SUMMARY
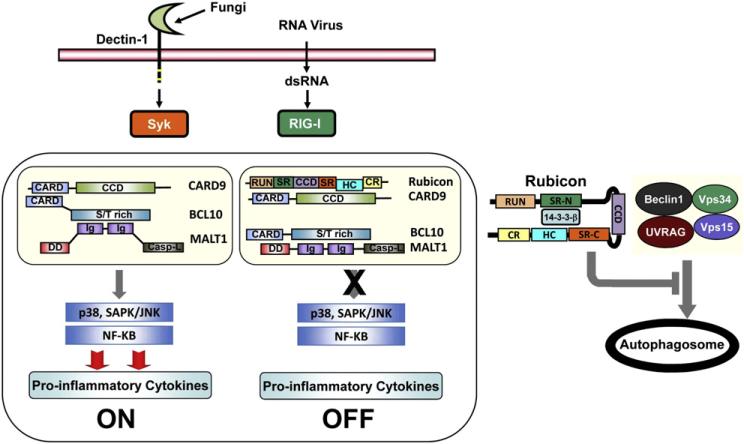
Assembly of a scaffold consisting of CARD9, BCL10, and MALT1 (CBM complex) is critical for effective signaling by multiple pattern recognition receptors (PRRs) including Dectin and RIG-I. The RUN domain Beclin-1-interacting cysteine-rich-containing Rubicon protein associates constitutively with the Beclin-UVRAG-Vps34 complex under normal conditions to regulate autophagy. Rubicon also interacts with the phagocytic NADPH-oxidase complex upon TLR stimulation to induce potent antimicrobial responses. Here, we show Rubicon is a physiological feedback inhibitor of CBM-mediated PRR signaling, preventing unbalanced proinflammatory responses. Upon Dectin-1- or RIG-I-mediated activation, Rubicon dynamically exchanges binding partners from 14-3-3β to CARD9 in a stimulation-specific and phosphorylation-dependent manner, disassembling the CBM signaling complex and ultimately terminating PRR-induced cytokine production. Remarkably, Rubicon's actions in the autophagy complex, phagocytosis complex, and CBM complex are functionally and genetically separable. Rubicon thus differentially targets signaling complexes, depending on environmental stimuli, and may function to coordinate various immune responses against microbial infection.
INTRODUCTION
The first step to mounting a protective immune response is the recognition of pathogens by cell surface receptors, called pattern recognition receptors (PRRs), located on professional phagocytes, dendritic cells, and nonimmune cells (Kumar et al., 2011; Palm and Medzhitov, 2009). PRRs include C-type lectin receptors, transmembrane Toll-like receptors (TLRs), NOD-like receptors (NLRs), and cytoplasmic RIG-I-like helicases (RLHs). After recognizing specific pathogen-associated molecular patterns, PRRs activate intracellular signaling pathways and stimulate inflammatory mediators. This triggers effector immune mechanisms (Puel et al., 2010), ultimately resulting in the elimination of the pathogen from the infected host. However, unbalanced, continuous production of inflammatory cytokines could lead to deleterious effects on host immunity such as autoimmune diseases. Not surprisingly, PRR-mediated innate immune responses are tightly regulated by several mechanisms, including PRR modification and degradation (Arimoto et al., 2007) or the expression of dominant-negative or alternatively spliced variants of PRRs and their downstream molecules (Janssens et al., 2002; Leung et al., 2007; Rosenstiel et al., 2006).
C-type lectin PRRs like Dectin, DC-SIGN, and mannose receptor interact with pathogens primarily through the recognition of distinct carbohydrates, such as mannose, fucose, or glucan structures (Drummond et al., 2011; Hara and Saito, 2009; Kerrigan and Brown, 2010; Reid et al., 2009; Ruland, 2008). Recognition by C-type lectins is important for the internalization of pathogens, which leads to the induction of intracellular signaling cascades to direct immune responses. Dectin-1 is a unique C-type lectin that recognizes β-glucan carbohydrates on various fungi, including Candida (C) albicans, and its activation induces both Th1 and Th17 immune responses that are essential for antifungal defenses (Drummond et al., 2011; Hara and Saito, 2009; Ruland, 2008). Engagement of Dectin-1 by fungal β-glucans leads to phosphorylation of the immunoreceptor tyrosine-based activation motif (ITAM)-like sequence within its cytoplasmic domain, and subsequent association of the Syk tyrosine kinase induces the assembly of a scaffold consisting of caspase recruitment domain 9 (CARD9), B cell lymphoma 10 (BCL10), and mucosa-associated lymphoid tissue lymphoma translocation gene 1 (MALT1) (Hara and Saito, 2009). On the other hand, Dectin-2 interacts with C. albicans through mannan structures present on both its yeast and hyphal forms (Bi et al., 2010; McGreal et al., 2006), indirectly activates Syk by associating with the FcRγ chain, and ultimately results in CARD9-dependent signal transduction (Drummond et al., 2011; Gringhuis et al., 2011; Saijo et al., 2010). Thus, Dectin constitutes a major fungal PRR that can couple to the Syk-CARD9 innate signaling pathway to activate monocyte lineage immune cells and regulate adaptive immune responses to fungal infections.
Two CARD-containing adaptor molecules, CARMA1 (CARD-containing MAGUK protein 1) and CARD9, play critical roles in the activation and regulation of both innate and adaptive immunity (Blonska and Lin, 2011; Hara and Saito, 2009). CARMA1 and CARD9 regulate the cell-type-specific activation of BCL10-MALT1-mediated activation of NF-κB and MAPK in lymphoid cells (e.g., T cells, B cells, and natural killer cells) and myeloid cells (e.g., macrophages and dendritic cells), respectively, called lymphoid-type CARMA1-BCL10-MALT1 (L-CBM) and myeloid-type CARD9-BCL10-MALT1 (M-CBM) (Hara and Saito, 2009). Myeloid CBM is essential in antifungal immunity and is implicated in mediating Dectin-Syk-induced NF-κB activation in response to C. albicans infection. Biochemical and genetic experiments demonstrate that the CBM signaling module mediates Dectin signaling for the activation of NF-κB and MAPK pathways (Blonska and Lin, 2011). In fact, dendritic cells and macrophages deficient in CARD9, BCL10, or MALT1 consistently show severe defects in pathogen-induced NF-κB activation and cytokine production, indicating that the CBM signaling module is key for antimicrobial innate immunity in myeloid cells. Furthermore, the CBM module is an essential component of RIG-I- and NLR-dependent proinflammatory responses (Gross et al., 2006; Poeck et al., 2010), and CARD9 also associates with the GDP-dissociation inhibitor LyGDI in phagosomes after bacterial and fungal infection (Underhill and Shimada, 2007; Wu et al., 2009), leading to reactive oxygen species (ROS) production and bacterial killing in macrophages. Thus, CARD9 is a key signaling molecule for efficient host microbe-elicited innate immunity.
Rubicon (RUN domain Beclin-1-interacting cysteine-rich-containing) was recently identified as a Beclin-1-binding partner that localizes to the late endosome/lysosome, and negatively regulates the maturation step of autophagy and the endocytic pathway (Matsunaga et al., 2009; Zhong et al., 2009). While Rubicon primarily associates with the Beclin-1-containing autophagy complex under normal and stressed conditions, we recently reported that Rubicon is also an essential positive regulator of the NADPH oxidase complex, (Yang et al., 2012 [accompanying paper, this issue of Cell Host & Microbe]). Upon microbial infection or TLR activation, Rubicon transiently interacts with p22phox of the NADPH oxidase complex to induce a potent antimicrobial burst of ROS and inflammatory cytokines (Yang et al., 2012). Thus, Rubicon regulates both autophagy and phagocytosis, depending on the environmental stimulus, and is perfectly positioned to coordinate different but related innate immune mechanisms in a phagocytic cell.
Most biological processes require both positive and negative regulatory mechanisms to maintain equilibrium. While the PRR pathway robustly induces host immune responses upon microbial infection, it must slow down and ultimately stop when it reaches a late stage to avoid excessive production of inflammatory cytokines, which could lead to deleterious effects on host immunity. Here we show that Rubicon differentially binds to 14-3-3β and CARD9 in a stimulation-specific, phosphorylation-dependent, and competitive manner, resulting in the disassembly and elimination of CBM complex signaling. Thus, Rubicon acts as a physiological brake for CBM-mediated immune responses, serving as an inhibitory feedback mechanism.
RESULTS
Temporal Interactions of Rubicon with 14-3-3β and CARD9
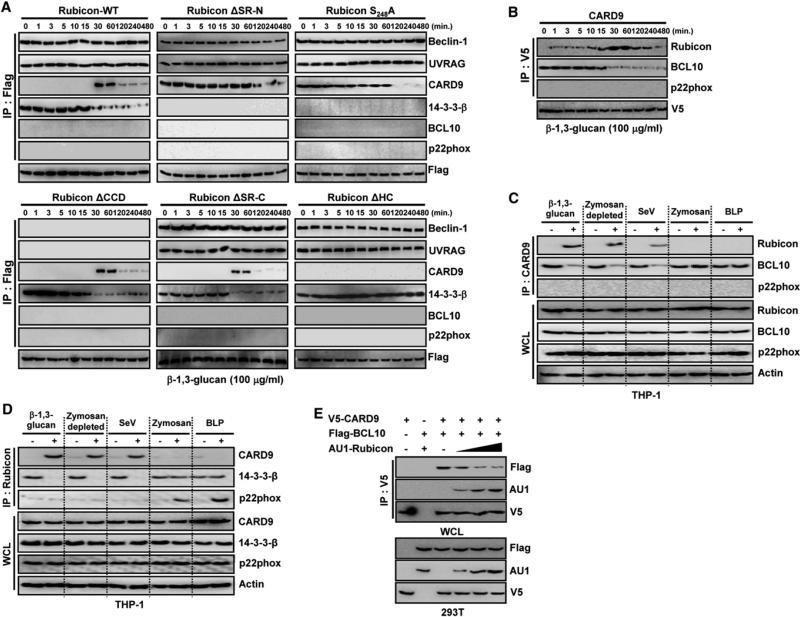
Rubicon contains a RUN domain, an N-terminal serine-rich region (SR-N), a coiled-coil domain (CCD), a C-terminal serine-rich region (SR-C), a helix-coil-rich region (H-C), and a cysteine-rich region (C-R) (Figure 1A). A yeast two-hybrid screen of the full length, the N-terminal region (aa 1–505), and the C-terminal region (aa 506–972) of Rubicon collectively revealed that Rubicon interacts with 14-3-3β and CARD9 through its N-terminal and C-terminal regions, respectively. Coimmunoprecipitation (coIP) showed that Rubicon interacted with endogenous 14-3-3β through its SR-N region (Figure 1B). The highly conserved 14-3-3 family members are phosphoserine/phosphothreonine binding proteins, and their high-affinity binding motifs are RSXpSXP (mode-1) and RXXXpSXP (mode-2), where pS and X represent phosphoserine and any amino acid, respectively (Morrison, 2009). A large-scale proteomics analysis of the human kinome (Oppermann et al., 2009) has shown that Rubicon undergoes multiple phosphorylations including the S248 residue within its SR-N region whose surrounding sequences (RSTpS248FP) fit very well with the mode-1 binding motif of 14-3-3. Indeed, the specific deletion (ΔSR-N) or point mutation (S248A) of Rubicon completely abolished its interaction with 14-3-3β, suggesting that this interaction appears to be S248-phosphorylation dependent (Figures 1B and 1C).
Figure 1. Rubicon Interactions with 14-3-3β and CARD9.
(A) Summary of the interactions of Rubicon WT and its mutants with Beclin-1, p22phox, 14-3-3β, and CARD9. The binding activities of Rubicon WT and mutants are summarized based on the results of Figures 1B, 2A, and 2B and Figures S1A and S1B.
(B) Differential Rubicon interactions with 14-3-3β and CARD9. Raw264.7 cells containing vector, Flag-Rubicon WT, or its mutant were stimulated with or without β-1,3-glucan for 30 min, followed by IP with αFlag and IB with α14-3-3β or αCARD9.
(C) S248A phosphorylation-dependent interaction of Rubicon with 14-3-3β. (Left) At 48 hr posttransfection with Flag-Rubicon WT or S248A mutant together with V5-14-3-3β, 293T cells were used for IP with αV5 and IB with αFlag. (Right) At 48 hr posttransfection with Flag-Rubicon WT or S248A mutant, Raw264.7 cells were stimulated with or without β-1,3-glucan for 30 min, followed by IP with αFlag and IB with α14-3-3β.
(D) Summary of CARD9 interaction with Rubicon. (Top panel) Schematic diagram of CARD9. (Bottom panel) At 48 hr posttransfection with V5-CARD9 or its deletion mutants together with Flag-Rubicon, 293T cells were used for IP with αFlag and IB with αV5.
(E) Rubicon-CARD9 interaction. At 48 hr posttransfection with V5-CARD9 and Flag-Rubicon, 293T cells were used for IP with αFlag and IB with αV5 or conversely for IP with αV5 and IB with αFlag.
(F) Rubicon mapping for CARD9 interaction. (Left) At 48 hr posttransfection with V5-CARD9 and mammalian GST alone or GST fusions with Rubicon full length (F), N terminus (N, aa 1–505), or C terminus (C, aa 506–972), 293T cells were used for GST-pull-down (PD) and IB with αV5. (Right) At 48 hr posttransfection with V5-CARD9 and mammalian GST alone or various GST-Rubicon fusions, 293T cells were used for GST-PD and IB with αV5. See also Figure S1.
Besides 14-3-3β binding, Rubicon also showed a robust interaction with CARD9: either the N-terminal CARD or the central CCD of CARD9 was sufficient to bind to Rubicon, and the C-terminal region of Rubicon is responsible for CARD9 binding (Figures 1D–1F). Computational sequence analysis predicted three helix-coiled repeats (HC1, aa 625–671; HC2, aa 672–709; and HC3, aa 710–760) within the Rubicon C-terminal region. A detailed mapping study using various mammalian glutathionine S-transferase (GST)-Rubicon fusions indicates that while each HC motif of Rubicon exhibited only a minimal binding affinity to CARD9, the GST-Rub-HC123 fusion carrying all three HC motifs bound CARD9 as strongly as GST-Rub-C carrying the C-terminal region (Figure 1F). These results indicate that Rubicon independently interacts with 14-3-3β and CARD9 through its N-terminal SR-N region and C-terminal HC region, respectively.
Dynamic Competition between 14-3-3β and CARD9 for Rubicon Binding
Under normal conditions, Rubicon efficiently interacted with endogenous 14-3-3β, whereas an interaction with endogenous CARD9 was not evident (Figure 1B). Furthermore, autophagy-inducing rapamycin and phagocytosis-inducing zymosan stimulations neither increased Rubicon's interaction with endogenous CARD9, nor did they affect its interaction with endogenous 14-3-3β (see Figures S1A and S1B available online). Since CARD9 plays a major role in the β-1,3-glucan-mediated activation of Dectin-1 pathway in myeloid cells (Figure S1C), we stimulated THP-1 cells containing Flag-Rubicon WT, ΔSR-N, S248A, or ΔHC mutant with β-1,3-glucan and purified the Rubicon complexes in a time course manner, followed by immunoblot with the specific antibodies. These showed that Rubicon (130,000) interacted with Vps15 (150,000), Vps34 (110,000), UVRAG (85,000), and Beclin-1 (60,000) autophagy proteins, and p22phox (22,000) and gp91phox (91,000) NADPH oxidase proteins (Figures S2A and S2B). Notably, coIP showed that Rubicon primarily interacted with 14-3-3β, but not CARD9, in the absence of stimulation, but this interaction markedly declined after 30 min of β-1,3-glucan stimulation concomitantly with an increase in the interaction with CARD9, which then declined after 120 min of stimulation (Figure S2B). The Rubicon ΔSR-N and S248A mutants that no longer bind 14-3-3β maintained a constant interaction with CARD9 until late time points after stimulation, whereas the Rubicon ΔHC mutant that no longer binds CARD9 had a constant interaction with 14-3-3β, indicating that the interactions of these mutants with CARD9 and 14-3-3β are thus independent of stimulation (Figure S2B). Furthermore, pretreatment with anti-Dectin-1 blocking 2A11 antibody suppressed the Rubion-CARD9 interaction induced by β-1,3-glucan stimulation, suggesting the signal-dependent Rubicon-CARD9 interaction (Figure S1D).
To further delineate this dynamic competition between 14-3-3β and CARD9 for Rubicon binding, Flag-Rubicon complexes were immunopurified upon β-1,3-glucan stimulation and immunoblotted with various antibodies. The interaction between Rubicon and 14-3-3β was initially evident but dramatically decreased after 30 min of β-1,3-glucan stimulation (Figure 2A and Figure S2C). In striking contrast, although Rubicon interaction with CARD9 was undetectable during the initial 30 min of β-1,3-glucan stimulation, it robustly increased after 30–60 min of stimulation and then declined thereafter (Figure 2A). These features were binding specific: the ΔSR-N and S284A mutants that lost 14-3-3β interaction continuously bound CARD9 with no β-1,3-glucan stimulation, but their CARD9 binding ultimately declined after 60 min of stimulation, and the ΔHC mutant that lost CARD9 interaction constantly associated with 14-3-3β regardless of β-1,3-glucan stimulation (Figure 2A). However, all three mutants (ΔSR-N, S284A, and ΔHC) showed similar Beclin-1 and p22phox binding kinetics to Rubicon WT, whereas ΔCCD (Beclin-1 binding deficient) and ΔSR-C (p22phox binding deficient) mutants showed similar 14-3-3β and CARD9 binding kinetics to WT (Figure 2A, Figures S1A and S1B). Importantly, these binding features and kinetics were stimulation specific, as rapamycin or zymosan stimulation did not lead to any detectable effects on 14-3-3β and CARD9 interactions (Figures S1A and S1B). These results collectively demonstrate that Rubicon dynamically changes binding partners from 14-3-3β to CARD9 in a specific-stimulation-dependent, competitive manner.
Figure 2. Differential Interactions of Rubicon with 14-3-3β and CARD9.
(A) Differential Rubicon interactions. Raw264.7 cells containing vector, Flag-Rubicon WT, or its mutant were stimulated with β-1,3-glucan for the indicated times, followed by IP with αFlag and IB with αBeclin-1, αUVRAG, αCARD9, α14-3-3β, αBCL10, αp22phox, or αFlag. The results of vector control cells are shown in Figure S4C, and the results of zymosan or rapamycin treatment are shown in Figures S1A and S1B.
(B) Differential CARD9 interactions upon β-1,3-glucan stimulation. Raw264.7 cells containing V5-CARD9 were stimulated with β-1,3-glucan for the indicated times, followed by IP with αV5 and IB with αRubicon, αBCL10, αp22phox, or αV5.
(C and D) Differential CARD9 interactions. THP-1 cells were stimulated with β-1,3-glucan, zymosan depleted by hot alkali treatment, SeV infection, zymosan, or BLP for 30 min, followed by IP with αCARD9 and IB with αRubicon, αBCL10, αp22phox, or αactin (C), or IP with αRubicon and IB with αCARD9, α14-3-3β, αp22phox, or αactin (D).
(E) Competition between Rubicon and BCL10 for CARD9 interaction. At 48 hr posttransfection with V5-CARD9 or Flag-BCL10 together with increasing amounts of AU1-Rubicon, 293T cells were used for IP with αV5 and IB with αFlag, αAU1, or αV5. See also Figure S2.
Rubicon Interaction Disassembles the CBM Signaling Complex
To examine the effect of Rubicon on the CBM complex, we purified radioactively labeled CARD9 complexes from mouse Raw264.7 cells containing V5-CARD9 upon stimulation with β-1,3-glucan and also performed the immunoblotting analysis of purified CARD9 complexes. As previously shown (Blonska and Lin, 2011; Hara and Saito, 2009; Ruland, 2008), CARD9 (65,000) was found to weakly associate with BCL10 (30,000) without stimulation, with this interaction detectably increasing at 15 min of β-1,3-glucan stimulation but decreasing thereafter (Figure 2B and Figures S2D and S2E). Surprisingly, unlike the CARD9-BCL10 interaction, the CARD9-Rubicon interaction was initially minimal but markedly increased after 30–60 min of β-1,3-glucan stimulation and declined afterward (Figure S2D). Since CARD9 also mediates RIG-I signaling during viral infection (Poeck et al., 2010), CARD9 complexes were immunopurified from THP-1 and Raw264.7 cells upon stimulation with live Sendai virus (SeV) infection for 30 min. Similar to stimulation with β-1,3-glucan or zymosan depleted by hot alkali treatment, SeV infection also induced the association of CARD9 with Rubicon and the concomitant dissociation of CARD9 with BCL10, whereas zymosan, bacterial lipoprotein (BLP), and lipopolysaccharide (LPS) stimulation did not do so (Figures 2C and 2D, Figures S2F and S2G). Finally, increasing levels of Rubicon overexpression disrupted the interaction between CARD9 and BCL10 in a dose-dependent manner, whereas this β-1,3-glucan-induced disruption of CARD9-BCL10 interaction was less pronounced upon the shRNA-mediated depletion of endogenous Rubicon expression (Figure 2E, Figures S2H and S2I). By striking contrast, Rubicon expression did not break the interaction between CARD9 and LyGDI under the same conditions (Figure S2J). These results show that CARD9 dynamically changes binding partners from BCL10 to Rubicon in a specific-stimulation-dependent manner, thus disassembling the CARD9-BCL10 signaling complex.
Rubicon Has a Negative Effect on CBM-Mediated Signal Transduction
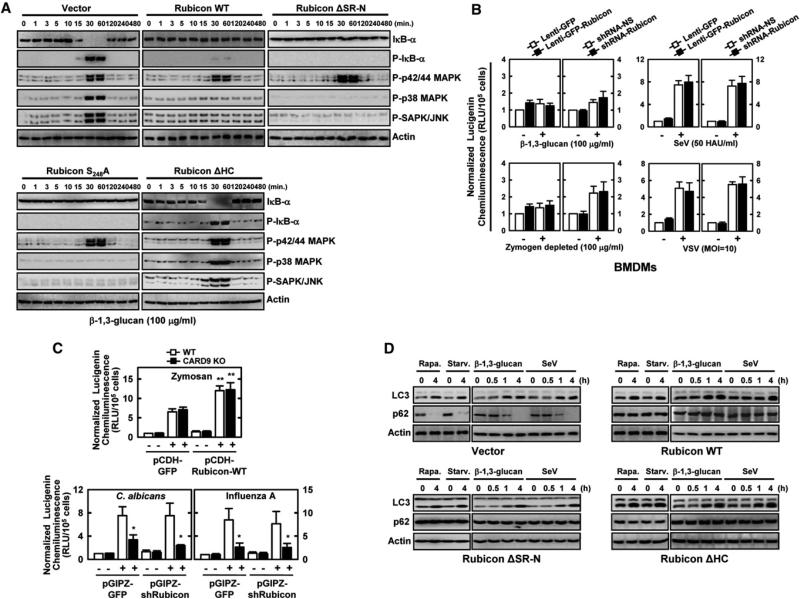
Consistent with Rubicon's ability to interact with CARD9 and disassemble the CBM complex, expression of Rubicon WT, ΔSR-N, or S284A mutant, but not ΔHC mutant, markedly suppressed p38/JNK/IκB phosphorylation and IκB degradation in Raw246.7 cells and mouse primary bone marrow-derived dendritic cells (BMDCs) induced by β-1,3-glucan stimulation, and NF-κB promoter activation in 293T cells; however, there was no significant effect on the phosphorylation of p42/44 MAPK (Figure 3A, Figures S2K and S3A). To measure Rubicon's effect on CARD9-mediated ROS production, Raw264.7 or primary mouse bone marrow-derived macrophage cells (BMDMs) were infected with recombinant lentiviruses or adenoviruses for Rubicon expression or Rubicon depletion, followed by stimulation with β-1,3-glucan, zymosan depleted by hot alkali treatment, and heat-killed C. albicans or infection with SeV or VSV. The resulting data demonstrate that neither Rubicon expression nor Rubicon depletion affected ROS production induced by various stimuli and infections (Figure 3B, Figures S3B to S3D). Furthermore, Rubicon expression significantly enhanced zymosan-induced ROS production in both wild-type and CARD9–/– BMDMs but showed nearly no effect on C. albicans- or influenza A virus-induced ROS production under the same conditions, consistent with the data showing Rubicon has no effect on the CARD9-LyGDI interaction (Figure 3C and Figures S3B–S3D). Finally, the loss of either 14-3-3β binding or CARD9 binding did not affect Rubicon's ability to suppress autophagosome maturation: similar to Rubicon WT, expression of the ΔSR-N or the ΔHC mutant, both capable of binding to Beclin-1, increased LC3-II and p62 protein levels upon stimulation with β-1,3-glucan or SeV, indicators of the autophagosome maturation suppression (Figure 3D). These results indicate that the Rubicon-CARD9 interaction specifically targets the CBM complex to suppress the activation of NF-κB and p38/JNK kinase activity without affecting CARD9-LyGDI complex formation and CARD9-mediated ROS production, as well as Rubicon-Beclin-1 complex formation and autophagosome maturation inhibition.
Figure 3. Rubicon Inhibits CBM-Mediated Signaling in a CARD9 Binding-Dependent Manner.
(A) Rubicon expression suppresses β-1,3-glucan-induced downstream signaling. Raw264.7 cells containing vector, Rubicon, or its mutants were stimulated with β-1,3-glucan for the indicated times and then subjected to IB analysis to detect the phosphorylated and total forms of p38, p42/p44 MAPK, JNK, or IκB-α.
(B) Rubicon expression levels show no effect on ROS production induced by Dectin-1-mediated stimulation. BMDMs infected with lentivirus-shRNA-NS or lentivirus-shRNA-Rubicon for Rubicon depletion or infected with lenti-GFP or lenti-GFP-Rubicon for Rubicon expression were analyzed for ROS production upon stimulation with β-1,3-glucan or zymosan depleted by hot alkali treatment (to remove its TLR-stimulating properties but keep its Dectin-1-stimulating activities), or infection with SeV or VSV.
(C) Rubicon expression levels show no effect on CARD9-mediated ROS production. WT or CARD9 KO BMDMs infected with lenti-GFP or lenti-GFP-Rubicon for Rubicon expression or infected with lentivirus-shRNA-NS or lentivirus-shRNA-Rubicon for Rubicon depletion were analyzed for ROS production upon stimulation with zymosan, C. albicans, or influenza A virus infection. Values are the mean ± SD of triplicate samples. *p < 0.05 and **p < 0.01 compared with WT control cultures.
(D) Rubicon suppresses autophagosome maturation. Raw246.7 cells expressing vector, Rubicon WT, ΔSR-N, or ΔHC mutant were treated with rapamycin, starvation, β-1,3-glucan, or SeV infection for the indicated times, and their cell lysates were used for IB with αLC3, αp62, or αactin. See also Figure S3.
Rubicon Has a Negative Effect on CBM-Mediated Inflammatory Cytokine Production
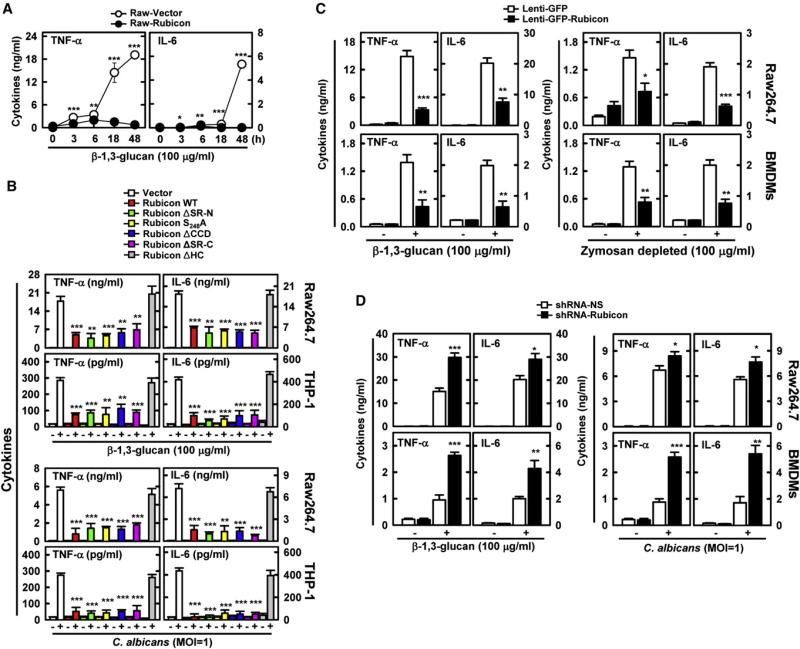
Rubicon expressing Raw264.7 cells produced markedly lower amounts of TNF-α and IL-6 than vector-containing control cells (Figure 4A, Figures S4A and S4B). To further delineate Rubicon's actions in CBM-mediated proinflammatory cytokine production, Raw264.7 and THP-1 cells expressing Rubicon mutants were evaluated for their activities in comparison to cells expressing WT upon various stimulations. Expression of the ΔSR-N, S284A, ΔCCD, or ΔSR-C mutant suppressed proinflammatory cytokine production as efficiently as WT, whereas expression of the ΔHC mutant had no effect (Figure 4B and Figure S4C). Consistently, lentivirus-mediated overexpression or shRNA-mediated depletion of Rubicon in mouse BMDMs or Raw264.7 cells significantly decreased or increased TNF-α and IL-6 production induced by β-1,3-glucan stimulation, respectively (Figures 4C and 4D, Figures S4D and S4E). Additionally, BMDCs were infected with recombinant adenoviruses (Ad-vector, Ad-Rubicon, or Ad-shRubicon), followed by β-1,3-glucan stimulation or SeV infection. This too showed that Rubicon expression markedly suppressed TNF-α and IL-6 production, whereas Rubicon depletion significantly enhanced it (Figure S4F). However, Rubicon expression neither showed any detectable effect on LPS (TLR4)- or CpG (TLR9)-stimulated cytokine productions in Raw264.7 cells nor affected cytokine productions in CARD9–/– BMDMs upon treatments with various stimuli including β-1,3-glucan, heat-killed Candida albicans, or viral infections (Figures S4G and S4H), indicating the specificity of Rubicon's action in Dectin1-CARD9-mediated signal transduction.
Figure 4. Negative Effect of Rubicon on Host Antifungal Immunity in a CARD9 Binding-Dependent Manner.
(A) Decrease of Dectin-1 pathway-induced cytokine productions by Rubicon. Raw264.7 cells containing vector or Rubicon were stimulated with β-1,3-glucan for ELISA. Additional data are shown in Figures S4A and S4B.
(B) Decrease of Dectin-1 pathway-induced cytokine productions by Rubicon. Raw264.7 cells and THP-1 cells containing vector, Rubicon WT, or its mutants were stimulated with β-1,3-glucan or C. albicans infection for cytokine production by ELISA. Additional data are shown in Figure S4C.
(C) Rubicon expression decreases cytokine productions induced by β-1,3-glucan or zymosan depleted by hot alkali treatment. Raw264.7 cells and BMDMs infected with lenti-GFP or lenti-GFP-Rubicon were stimulated with β-1,3-glucan or zymosan depleted by hot alkali treatment for cytokine ELISA.
(D) Depletion of Rubicon expression increases Dectin-1 pathway-induced cytokine productions. Raw264.7 cells and BMDMs lentivirus-shRNA-NS or lentivirus-shRNA-Rubicon were stimulated with β-1,3-glucan or C. albicans infection for cytokine ELISA. The data are the mean ± SD of values from three experiments.
*p < 0.05; **p < 0.01; ***p < 0.001 compared with the control. See also Figure S4.
Rubicon Has a Negative Effect on Host Antifungal Activity
Corresponding with the levels of proinflammatory cytokine production, the viability and growth rate of intracellular C. albicans were increased in Rubicon-expressing cells and decreased in Rubicon-depleted cells compared to vector-containing control cells (Figures 5A and 5B). Furthermore, the expression of ΔSR-N, S284A, ΔCCD, or ΔSR-C mutant increased the viability of intracellular C. albicans as efficiently as that of WT, whereas the expression of the ΔHC mutant showed no effects under the same conditions (Figure 5B).
Figure 5. Negative Effect of Rubicon on Host Antifungal Activity.
(A) Rubicon expression suppresses intracellular fungal killing activity. Raw246.7 or BMDMs infected with lenti-GFP or lenti-GFP-Rubicon for Rubicon expression or infected with lentivirus-shRNA-NS or lentivirus-shRNA-Rubicon for Rubicon depeletion were infected with C. albicans for the indicated times and then lysed to determine intracellular C. albicans loads.
(B) Rubicon suppresses intracellular fungal killing activity in a CARD9-binding-dependent manner. Raw246.7 cells expressing vector, Rubicon WT, or its mutants were infected with C. albicans for the indicated times and then lysed to determine intracellular C. albicans loads.
(C–E) Alteration of Rubicon gene expression affects mouse mortality after C. albicans infection. At 48 hr postinjection with Ad-vector (1 × 1013 pfu/kg), Ad-shRubicon (1 × 1012 pfu/kg), or Ad-Rubicon (1 × 1013 pfu/kg) twice i.v. via tail vein, mice were infected with C. albicans (1 × 107 CFU per mouse), and mortality was measured for n = 10 mice per group (C). Serum cytokine levels (D) or C. albicans loads of infected mice (n = 5 per group) in liver, spleen, kidney, and lung (E) were determined at 4 days p.i. with C. albicans (1 × 106 CFU). Additional data including Rubicon expression in mice are shown in Figures S5A and S5B. CFU, colony-forming units.
(F–I) Doxycycline-inducible, macrophage-specific Rubicon expression affects mouse mortality after C. albicans infection. After 7 days of doxycycline treatment, SRA-rtTA/Flag-Rubicon double transgenic mice were infected with C. albicans (1 × 107 CFU per mouse), and mortality was measured for n = 6 mice per group (F). C. albicans loads of infected mice (n = 6 per group) in liver and spleen (G), serum cytokine levels (H), and H&E staining of spleens (I) were determined at 4 days p.i. with C. albicans (1 × 106 CFU). Additional data including Rubicon expression in mice are shown in Figures S5C–S5E. The data are the mean ± SD of values from three experiments. *p < 0.05; **p < 0.01; ***p < 0.001 compared with the control. Scale bars, 200 μm. See also Figure S5.
In order to assess whether the depletion or expression of Rubicon affects in vivo host responses to C. albicans infection, recombinant adenoviruses (Ad-vector, Ad-shRubicon, or Ad-Rubicon) were injected intravenously via the tail vein twice and subsequently challenged intravenously with a lethal C. albicans dose (1 × 107 CFU per mouse) (Figures S5A and S5B). Mice infected with Ad-vector showed a median survival of 8 days, mice infected with Ad-Rubicon died detectably sooner (median survival, 6 days), and mice infected with Ad-shRubicon showed an increased survival rate (50% survival) (Figure 5C). To determine whether these effects were due to impaired and enhanced fungal clearance in the Rubicon-expressing and Rubicon-depleted mice, respectively, we measured the C. albicans loads and serum cytokine levels in the liver, spleen, kidney, and lung at 4 days after infection with a lower C. albicans dose (1 × 106 CFU per mouse). Under these conditions, Rubicon-expressing mice and Rubicon-depleted mice had, respectively, ~100-fold higher and ~50- to 100-fold lower C. albicans loads in those organs compared to mice infected with Ad-vector (Figure 5E). Correlated with the C. albicans loads, the serum levels of TNF-α and IL-6 were lower in Rubicon-expressing mice and higher in Rubicon-depleted mice than in mice infected with Ad-vector (Figure 5D).
To further investigate Rubicon's effect on in vivo host responses to C. albicans infection, we generated double transgenic mice SRA-rtTA/Flag-Rubicon that have both reverse tetracycline transactivator (rtTA) under the control of macrophage-specific promoter, a modified human scavenger receptor A (SRA) promoter (Puel et al., 2010), and Flag-tagged Rubicon under the control of tetracycline-inducible promoter. Immunoblot and confocal microscopy analyses of splenocytes isolated from SRA-rtTA/Flag-Rubicon mice showed the macrophage-specific, inducible expression of Rubicon (Figures S5C and S5D). At 7 days posttreatment of doxycycline, mice were subsequently challenged intravenously with C. albicans (1 × 107 CFU per mouse). Untreated SRA-rtTA/Flag-Rubicon mice showed a median survival of ~7 days with 40% survival rate, but doxycycline-treated mice died noticeably sooner (median survival, 2 days) (Figure 5F). In addition, untreated mice had ~100- to 300-fold lower C. albicans loads in spleen and liver, respectively, compared to doxycycline-treated mice (Figure 5G). Correlated with the C. albicans loads, untreated mice showed much higher serum TNF-α and IL-6 levels and more severe infection-induced splenomegaly and inflammation than doxycycline-treated mice (Figures 5H and 5I and Figure S5E). These results unambiguously show that host defenses against fungal infection are substantially affected by the levels of Rubicon expression.
Rubicon Negatively Affects Host Antiviral Activity
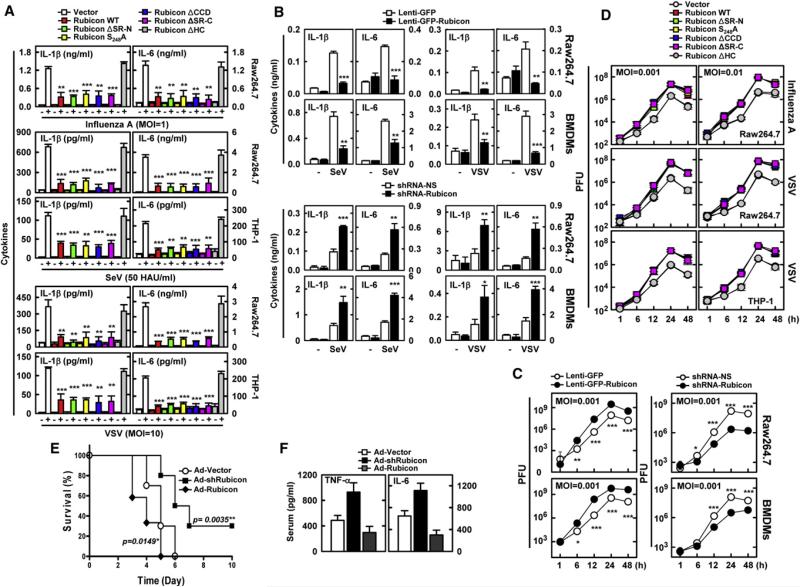
CARD9 also functions as an adaptor molecule to deliver the virus infection-mediated intracellular RIG-I signaling (Poeck et al., 2010). Rubicon expression markedly suppressed the phosphorylation of p38, JNK, and IκB, and the degradation of IκB induced by SeV infection (Figure S6A), and thereby led to significant decreases in TNF-α, IL-6, and IL-1β production (Figures 6A and 6B, Figures S6B–S6D). Conversely, the shRNA-mediated depletion of endogenous Rubicon expression led to detectable increases of TNF-α, IL-6, and IL-1β production upon SeV or vesicular stomatitis virus (VSV) infection (Figure 6B and Figure S6E). Consistent with the inflammatory cytokine productions, the replication rate of VSV was increased in Rubicon-expressing cells and decreased in Rubicon-depleted cells compared to vector-containing cells (Figure 6C, Figures S6F and S6G). Furthermore, Raw264.7 or THP-1 cells stably expressing Rubicon mutants were evaluated for viral infection-induced proinflammatory cytokine production. Expression of the ΔSR-N, S284A, ΔCCD, or ΔSR-C mutant suppressed proinflammatory cytokine production as efficiently as that of WT, whereas expression of the ΔHC mutant showed no significant effects (Figure 6A and Figure S6C). Correlated with their effects on proinflammatory cytokine production, expression of the Rubicon WT, ΔSR-N, and S284A mutants led to the increased replications of influenza A virus A/PR/8/34 and VSV, whereas expression of the ΔHC mutant showed no effects (Figure 6D and Figure S6H).
Figure 6. Negative Effect of Rubicon on Host Antiviral Activity.
(A) Decrease of virus infection-induced cytokine production by Rubicon. Raw264.7 cells containing vector, Rubicon WT, or its mutants were infected with influenza A virus PR8, SeV, or GFP-VSV for cytokine ELISA. Additional data of THP-1 cells are shown in Figures S6C.
(B) Rubicon expression levels affect virus infection-induced cytokine production. Raw264.7 cells and BMDMs infected with lenti-GFP or lenti-GFP-Rubicon for Rubicon expression or infected with lentivirus-shRNA-NS or lentivirus-shRNA-Rubicon for Rubicon depeletion were infected with SeV or VSV for cytokine ELISA. Additional data are shown in Figures S6D and S6E.
(C) Rubicon expression levels affect intracellular virus-killing activity. Raw246.7 or BMDMs infected with lenti-GFP or lenti-GFP-Rubicon for Rubicon expression or infected with lentivirus-shRNA-NS or lentivirus-shRNA-Rubicon for Rubicon depeletion were infected with VSV for the indicated times, the culture medium was harvested, and virus loads were determined by plaque assay on Vero cells. Additional data are shown in Figures S6F and S6G.
(D) Rubicon expression suppresses intracellular virus-killing activity in a CARD9-binding-dependent manner. Raw246.7 cells expressing vector, Rubicon WT, or its mutants were infected with influenza A virus PR8 strain or VSV at various mois for the indicated times, and virus loads were determined by plaque assay on MDCK or Vero cells, respectively. Additional data are shown in Figure S6H.
(E and F) Alteration of Rubicon gene expression affects mouse mortality after influenza A virus infection. At 48 hr postinjection with Ad-vector (1 × 1013 pfu/kg), AdshRubicon (1 × 1012 pfu/kg), or Ad-Rubicon (1 × 1013 pfu/kg) twice i.v. via tail vein, mice were infected with influenza A virus PR8 strain (1 × 105 PFU per mouse) and mortality was measured for n = 10 mice per group (E). (F) Serum cytokine levels of infected mice (n = 5 per group) were determined at 3 days p.i. with influenza A virus PR8 strain (5 × 104 PFU). Additional data including body weight loss and lung virus loads of the infected mice are shown in Figures S6I and S6J. PFU, plaque-forming units. The data are the mean ± SD of values from three experiments. *p < 0.05; **p < 0.01; ***p < 0.001 compared with the control. See also Figure S6.
In order to assess whether Rubicon expression levels affect in vivo host responses to influenza A virus infection, a similar approach to that described in Figure 5 utilizing recombinant Rubicon adenoviruses was used, and mice were challenged intranasally with a lethal dose of influenza A virus A/PR/8/34 strain (1 × 105 PFU per mouse). Mice infected with Ad-vector showed rapid loss of weight and a median survival of 5 days, mice infected with Ad-Rubicon lost weight faster and died detectably sooner (median survival, 4 days), and mice infected with Ad-shRubicon showed a noticeably delayed mortality rate and increased survival rate (30% survival) (Figure 6E and Figure S6I). Rubicon-expressing mice and Rubicon-depleted mice had, respectively, ~100-fold higher and ~100-fold lower virus loads in their lungs compared to mice infected with Ad-vector (Figure S6J). In correlation with the influenza A virus loads, the serum levels of TNF-α and IL-6 were lower in Rubicon-expressing mice and higher in Rubicon-depleted mice than in mice infected with Ad-vector (Figure 6F). Thus, host defenses against viral infection are substantially affected by the levels of Rubicon expression.
DISCUSSION
In this report, we present Rubicon as a specific feedback inhibitor of CARD9-mediated PRR-signal transduction, preventing unbalanced proinflammatory responses.
Changing Partners
Similar to its interaction with the Beclin-1 complex and the NADPH oxidase complex upon autophagy and TLR stimulation, respectively, Rubicon also changes its binding partners from 14-3-3β to CARD9 in a Dectin-1- or RIG-I-stimulation-dependent, competitive manner, thereby disassembling the CBM signaling complex and ultimately stopping PRR-induced inflammatory cytokine production. The obvious question is what types of molecular mechanisms specifically induce Rubicon's transfer from 14-3-3β binding to CARD9 binding. Rubicon contains two serine-rich regions, SR-N and SR-C, in its central region that appear to be responsible for “latent” binding to 14-3-3β and “inducible” binding to p22phox, respectively. Indeed, the specific deletion (ΔSR-N) or point mutation (S248A) of Rubicon completely abolished its interaction with 14-3-3β (Figures 1B and 1C). Based on these, we hypothesize that Rubicon's interaction with 14-3-3β, which occurs constitutively and during the initial period of stimulation, sterically hinders a CARD9 interaction, permitting CBM signal transduction, NF-κB activation, and proinflammatory cytokine production (Figure 7). At the late stage of signaling, however, Rubicon may lose S248-phosphorylation, dissociating from 14-3-3β to allow a stable but transient interaction with CARD9, thus disassembling the CBM signaling module and thereby terminating PRR-mediated proinflammatory cytokine production (Figure 7). However, it is also possible that Rubicon interaction with 14-3-3β may result in the recruitment of other proteins and/or alteration in its subcellular localization, which prevents the interaction with CARD9. Nevertheless, by changing partners between 14-3-3β and CARD9 in a specific-stimulation- and time-dependent manner, Rubicon functions as the regulatory “off” switch of Dectin-1-and RIG-I-mediated inflammatory signal transduction in this model.
Figure 7. Hypothetical Model of Rubicon-Mediated Feedback Inhibition of CBM Signal Transduction.
Please see the Discussion for detail.
Several of the data presented here support our hypothesis. First, Rubicon's interaction with 14-3-3β was initially evident but dramatically decreased after 30 min of β-1,3-glucan stimulation. By striking contrast, Rubicon interaction with CARD9 was undetectable during the initial 30 min of stimulation, but robustly increased after 30–60 min of stimulation, and then declined thereafter (Figures 2A and 2B and Figure S2A). These features are also binding specific: the ΔSR-N and S284A mutants that lost 14-3-3β interaction continuously bound CARD9 with no β-1,3-glucan stimulation conditions, and the ΔHC mutant that lost CARD9 interaction continuously associated with 14-3-3β regardless of β-1,3-glucan stimulation. Second, as seen with Rubicon, CARD9 also shows differential binding partners and kinetics: it associates with BCL10 without stimulation, with these interactions detectably increasing at 15 min of β-1,3-glucan stimulation but drastically decreasing thereafter (Figure S2D). Contrastingly, the CARD9-Rubicon interaction is initially minimal, but markedly increases after 30–60 min of β-1,3-glucan stimulation and declines afterward (Figure S2D). Third, the unique binding partners and kinetics of both Rubicon and CARD9 are also stimulation specific for only β-1,3-glucan stimulation or SeV infection, as neither rapamycin nor zymosan stimulation modified their interactions. These results collectively demonstrate that Rubicon dynamically changes binding partners from 14-3-3β to CARD9 in a specific-stimulation-dependent, competitive manner.
Inhibitory Feedback Regulation of CARD9 Signaling
Positive and negative regulatory mechanisms are required to maintain most biological processes at equilibrium. While a robust antimicrobial response induced by the PRR pathway is essential for controling microbial infection, this response must be metered and eventually eliminated when it reaches a late stage, to prevent damage from the excessive production of inflammatory cytokines. Since CARD9 is a central molecule for transducing signals from multiple PRRs to induce immunity to various pathogens, it is important to understand how the deregulation of CARD9 signaling might contribute to immune cell-mediated diseases. Loss-of-function mutations in the CARD9 gene causes susceptibility to infections, as CARD9–/– mice are highly vulnerable to fungal and bacterial pathogen infections (Hsu et al., 2007; Ruland, 2008; Werninghaus et al., 2009; Wu et al., 2009). Furthermore, a CARD9 mutation in humans is associated with an increased susceptibility to chronic mucocutaneous candidiasis (Glocker et al., 2009). On the other hand, aberrant activation of CARD9, either through genetic mutations or via environmental factors, may result in pathological immune cell activation, causing inflammatory diseases or certain cancers (Ruland, 2008). For instance, a single CARD9 nucleotide polymorphism is associated with inflammatory bowel disease (Zhernakova et al., 2008), and CARD9 overexpression is found in gastric B cell lymphoma specimens (Zhernakova et al., 2008; Zhou et al., 2006). To avoid aberrant activation of CARD9 signaling activity and excessive production of inflammatory cytokines, the host immune system employs Rubicon to target 14-3-3β and CARD9 in a specific-stimulation-dependent manner, in order to disassemble and halt CBM complex-mediated signaling activity. Thus, Rubicon serves as an inhibitory feedback regulator of CBM-mediated immune responses by acting as a physiological brake.
Rubicon's Roles in Autophagy-, Phagocytosis-, and PRR-Mediated Signal Transduction
The initial innate immunity mechanisms of autophagy, phagocytosis, and PRRs have been shown to efficiently collaborate to generate effective host responses against microbial infections (Kumar et al., 2011; Palm and Medzhitov, 2009; Reid et al., 2009). As seen in the cooperation between PRRs and phagocytosis and between PRRs and autophagy, phagocytosis and autophagy also collaborate as part of the host's first line of immune defense against microbial invasions (Deretic and Levine, 2009). Specifically, autophagy facilitates phagocytosis by promoting phagosome maturation and rapid acidification, and by preventing pathogens from escaping into the cytosol (Huang et al., 2009; Sanjuan et al., 2007; Tal et al., 2009; Travassos et al., 2010; Virgin and Levine, 2009). While Rubicon was initially identified as an autophagy gatekeeper, it is involved in three different ancient innate immune machineries, autophagy, phagocytosis, and PRRs, by targeting them depending on the environmental stimuli and ultimately generating an intracellular immune milieu against microbial infection. To efficiently do this, Rubicon acts on the Beclin-1-containing autophagy complex, on the NADPH-oxidase-containing phagocytosis complex, and on the CARD9-containing signaling complex in a functionally and genetically separable manner. Although the precise contributions of Rubicon to the control of host immunity are only beginning to be defined, it is clear that Rubicon is an important regulator of innate immune responses against various microbial infections.
EXPERIMENTAL PROCEDURES
Yeast Two-Hybrid Screen
Yeast transformation with library cDNA was performed as recommended by the manufacturers.
Briefly, yeast strain Y187 bearing Gal4-Rubicon full-length or Gal4-N-terminal region (aa 1–505), or Gal4-C-terminal region (aa 506–972) plasmid, was grown overnight in synthetic dropout (SD)/-Trp medium to a density of approximately 107 cells/ml, then diluted in 1 l of warmed YPD to an optical density (OD600) of 0.2–0.3 and grown to exponential stage. Cells were harvested and washed with 100 ml of water twice and TE buffer once. The pellets were resuspended in 8 ml of 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, 0.1 M Li-acetate (LiOAc), and the suspension was mixed with 1 mg of transforming DNA and 20 mg of single-stranded salmon sperm DNA, after which 60 ml of a solution of 40% polyethyleneglycol-4000 in Tris-EDTA-LiOAc was added and mixed thoroughly, followed by incubation at 30° C with agitation for 30 min. After a heat pulse at 42° C for 15 min, cells were pelleted, washed with 50 ml of Tris-EDTA, and plated on selective medium. Library screening and recovery of plasmids were performed according to the manufacturer's instructions (Clontech).
Quantification of C. albicans Growth
To ensure the reliable quantification of intracellular yeast cells, colony-forming units (CFUs) were utilized. Cells were infected for 2 hr with C. albicans at different mois. Then cells were washed three times with PBS, and fresh medium was added. After various periods of incubation, cells were lysed with 0.3% saponin to release the intracellular yeast cells, and the lysates of infected cells were then resuspended vigorously, transferred to screw-capped tubes, and sonicated in a preheated 30° C water bath sonicator (Elma) for 5 min. Aliquots of the sonicates were then diluted 10-fold in YPD broth medium. Four dilutions of each sample were plated separately on YPD agar plates and incubated at 30° C with 5% CO2 for 1 day. CFU was counted, and the intracellular yeast cells were calculated in comparison with CFU of yeast grown in the same conditions without cells. For statistical reasons, dilutions that yielded 50–200 plaques per well were used for calculation of the titer.
Quantification of VSV or Influenza A Virus Growth
The titer or concentration of infectious virus particles in solution or cell culture medium was determined by plaque assay. For VSV, Vero cells were grown until a monolayer of cells was formed. Serially diluted virus solution (0.1 ml) was added to each well and incubated for 1 hr at RT in a rocker. Virus solutions were then removed, and cells were covered with overlay medium (1:1 mixture of 1.5% methyl cellulose agar and MEM without FBS). After 5 min, mixtures were solidified and placed in a 37° C incubator for 24–36 hr.
For influenza A virus, MDCK cell monolayers were inoculated with 0.1 ml of virus appropriately diluted in PBS supplemented with 0.2% bovine serum albumin for 60 min at room temperature. At the completion of the 60 min incubation, cells were mixed the 2% Oxoid agar with the remaining premixed components of the agar overlay (2× MEM without FBS, 1% DEAE dextran, 5% NaHCO3, 1 mg/ml trypsin-TPCK) and incubated for at least 3 days, or until virus plaques were clearly visible. Then crystal violet stain was added to each well and incubated overnight at RT for counting plagues, and the titer of the solution was calculated.
Production of Lentiviral shRubicon or Rubicon
For silencing of human Rubicon, oligonucleotide sequences for shRNA interference with Rubicon expression are bp 513–536 of 5′-GAUCGAUGCGUCCAUGUUU-3′, followed by a 9 nt noncomplementary spacer (TTCAAGAGA) and the reverse complement of the initial 19 nt sequence. These dsDNA oligonucleotides were cloned into the pGIPZ lentiviral vector (Open Biosystems). Lentiviruses were produced by transient transfection using packaging plasmids (psPAX2 and pMD2.VSV-G purchased from Addgene) after Lipofectamine 2000 mediated transient transfection into 293T cells. Control vector was constructed by inserting sequences with limited homology to the Rubicon sequences. DNA fragments corresponding to the coding sequences of the Rubicon genes were amplified by PCR and subcloned into pCDH-CMV vector (System Biosciences).
Transduction of Lentiviral shRubicon or Rubicon
Viral-containing media were collected 72 hr posttransfection and harvested for the viral particles by passing the supernatants through a 0.45 μm filter. The supernatants were used to infect 2 × 105 cells in 6-well plates in the presence of 8 μg/ml Polybrene. For lentivirus infection, cells (5 × 105 cells/ml) in DMEM + 10% FBS were seeded in 24-well plates. After 24 hr, cells were infected with lentiviral vectors at various mois in the presence of 8 μg/ml Polybrene. On the following day, the medium was freshly replaced, and cells were incubated for additional 4 days for Rubicon knockdown. A parallel experiment using a GFP encoding lentivirus indicated that a minimum of 80% of Raw264.7 and BMDMs cells were transduced by lentiviruses. Titration of the lentiviral vectors was determined using 293T cells. Briefly, approximately 2 × 105 cells were plated in each well of a 6-well plate. On the following day, cells were infected with viral supernatants in the presence of 8 μg/ml Polybrene. After 24 hr, medium was removed and replaced with fresh medium containing 5 μg/ml puromycin. On day 14, cells were stained with crystal violet for 15 min, and colonies were counted using a cutoff of 50 viable cells.
Construction of Adenoviral shRubicon or Rubicon
An adenovirus expressing short hairpin RNA (shRNA) to the Rubicon gene was constructed using the AdEasy system (Stategene). The shRNA oligonucleotides sequences were as follows: 5′-GATCCCCGATAGACAGTATATCAGAATTCAAGAGATTCTGATATACTGTCTATCTTTTTA-3′, 5′-GGGCTATCTGTCATATAGTCTTAAGTTCTCTAAGACTATATGACAGATAGAAAAATTCGA-3′. These dsDNA oligonucleotides were cloned into the pSuper vector between the BglII and HindIII restriction sites containing the human H1 promoter. The double-strand shRNA oligonucleotides containing the termination signal were inserted at the 3′ end of the human H1 promoter and subcloned into the pShuttle vector NotI and HindIII restriction sites. Control vector was constructed by inserting a sequence that expresses an shRNA with limited homology to the Rubicon sequences. DNA fragments corresponding to the coding sequences of the Rubicon genes were amplified by PCR and subcloned into the pShuttle-CMV vector between the NotI and EcoRV restriction sites.
Adenovirus Production
Recombinant adenoviruses were constructed using AdEasy system: digested adenovirus vectors with the Pac I were transfected into the AD-293 producer cells in a 6-well plate and cultured with fresh media until cytopathic effect was observed. When 80% CPE were observed, recombinant adenoviruses were harvested by repeatedly freezing at –80° C and thawing at 37° C four times. Cell lysates were then centrifuged at 2000 g for 30 min at 25° C, and the supernatants containing recombinant adenovirus particles were stored at –80° C.
All adenoviruses were propagated in AD-293 cells, purified, and concentrated by BD Adeno-X purification kit. The typical titers were in the range of 1012–1013 plaque-forming units (pfu)/mL as determined via plaque assay using 1.25% SeaPlaque GTG agarose (BioWhittaker Molecular Applications) overlay. A sterile carrier solution phosphate-buffered saline was used for control injections and dilution of the viruses.
Supplementary Material
ACKNOWLEDGMENTS
This work was partly supported by CA82057, CA31363, CA115284, DE019085, AI073099, AI083025, HL110609 the GRL Program (K20815000001) from the National Research Foundation of Korea, the Hastings Foundation, and the Fletcher Jones Foundation (J.U.J.); and by the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Science, and Technology (2011-0014785, J.S.L. and J.U.J.). We thank Drs. Richard Bennett, Will DePaolo, and Yi Luo for reagents and Stacy Lee for manuscript preparation. Finally, we thank all of J.U.J.'s lab members for their discussions.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures and Supplemental Experimental Procedures and can be found with this article online at doi:10.1016/j.chom. 2012.01.019.
REFERENCES
- Arimoto K, Takahashi H, Hishiki T, Konishi H, Fujita T, Shimotohno K. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc. Natl. Acad. Sci. USA. 2007;104:7500–7505. doi: 10.1073/pnas.0611551104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi L, Gojestani S, Wu W, Hsu YM, Zhu J, Ariizumi K, Lin X. CARD9 mediates dectin-2-induced IkappaBalpha kinase ubiquitination leading to activation of NF-kappaB in response to stimulation by the hyphal form of Candida albicans. J. Biol. Chem. 2010;285:25969–25977. doi: 10.1074/jbc.M110.131300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blonska M, Lin X. NF-kappaB signaling pathways regulated by CARMA family of scaffold proteins. Cell Res. 2011;21:55–70. doi: 10.1038/cr.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond RA, Saijo S, Iwakura Y, Brown GD. The role of Syk/CARD9 coupled C-type lectins in antifungal immunity. Eur. J. Immunol. 2011;41:276–281. doi: 10.1002/eji.201041252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glocker EO, Hennigs A, Nabavi M, Schaffer AA, Woellner C, Salzer U, Pfeifer D, Veelken H, Warnatz K, Tahami F, et al. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N. Engl. J. Med. 2009;361:1727–1735. doi: 10.1056/NEJMoa0810719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gringhuis SI, Wevers BA, Kaptein TM, van Capel TM, Theelen B, Boekhout T, de Jong EC, Geijtenbeek TB. Selective C-Rel activation via Malt1 controls anti-fungal T(H)-17 immunity by dectin-1 and dectin-2. PLoS Pathog. 2011;7:e1001259. doi: 10.1371/journal.ppat.1001259. 10.1371/journal.ppat.1001259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross O, Gewies A, Finger K, Schafer M, Sparwasser T, Peschel C, Forster I, Ruland J. Card9 controls a non-TLR signalling pathway for innate anti-fungal immunity. Nature. 2006;442:651–656. doi: 10.1038/nature04926. [DOI] [PubMed] [Google Scholar]
- Hara H, Saito T. CARD9 versus CARMA1 in innate and adaptive immunity. Trends Immunol. 2009;30:234–242. doi: 10.1016/j.it.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Hsu YM, Zhang Y, You Y, Wang D, Li H, Duramad O, Qin XF, Dong C, Lin X. The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nat. Immunol. 2007;8:198–205. doi: 10.1038/ni1426. [DOI] [PubMed] [Google Scholar]
- Huang J, Canadien V, Lam GY, Steinberg BE, Dinauer MC, Magalhaes MA, Glogauer M, Grinstein S, Brumell JH. Activation of antibacterial autophagy by NADPH oxidases. Proc. Natl. Acad. Sci. USA. 2009;106:6226–6231. doi: 10.1073/pnas.0811045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens S, Burns K, Tschopp J, Beyaert R. Regulation of interleukin-1- and lipopolysaccharide-induced NF-kappaB activation by alternative splicing of MyD88. Curr. Biol. 2002;12:467–471. doi: 10.1016/s0960-9822(02)00712-1. [DOI] [PubMed] [Google Scholar]
- Kerrigan AM, Brown GD. Syk-coupled C-type lectin receptors that mediate cellular activation via single tyrosine based activation motifs. Immunol. Rev. 2010;234:335–352. doi: 10.1111/j.0105-2896.2009.00882.x. [DOI] [PubMed] [Google Scholar]
- Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int. Rev. Immunol. 2011;30:16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- Leung E, Hong J, Fraser A, Krissansen GW. Splicing of NOD2 (CARD15) RNA transcripts. Mol. Immunol. 2007;44:284–294. doi: 10.1016/j.molimm.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- McGreal EP, Rosas M, Brown GD, Zamze S, Wong SY, Gordon S, Martinez-Pomares L, Taylor PR. The carbohydrate-recognition domain of Dectin-2 is a C-type lectin with specificity for high mannose. Glycobiology. 2006;16:422–430. doi: 10.1093/glycob/cwj077. [DOI] [PubMed] [Google Scholar]
- Morrison DK. The 14-3-3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol. 2009;19:16–23. doi: 10.1016/j.tcb.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppermann FS, Gnad F, Olsen JV, Hornberger R, Greff Z, Keri G, Mann M, Daub H. Large-scale proteomics analysis of the human kinome. Mol. Cell. Proteomics. 2009;8:1751–1764. doi: 10.1074/mcp.M800588-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm NW, Medzhitov R. Pattern recognition receptors and control of adaptive immunity. Immunol. Rev. 2009;227:221–233. doi: 10.1111/j.1600-065X.2008.00731.x. [DOI] [PubMed] [Google Scholar]
- Poeck H, Bscheider M, Gross O, Finger K, Roth S, Rebsamen M, Hannesschlager N, Schlee M, Rothenfusser S, Barchet W, et al. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1 beta production. Nat. Immunol. 2010;11:63–69. doi: 10.1038/ni.1824. [DOI] [PubMed] [Google Scholar]
- Puel A, Picard C, Cypowyj S, Lilic D, Abel L, Casanova JL. Inborn errors of mucocutaneous immunity to Candida albicans in humans: a role for IL-17 cytokines? Curr. Opin. Immunol. 2010;22:467–474. doi: 10.1016/j.coi.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DM, Gow NA, Brown GD. Pattern recognition: recent insights from Dectin-1. Curr. Opin. Immunol. 2009;21:30–37. doi: 10.1016/j.coi.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenstiel P, Huse K, Till A, Hampe J, Hellmig S, Sina C, Billmann S, von Kampen O, Waetzig GH, Platzer M, et al. A short isoform of NOD2/CARD15, NOD2-S, is an endogenous inhibitor of NOD2/receptor-interacting protein kinase 2-induced signaling pathways. Proc. Natl. Acad. Sci. USA. 2006;103:3280–3285. doi: 10.1073/pnas.0505423103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruland J. CARD9 signaling in the innate immune response. Ann. N Y Acad. Sci. 2008;1143:35–44. doi: 10.1196/annals.1443.024. [DOI] [PubMed] [Google Scholar]
- Saijo S, Ikeda S, Yamabe K, Kakuta S, Ishigame H, Akitsu A, Fujikado N, Kusaka T, Kubo S, Chung SH, et al. Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity. 2010;32:681–691. doi: 10.1016/j.immuni.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc. Natl. Acad. Sci. USA. 2009;106:2770–2775. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, Yuan L, Soares F, Chea E, Le Bourhis L, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat. Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- Underhill DM, Shimada T. A pair of 9s: it's in the CARDs. Nat. Immunol. 2007;8:122–124. doi: 10.1038/ni0207-122. [DOI] [PubMed] [Google Scholar]
- Virgin HW, Levine B. Autophagy genes in immunity. Nat. Immunol. 2009;10:461–470. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werninghaus K, Babiak A, Gross O, Holscher C, Dietrich H, Agger EM, Mages J, Mocsai A, Schoenen H, Finger K, et al. Adjuvanticity of a synthetic cord factor analogue for subunit Mycobacterium tuberculosis vaccination requires FcRgamma-Syk-Card9-dependent innate immune activation. J. Exp. Med. 2009;206:89–97. doi: 10.1084/jem.20081445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Hsu YM, Bi L, Songyang Z, Lin X. CARD9 facilitates microbe-elicited production of reactive oxygen species by regulating the LyGDI-Rac1 complex. Nat. Immunol. 2009;10:1208–1214. doi: 10.1038/ni.1788. [DOI] [PubMed] [Google Scholar]
- Yang C-S, Lee J-S, Rodgers M, Min C-K, Lee J-Y, Kim HJ, Lee K-H, Kim C-J, Oh B, Zandi E, et al. The autophagy protein Rubicon mediates phagocytic NADPH oxidase activation in response to microbial infection or TLR stimulation. Cell Host Microbe. 2012;11(this issue):264–276. doi: 10.1016/j.chom.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhernakova A, Festen EM, Franke L, Trynka G, van Diemen CC, Monsuur AJ, Bevova M, Nijmeijer RM, van 't Slot R, Heijmans R, et al. Genetic analysis of innate immunity in Crohn's disease and ulcerative colitis identifies two susceptibility loci harboring CARD9 and IL18RAP. Am. J. Hum. Genet. 2008;82:1202–1210. doi: 10.1016/j.ajhg.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Ye H, Martin-Subero JI, Hamoudi R, Lu YJ, Wang R, Siebert R, Shipley J, Isaacson PG, Dogan A, et al. Distinct comparative genomic hybridisation profiles in gastric mucosa-associated lymphoid tissue lymphomas with and without t(11;18)(q21;q21). Br. J. Haematol. 2006;133:35–42. doi: 10.1111/j.1365-2141.2006.05969.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.