C, Torroni, A, Sherman, SL and Warren, ST (1995). Normal variation at the myotonic dystrophy locus in global human populations. Am J Hum Genet 56: 123–130. [PMC free article] [PubMed] [Google Scholar]
- Rau, F, Freyermuth, F, Fugier, C, Villemin, JP, Fischer, MC, Jost, Bet al. (2011). Misregulation of miR-1 processing is associated with heart defects in myotonic dystrophy. Nat Struct Mol Biol 18: 840–845. [DOI] [PubMed] [Google Scholar]
- Mahadevan, MS, Yadava, RS, Yu, Q, Balijepalli, S, Frenzel-McCardell, CD, Bourne, TDet al. (2006). Reversible model of RNA toxicity and cardiac conduction defects in myotonic dystrophy. Nat Genet 38: 1066–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulders, SA, van Engelen, BG, Wieringa, B and Wansink, DG (2010). Molecular therapy in myotonic dystrophy: focus on RNA gain-of-function. Hum Mol Genet 19(R1): R90–R97. [DOI] [PubMed] [Google Scholar]
- Mulders, SA, van den Broek, WJ, Wheeler, TM, Croes, HJ, van Kuik-Romeijn, P, de Kimpe, SJet al. (2009). Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proc Natl Acad Sci USA 106: 13915–13920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler, TM, Sobczak, K, Lueck, JD, Osborne, RJ, Lin, X, Dirksen, RTet al. (2009). Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science 325: 336–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, JE, Bennett, CF and Cooper, TA (2012). RNase H-mediated degradation of toxic RNA in myotonic dystrophy type 1. Proc Natl Acad Sci USA 109: 4221–4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- François, V, Klein, AF, Beley, C, Jollet, A, Lemercier, C, Garcia, Let al. (2011). Selective silencing of mutated mRNAs in DM1 by using modified hU7-snRNAs. Nat Struct Mol Biol 18: 85–87. [DOI] [PubMed] [Google Scholar]
- Tashiro, F and Ueno, Y (1978). Ribonuclease H from rat liver. I. Partial purification and characterization of nuclear ribonuclease H1. J Biochem 84: 385–393. [DOI] [PubMed] [Google Scholar]
- Kurreck, J (2003). Antisense technologies. Improvement through novel chemical modifications. Eur J Biochem 270: 1628–1644. [DOI] [PubMed] [Google Scholar]
- Eisen, JS and Smith, JC (2008). Controlling morpholino experiments: don't stop making antisense. Development 135: 1735–1743. [DOI] [PubMed] [Google Scholar]
- Morita, K, Hasegawa, C, Kaneko, M, Tsutsumi, S, Sone, J, Ishikawa, Tet al. (2002). 2'-O,4'-C-ethylene-bridged nucleic acids (ENA): highly nuclease-resistant and thermodynamically stable oligonucleotides for antisense drug. Bioorg Med Chem Lett 12: 73–76. [DOI] [PubMed] [Google Scholar]
- Krol, J, Fiszer, A, Mykowska, A, Sobczak, K, de Mezer, M and Krzyzosiak, WJ (2007). Ribonuclease dicer cleaves triplet repeat hairpins into shorter repeats that silence specific targets. Mol Cell 25: 575–586. [DOI] [PubMed] [Google Scholar]
- Schroder, W, Cloonan, N, Bushell, G and Sculley, T (2004). Alternative polyadenylation and splicing of mRNAs transcribed from the human Sin1 gene. Gene 339: 17–23. [DOI] [PubMed] [Google Scholar]
- Furling, D, Lemieux, D, Taneja, K and Puymirat, J (2001). Decreased levels of myotonic dystrophy protein kinase (DMPK) and delayed differentiation in human myotonic dystrophy myoblasts. Neuromuscul Disord 11: 728–735. [DOI] [PubMed] [Google Scholar]
- Kozlowski, P, de Mezer, M and Krzyzosiak, WJ (2010). Trinucleotide repeats in human genome and exome. Nucleic Acids Res 38: 4027–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerwins, P, Blank, JL and Johnson, GL (1997). Cloning of a novel mitogen-activated protein kinase kinase kinase, MEKK4, that selectively regulates the c-Jun amino terminal kinase pathway. J Biol Chem 272: 8288–8295. [DOI] [PubMed] [Google Scholar]
- Groenen, PJ, Wansink, DG, Coerwinkel, M, van den Broek, W, Jansen, G and Wieringa, B (2000). Constitutive and regulated modes of splicing produce six major myotonic dystrophy protein kinase (DMPK) isoforms with distinct properties. Hum Mol Genet 9: 605–616. [DOI] [PubMed] [Google Scholar]
- Tiscornia, G and Mahadevan, MS (2000). Myotonic dystrophy: the role of the CUG triplet repeats in splicing of a novel DMPK exon and altered cytoplasmic DMPK mRNA isoform ratios. Mol Cell 5: 959–967. [DOI] [PubMed] [Google Scholar]
- Bhindi, R, Fahmy, RG, Lowe, HC, Chesterman, CN, Dass, CR, Cairns, MJet al. (2007). Brothers in arms: DNA enzymes, short interfering RNA, and the emerging wave of small-molecule nucleic acid-based gene-silencing strategies. Am J Pathol 171: 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry, SP, Giclas, PC, Leeds, J, Pangburn, M, Auletta, C, Levin, AAet al. (1997). Activation of the alternative pathway of complement by a phosphorothioate oligonucleotide: potential mechanism of action. J Pharmacol Exp Ther 281: 810–816. [PubMed] [Google Scholar]
- Rudin, CM, Holmlund, J, Fleming, GF, Mani, S, Stadler, WM, Schumm, Pet al. (2001). Phase I Trial of ISIS 5132, an antisense oligonucleotide inhibitor of c-raf-1, administered by 24-hour weekly infusion to patients with advanced cancer. Clin Cancer Res 7: 1214–1220. [PubMed] [Google Scholar]
- Mooers, BH, Logue, JS and Berglund, JA (2005). The structural basis of myotonic dystrophy from the crystal structure of CUG repeats. Proc Natl Acad Sci USA 102: 16626–16631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi, M (2006). ENA oligonucleotides as therapeutics. Curr Opin Mol Ther 8: 144–149. [PubMed] [Google Scholar]
- Robb, GB, Brown, KM, Khurana, J and Rana, TM (2005). Specific and potent RNAi in the nucleus of human cells. Nat Struct Mol Biol 12: 133–137. [DOI] [PubMed] [Google Scholar]
- Sobczak, K, Wheeler, TM, Wang, W and Thornton, CA (2013). RNA interference targeting CUG repeats in a mouse model of myotonic dystrophy. Mol Ther 21: 380–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wansink, DG and Wieringa, B (2003). Transgenic mouse models for myotonic dystrophy type 1 (DM1). Cytogenet Genome Res 100: 230–242. [DOI] [PubMed] [Google Scholar]
- Smith, PJ, Zhang, C, Wang, J, Chew, SL, Zhang, MQ and Krainer, AR (2006). An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum Mol Genet 15: 2490–2508. [DOI] [PubMed] [Google Scholar]
- Fiszer, A, Olejniczak, M, Switonski, PM, Wroblewska, JP, Wisniewska-Kruk, J, Mykowska, Aet al. (2012). An evaluation of oligonucleotide-based therapeutic strategies for polyQ diseases. BMC Mol Biol 13: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma, MK and Parker, R (2007). RNA quality control in eukaryotes. Cell 131: 660–668. [DOI] [PubMed] [Google Scholar]
- Nakamori, M, Gourdon, G and Thornton, CA (2011). Stabilization of expanded (CTG)•(CAG) repeats by antisense oligonucleotides. Mol Ther 19: 2222–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, YL and Rana, TM (2003). siRNA function in RNAi: a chemical modification analysis. RNA 9: 1034–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarzguioui, M, Holen, T, Babaie, E and Prydz, H (2003). Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res 31: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch, DA, Jensen, S, Liu, Y, Kaur, K, Arar, K, White, MAet al. (2003). RNA interference in mammalian cells by chemically-modified RNA. Biochemistry 42: 7967–7975. [DOI] [PubMed] [Google Scholar]
- Sun, X, Rogoff, HA and Li, CJ (2008). Asymmetric RNA duplexes mediate RNA interference in mammalian cells. Nat Biotechnol 26: 1379–1382. [DOI] [PubMed] [Google Scholar]
- Chu, CY and Rana, TM (2008). Potent RNAi by short RNA triggers. RNA 14: 1714–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerton, JE (2007). Morpholino, siRNA, and S-DNA compared: impact of structure and mechanism of action on off-target effects and sequence specificity. Curr Top Med Chem 7: 651–660. [DOI] [PubMed] [Google Scholar]
- Wansink, DG, van Herpen, RE, Coerwinkel-Driessen, MM, Groenen, PJ, Hemmings, BA and Wieringa, B (2003). Alternative splicing controls myotonic dystrophy protein kinase structure, enzymatic activity, and subcellular localization. Mol Cell Biol 23: 5489–5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J, Matsui, M, Gagnon, KT, Schwartz, JC, Gabillet, S, Arar, Ket al. (2009). Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat Biotechnol 27: 478–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seznec, H, Lia-Baldini, AS, Duros, C, Fouquet, C, Lacroix, C, Hofmann-Radvanyi, Het al. (2000). Transgenic mice carrying large human genomic sequences with expanded CTG repeat mimic closely the DM CTG repeat intergenerational and somatic instability. Hum Mol Genet 9: 1185–1194. [DOI] [PubMed] [Google Scholar]
- Ye, J, Coulouris, G, Zaretskaya, I, Cutcutache, I, Rozen, S and Madden, TL (2012). Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owczarzy, R, Tataurov, AV, Wu, Y, Manthey, JA, McQuisten, KA, Almabrazi, HGet al. (2008). IDT SciTools: a suite for analysis and design of nucleic acid oligomers. Nucleic Acids Res 36(Web Server issue): W163–W169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin, SA, Benes, V, Garson, JA, Hellemans, J, Huggett, J, Kubista, Met al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55: 611–622. [DOI] [PubMed] [Google Scholar]
- Livak, KJ and Schmittgen, TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Vandesompele, J, De Preter, K, Pattyn, F, Poppe, B, Van Roy, N, De Paepe, Aet al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, RZ, Baker, B, Chappell, A, Geary, RS, Cheung, E and Levin, AA (2002). Development of an ultrasensitive noncompetitive hybridization-ligation enzyme-linked immunosorbent assay for the determination of phosphorothioate oligodeoxynucleotide in plasma. Anal Biochem 304: 19–25. [DOI] [PubMed] [Google Scholar]
- van den Broek, WJ, Nelen, MR, Wansink, DG, Coerwinkel, MM, te Riele, H, Groenen, PJet al. (2002). Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum Mol Genet 11: 191–198. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
AON stability in cell extract.
Concentration–response curve of PS142 including raw data.
Differential uptake and localization of AONs in DM500 myoblasts.
Schematic representation of triplet repeat transcripts and segments thereof, analyzed by RT-(q)PCR.
Expression of mouse transcripts, carrying a small (CUG)n repeat, after transfecting DM500 myotubes with a selection of AONs.
Control experiments to exclude perturbing effects of AONs during RT-(q)PCR quantification.
(CAG)n oligos can modulate alternative splicing of the (CUG)n-containing exon in the MAP3K4 primary transcript.
Test tube experiments using in vitro-synthesized RNA bearing a (CUG)90 or (CAG)90 tract.
Complement activation assay.
siRNAs (RNA duplexes) used in this study.
PCR primers used in this study (5′ -> 3′).