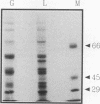
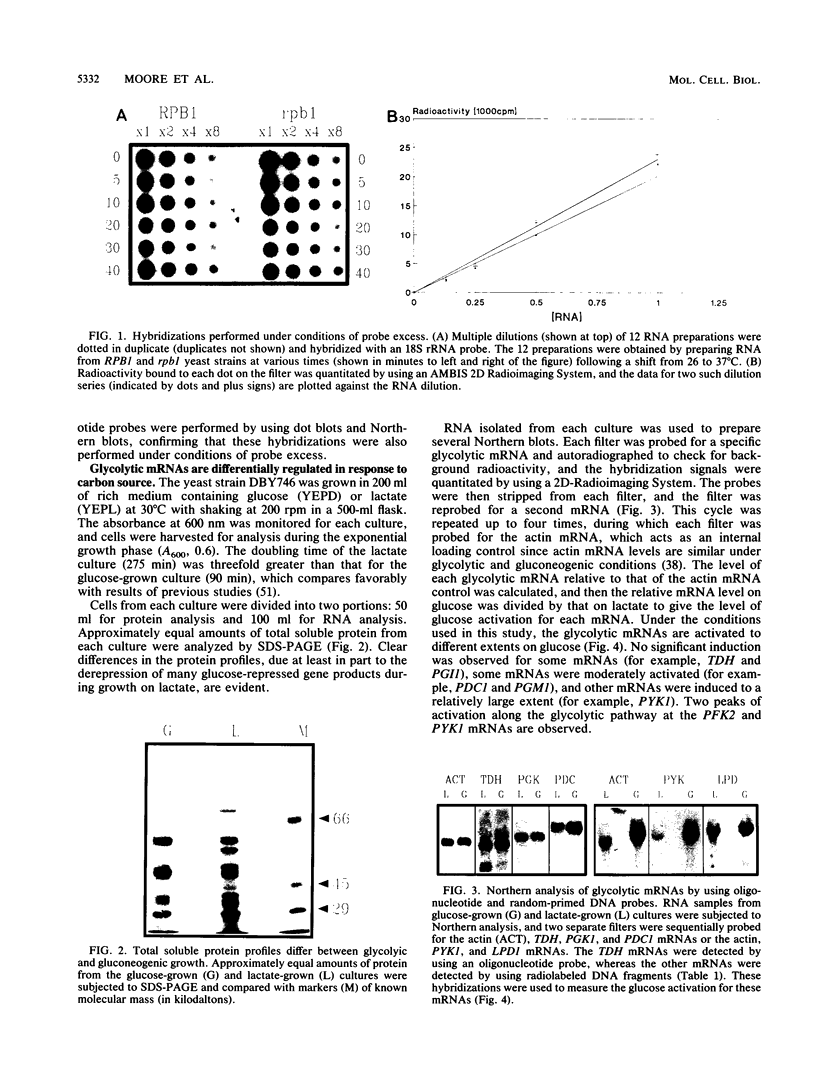
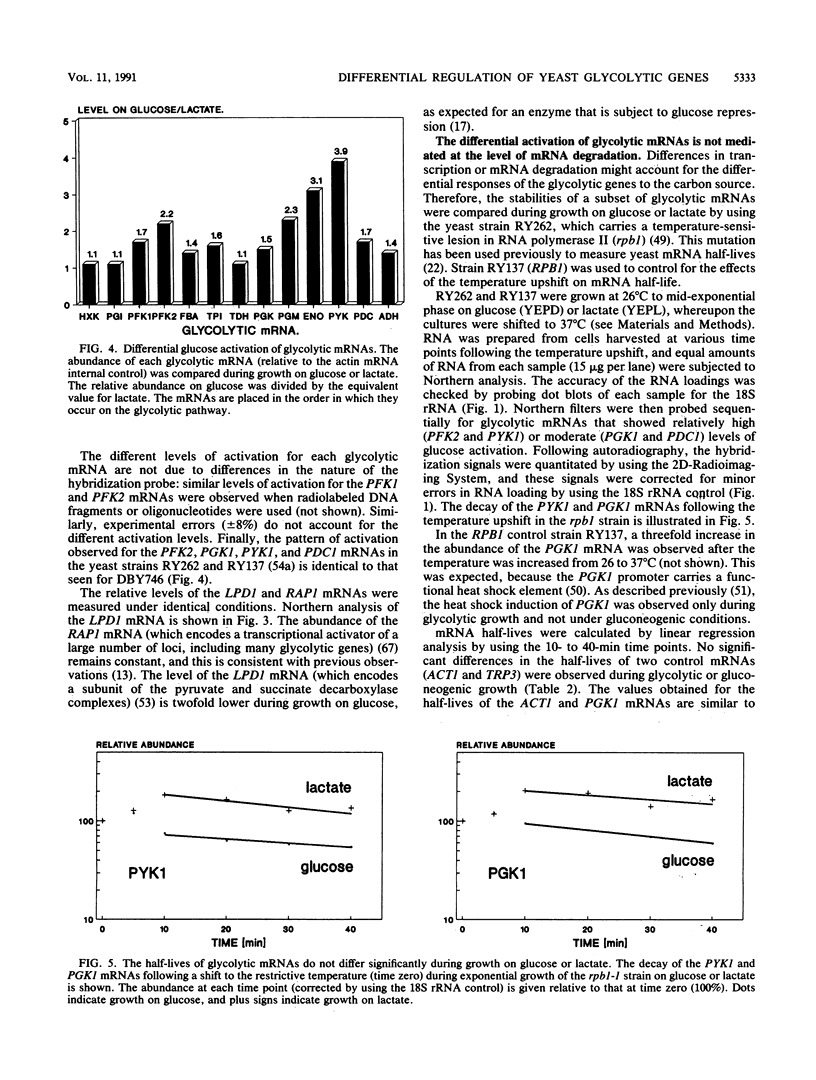
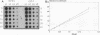Abstract
The regulation of glycolytic genes in response to carbon source in the yeast Saccharomyces cerevisiae has been studied. When the relative levels of each glycolytic mRNA were compared during exponential growth on glucose or lactate, the various glycolytic mRNAs were found to be induced to differing extents by glucose. No significant differences in the stabilities of the PFK2, PGK1, PYK1, or PDC1 mRNAs during growth on glucose or lactate were observed. PYK::lacZ and PGK::lacZ fusions were integrated independently into the yeast genome at the ura3 locus. The manner in which these fusions were differentially regulated in response to carbon source was similar to that of their respective wild-type loci. Therefore, the regulation of glycolytic mRNA levels is mediated at the transcriptional level. When the mRNAs are ordered with respect to the glycolytic pathway, two peaks of maximal induction are observed at phosphofructokinase and pyruvate kinase. These enzymes (i) catalyze the two essentially irreversible steps on the pathway, (ii) are the two glycolytic enzymes that are circumvented during gluconeogenesis and hence are specific to glycolysis, and (iii) are encoded by mRNAs that we have shown previously to be coregulated at the translational level in S. cerevisiae (P. A. Moore, A. J. Bettany, and A. J. P. Brown, NATO ASI Ser. Ser. H Cell Biol. 49:421-432, 1990). This differential regulation of glycolytic mRNA levels might therefore have a significant influence upon glycolytic flux in S. cerevisiae.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aguilera A., Zimmermann F. K. Isolation and molecular analysis of the phosphoglucose isomerase structural gene of Saccharomyces cerevisiae. Mol Gen Genet. 1986 Jan;202(1):83–89. doi: 10.1007/BF00330521. [DOI] [PubMed] [Google Scholar]
- Alber T., Kawasaki G. Nucleotide sequence of the triose phosphate isomerase gene of Saccharomyces cerevisiae. J Mol Appl Genet. 1982;1(5):419–434. [PubMed] [Google Scholar]
- Baker H. V. Glycolytic gene expression in Saccharomyces cerevisiae: nucleotide sequence of GCR1, null mutants, and evidence for expression. Mol Cell Biol. 1986 Nov;6(11):3774–3784. doi: 10.1128/mcb.6.11.3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggs J. D. Transformation of yeast by a replicating hybrid plasmid. Nature. 1978 Sep 14;275(5676):104–109. doi: 10.1038/275104a0. [DOI] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase. J Biol Chem. 1982 Mar 25;257(6):3018–3025. [PubMed] [Google Scholar]
- Bettany A. J., Moore P. A., Cafferkey R., Bell L. D., Goodey A. R., Carter B. L., Brown A. J. 5'-secondary structure formation, in contrast to a short string of non-preferred codons, inhibits the translation of the pyruvate kinase mRNA in yeast. Yeast. 1989 May-Jun;5(3):187–198. doi: 10.1002/yea.320050308. [DOI] [PubMed] [Google Scholar]
- Brindle P. K., Holland J. P., Willett C. E., Innis M. A., Holland M. J. Multiple factors bind the upstream activation sites of the yeast enolase genes ENO1 and ENO2: ABFI protein, like repressor activator protein RAP1, binds cis-acting sequences which modulate repression or activation of transcription. Mol Cell Biol. 1990 Sep;10(9):4872–4885. doi: 10.1128/mcb.10.9.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A. J. Messenger RNA stability in yeast. Yeast. 1989 Jul-Aug;5(4):239–257. doi: 10.1002/yea.320050405. [DOI] [PubMed] [Google Scholar]
- Buchman A. R., Lue N. F., Kornberg R. D. Connections between transcriptional activators, silencers, and telomeres as revealed by functional analysis of a yeast DNA-binding protein. Mol Cell Biol. 1988 Dec;8(12):5086–5099. doi: 10.1128/mcb.8.12.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke R. L., Tekamp-Olson P., Najarian R. The isolation, characterization, and sequence of the pyruvate kinase gene of Saccharomyces cerevisiae. J Biol Chem. 1983 Feb 25;258(4):2193–2201. [PubMed] [Google Scholar]
- Butler G., McConnell D. J. Identification of an upstream activation site in the pyruvate decarboxylase structural gene (PDC1) of Saccharomyces cerevisiae. Curr Genet. 1988 Nov;14(5):405–412. doi: 10.1007/BF00521261. [DOI] [PubMed] [Google Scholar]
- Chambers A., Tsang J. S., Stanway C., Kingsman A. J., Kingsman S. M. Transcriptional control of the Saccharomyces cerevisiae PGK gene by RAP1. Mol Cell Biol. 1989 Dec;9(12):5516–5524. doi: 10.1128/mcb.9.12.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan A. M., Donahue T. F. Sequence and structural features associated with translational initiator regions in yeast--a review. Gene. 1987;59(1):1–18. doi: 10.1016/0378-1119(87)90261-7. [DOI] [PubMed] [Google Scholar]
- Clifton D., Fraenkel D. G. The gcr (glycolysis regulation) mutation of Saccharomyces cerevisiae. J Biol Chem. 1981 Dec 25;256(24):13074–13078. [PubMed] [Google Scholar]
- Della Seta F., Ciafré S. A., Marck C., Santoro B., Presutti C., Sentenac A., Bozzoni I. The ABF1 factor is the transcriptional activator of the L2 ribosomal protein genes in Saccharomyces cerevisiae. Mol Cell Biol. 1990 May;10(5):2437–2441. doi: 10.1128/mcb.10.5.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis C. L., Ferguson J., Young E. T. mRNA levels for the fermentative alcohol dehydrogenase of Saccharomyces cerevisiae decrease upon growth on a nonfermentable carbon source. J Biol Chem. 1983 Jan 25;258(2):1165–1171. [PubMed] [Google Scholar]
- Entian K. D., Fröhlich K. U. Saccharomyces cerevisiae mutants provide evidence of hexokinase PII as a bifunctional enzyme with catalytic and regulatory domains for triggering carbon catabolite repression. J Bacteriol. 1984 Apr;158(1):29–35. doi: 10.1128/jb.158.1.29-35.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entian K. D. Glucose repression: a complex regulatory system in yeast. Microbiol Sci. 1986 Dec;3(12):366–371. [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Heinisch J., Ritzel R. G., von Borstel R. C., Aguilera A., Rodicio R., Zimmermann F. K. The phosphofructokinase genes of yeast evolved from two duplication events. Gene. 1989 May 30;78(2):309–321. doi: 10.1016/0378-1119(89)90233-3. [DOI] [PubMed] [Google Scholar]
- Herrick D., Parker R., Jacobson A. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1990 May;10(5):2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland J. P., Brindle P. K., Holland M. J. Sequences within an upstream activation site in the yeast enolase gene ENO2 modulate repression of ENO2 expression in strains carrying a null mutation in the positive regulatory gene GCR1. Mol Cell Biol. 1990 Sep;10(9):4863–4871. doi: 10.1128/mcb.10.9.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland J. P., Holland M. J. The primary structure of a glyceraldehyde-3-phosphate dehydrogenase gene from Saccharomyces cerevisiae. J Biol Chem. 1979 Oct 10;254(19):9839–9845. [PubMed] [Google Scholar]
- Holland J. P., Labieniec L., Swimmer C., Holland M. J. Homologous nucleotide sequences at the 5' termini of messenger RNAs synthesized from the yeast enolase and glyceraldehyde-3-phosphate dehydrogenase gene families. The primary structure of a third yeast glyceraldehyde-3-phosphate dehydrogenase gene. J Biol Chem. 1983 Apr 25;258(8):5291–5299. [PubMed] [Google Scholar]
- Holland M. J., Holland J. P., Thill G. P., Jackson K. A. The primary structures of two yeast enolase genes. Homology between the 5' noncoding flanking regions of yeast enolase and glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem. 1981 Feb 10;256(3):1385–1395. [PubMed] [Google Scholar]
- Holland M. J., Yokoi T., Holland J. P., Myambo K., Innis M. A. The GCR1 gene encodes a positive transcriptional regulator of the enolase and glyceraldehyde-3-phosphate dehydrogenase gene families in Saccharomyces cerevisiae. Mol Cell Biol. 1987 Feb;7(2):813–820. doi: 10.1128/mcb.7.2.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki G., Fraenkel D. G. Cloning of yeast glycolysis genes by complementation. Biochem Biophys Res Commun. 1982 Oct 15;108(3):1107–1122. doi: 10.1016/0006-291x(82)92114-3. [DOI] [PubMed] [Google Scholar]
- Kellermann E., Seeboth P. G., Hollenberg C. P. Analysis of the primary structure and promoter function of a pyruvate decarboxylase gene (PDC1) from Saccharomyces cerevisiae. Nucleic Acids Res. 1986 Nov 25;14(22):8963–8977. doi: 10.1093/nar/14.22.8963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopetzki E., Entian K. D., Mecke D. Complete nucleotide sequence of the hexokinase PI gene (HXK1) of Saccharomyces cerevisiae. Gene. 1985;39(1):95–101. doi: 10.1016/0378-1119(85)90113-1. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Legrain P., Rosbash M. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell. 1989 May 19;57(4):573–583. doi: 10.1016/0092-8674(89)90127-x. [DOI] [PubMed] [Google Scholar]
- Lehrach H., Diamond D., Wozney J. M., Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977 Oct 18;16(21):4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- Lillehaug J. R., Kleppe K. Effect of salts and polyamines on T4 polynucleotide kinase. Biochemistry. 1975 Mar 25;14(6):1225–1229. doi: 10.1021/bi00677a021. [DOI] [PubMed] [Google Scholar]
- Lindquist S. Regulation of protein synthesis during heat shock. Nature. 1981 Sep 24;293(5830):311–314. doi: 10.1038/293311a0. [DOI] [PubMed] [Google Scholar]
- Maitra P. K., Lobo Z. A kinetic study of glycolytic enzyme synthesis in yeast. J Biol Chem. 1971 Jan 25;246(2):475–488. [PubMed] [Google Scholar]
- Mayer S. A., Dieckmann C. L. The yeast CBP1 gene produces two differentially regulated transcripts by alternative 3'-end formation. Mol Cell Biol. 1989 Oct;9(10):4161–4169. doi: 10.1128/mcb.9.10.4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlister L., Holland M. J. Targeted deletion of a yeast enolase structural gene. Identification and isolation of yeast enolase isozymes. J Biol Chem. 1982 Jun 25;257(12):7181–7188. [PubMed] [Google Scholar]
- McNally T., Purvis I. J., Fothergill-Gilmore L. A., Brown A. J. The yeast pyruvate kinase gene does not contain a string of non-preferred codons: revised nucleotide sequence. FEBS Lett. 1989 Apr 24;247(2):312–316. doi: 10.1016/0014-5793(89)81359-6. [DOI] [PubMed] [Google Scholar]
- McNeil J. B., Dykshoorn P., Huy J. N., Small S. The DNA-binding protein RAP1 is required for efficient transcriptional activation of the yeast PYK glycolytic gene. Curr Genet. 1990 Dec;18(5):405–412. doi: 10.1007/BF00309909. [DOI] [PubMed] [Google Scholar]
- Mellor J., Dobson M. J., Roberts N. A., Tuite M. F., Emtage J. S., White S., Lowe P. A., Patel T., Kingsman A. J., Kingsman S. M. Efficient synthesis of enzymatically active calf chymosin in Saccharomyces cerevisiae. Gene. 1983 Sep;24(1):1–14. doi: 10.1016/0378-1119(83)90126-9. [DOI] [PubMed] [Google Scholar]
- Moore P. A., Bettany A. J., Brown A. J. Multiple copies of the pyruvate kinase gene affect yeast cell growth. J Gen Microbiol. 1990 Dec;136(12):2359–2366. doi: 10.1099/00221287-136-12-2359. [DOI] [PubMed] [Google Scholar]
- Moore P. A., Bettany A. J., Brown J. P. Expression of a yeast glycolytic gene is subject to dosage limitation. Gene. 1990 Apr 30;89(1):85–92. doi: 10.1016/0378-1119(90)90209-a. [DOI] [PubMed] [Google Scholar]
- Mountain H. A., Sudbery P. E. Regulation of the Saccharomyces cerevisiae WHI2 gene. J Gen Microbiol. 1990 Apr;136(4):727–732. doi: 10.1099/00221287-136-4-727. [DOI] [PubMed] [Google Scholar]
- Nishizawa M., Suzuki Y., Nogi Y., Matsumoto K., Fukasawa T. Yeast Gal11 protein mediates the transcriptional activation signal of two different transacting factors, Gal4 and general regulatory factor I/repressor/activator site binding protein 1/translation upstream factor. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5373–5377. doi: 10.1073/pnas.87.14.5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M., Scafe C., Sexton J., Young R. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol. 1987 May;7(5):1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper P. W., Curran B., Davies M. W., Hirst K., Lockheart A., Ogden J. E., Stanway C. A., Kingsman A. J., Kingsman S. M. A heat shock element in the phosphoglycerate kinase gene promoter of yeast. Nucleic Acids Res. 1988 Feb 25;16(4):1333–1348. doi: 10.1093/nar/16.4.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper P. W., Curran B., Davies M. W., Hirst K., Lockheart A., Seward K. Catabolite control of the elevation of PGK mRNA levels by heat shock in Saccharomyces cerevisiae. Mol Microbiol. 1988 May;2(3):353–361. doi: 10.1111/j.1365-2958.1988.tb00039.x. [DOI] [PubMed] [Google Scholar]
- Purvis I. J., Loughlin L., Bettany A. J., Brown A. J. Translation and stability of an Escherichia coli beta-galactosidase mRNA expressed under the control of pyruvate kinase sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 1987 Oct 12;15(19):7963–7974. doi: 10.1093/nar/15.19.7963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J., Reid G. A., Dawes I. W. The nucleotide sequence of the LPD1 gene encoding lipoamide dehydrogenase in Saccharomyces cerevisiae: comparison between eukaryotic and prokaryotic sequences for related enzymes and identification of potential upstream control sites. J Gen Microbiol. 1988 May;134(5):1131–1139. doi: 10.1099/00221287-134-5-1131. [DOI] [PubMed] [Google Scholar]
- Santangelo G. M., Tornow J. Efficient transcription of the glycolytic gene ADH1 and three translational component genes requires the GCR1 product, which can act through TUF/GRF/RAP binding sites. Mol Cell Biol. 1990 Feb;10(2):859–862. doi: 10.1128/mcb.10.2.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago T. C., Purvis I. J., Bettany A. J., Brown A. J. The relationship between mRNA stability and length in Saccharomyces cerevisiae. Nucleic Acids Res. 1986 Nov 11;14(21):8347–8360. doi: 10.1093/nar/14.21.8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaff I., Green J. B., Gozalbo D., Hohmann S. A deletion of the PDC1 gene for pyruvate decarboxylase of yeast causes a different phenotype than previously isolated point mutations. Curr Genet. 1989 Feb;15(2):75–81. doi: 10.1007/BF00435452. [DOI] [PubMed] [Google Scholar]
- Schmitt H. D., Ciriacy M., Zimmermann F. K. The synthesis of yeast pyruvate decarboxylase is regulated by large variations in the messenger RNA level. Mol Gen Genet. 1983;192(1-2):247–252. doi: 10.1007/BF00327674. [DOI] [PubMed] [Google Scholar]
- Schwelberger H. G., Kohlwein S. D., Paltauf F. Molecular cloning, primary structure and disruption of the structural gene of aldolase from Saccharomyces cerevisiae. Eur J Biochem. 1989 Mar 15;180(2):301–308. doi: 10.1111/j.1432-1033.1989.tb14648.x. [DOI] [PubMed] [Google Scholar]
- Scott E. W., Allison H. E., Baker H. V. Characterization of TPI gene expression in isogeneic wild-type and gcr1-deletion mutant strains of Saccharomyces cerevisiae. Nucleic Acids Res. 1990 Dec 11;18(23):7099–7107. doi: 10.1093/nar/18.23.7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachelek C., Stachelek J., Swan J., Botstein D., Konigsberg W. Identification, cloning and sequence determination of the genes specifying hexokinase A and B from yeast. Nucleic Acids Res. 1986 Jan 24;14(2):945–963. doi: 10.1093/nar/14.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekamp-Olson P., Najarian R., Burke R. L. The isolation, characterization and nucleotide sequence of the phosphoglucoisomerase gene of Saccharomyces cerevisiae. Gene. 1988 Dec 15;73(1):153–161. doi: 10.1016/0378-1119(88)90321-6. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornow J., Santangelo G. M. Efficient expression of the Saccharomyces cerevisiae glycolytic gene ADH1 is dependent upon a cis-acting regulatory element (UASRPG) found initially in genes encoding ribosomal proteins. Gene. 1990 May 31;90(1):79–85. doi: 10.1016/0378-1119(90)90441-s. [DOI] [PubMed] [Google Scholar]
- Tuite M. F., Dobson M. J., Roberts N. A., King R. M., Burke D. C., Kingsman S. M., Kingsman A. J. Regulated high efficiency expression of human interferon-alpha in Saccharomyces cerevisiae. EMBO J. 1982;1(5):603–608. doi: 10.1002/j.1460-2075.1982.tb01215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdier J. M. Regulatory DNA-binding proteins in yeast: an overview. Yeast. 1990 Jul-Aug;6(4):271–297. doi: 10.1002/yea.320060402. [DOI] [PubMed] [Google Scholar]
- Watson H. C., Walker N. P., Shaw P. J., Bryant T. N., Wendell P. L., Fothergill L. A., Perkins R. E., Conroy S. C., Dobson M. J., Tuite M. F. Sequence and structure of yeast phosphoglycerate kinase. EMBO J. 1982;1(12):1635–1640. doi: 10.1002/j.1460-2075.1982.tb01366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M. F., Fothergill-Gilmore L. A. Sequence of the gene encoding phosphoglycerate mutase from Saccharomyces cerevisiae. FEBS Lett. 1988 Mar 14;229(2):383–387. doi: 10.1016/0014-5793(88)81161-x. [DOI] [PubMed] [Google Scholar]
- Wills C. Regulation of sugar and ethanol metabolism in Saccharomyces cerevisiae. Crit Rev Biochem Mol Biol. 1990;25(4):245–280. doi: 10.3109/10409239009090611. [DOI] [PubMed] [Google Scholar]
- Yoshino M., Murakami K. AMP deaminase reaction as a control system of glycolysis in yeast. Activation of phosphofructokinase and pyruvate kinase by the AMP deaminase-ammonia system. J Biol Chem. 1982 Mar 25;257(6):2822–2828. [PubMed] [Google Scholar]