Abstract
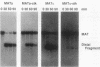
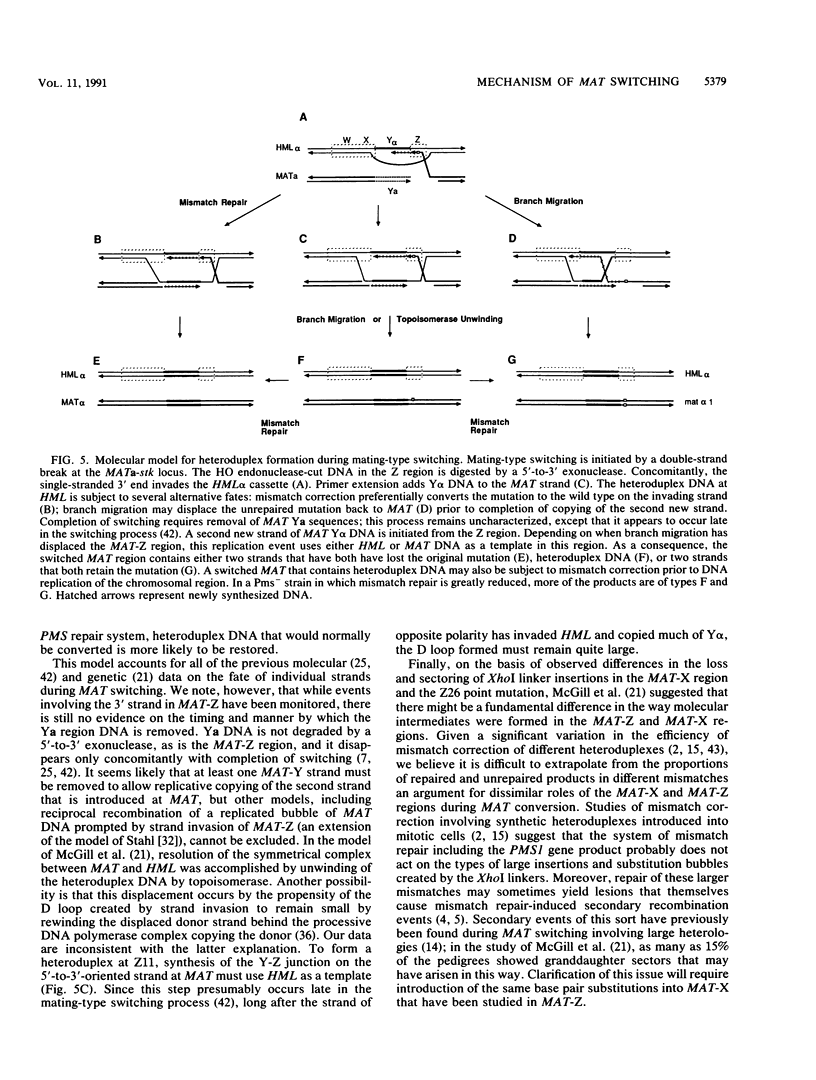
We sequenced two alleles of the MATa locus of Saccharomyces cerevisiae that reduce homothallic switching and confer viability to HO rad52 strains. Both the MATa-stk (J. E. Haber, W. T. Savage, S. M. Raposa, B. Weiffenbach, and L. B. Rowe, Proc. Natl. Acad. Sci. USA 77:2824-2828, 1980) and MATa-survivor (R. E. Malone and D. Hyman, Curr. Genet. 7:439-447, 1983) alleles result from a T----A base change at position Z11 of the MAT locus. These strains also contain identical base substitutions at HMRa, so that the mutation is reintroduced when MAT alpha switches to MATa. Mating-type switching in a MATa-stk strain relative to a MATa Z11T strain is reduced at least 50-fold but can be increased by expression of HO from a galactose-inducible promoter. We confirmed by Southern analysis that the Z11A mutation reduced the efficiency of double-strand break formation compared with the Z11T variant; the reduction was more severe in MAT alpha than in MATa. In MAT alpha, the Z11A mutation also creates a mat alpha 1 (sterile) mutation that distinguishes switches of MATa-stk to either MAT alpha or mat alpha 1-stk. Pedigree analysis of cells induced to switch in G1 showed that MATa-stk switched frequently (23% of the time) to produce one mat alpha 1-stk and one MAT alpha progeny. This postswitching segregation suggests that Z11 was often present in heteroduplex DNA that was not mismatch repaired. When mismatch repair was prevented by deletion of the PMS1 gene, there was an increase in the proportion of mat alpha 1-stk/MAT alpha sectors (59%) and in pairs of switched cells that both retained the stk mutation (27%). We conclude that at least one strand of DNA only 4 bp from the HO cut site is not degraded in most of the gene conversion events that accompany MAT switching.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Astell C. R., Ahlstrom-Jonasson L., Smith M., Tatchell K., Nasmyth K. A., Hall B. D. The sequence of the DNAs coding for the mating-type loci of Saccharomyces cerevisiae. Cell. 1981 Nov;27(1 Pt 2):15–23. doi: 10.1016/0092-8674(81)90356-1. [DOI] [PubMed] [Google Scholar]
- Bishop D. K., Williamson M. S., Fogel S., Kolodner R. D. The role of heteroduplex correction in gene conversion in Saccharomyces cerevisiae. Nature. 1987 Jul 23;328(6128):362–364. doi: 10.1038/328362a0. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Borts R. H., Haber J. E. Meiotic recombination in yeast: alteration by multiple heterozygosities. Science. 1987 Sep 18;237(4821):1459–1465. doi: 10.1126/science.2820060. [DOI] [PubMed] [Google Scholar]
- Borts R. H., Leung W. Y., Kramer W., Kramer B., Williamson M., Fogel S., Haber J. E. Mismatch repair-induced meiotic recombination requires the pms1 gene product. Genetics. 1990 Mar;124(3):573–584. doi: 10.1093/genetics/124.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant F. R., Johnson K. A., Benkovic S. J. Elementary steps in the DNA polymerase I reaction pathway. Biochemistry. 1983 Jul 19;22(15):3537–3546. doi: 10.1021/bi00284a001. [DOI] [PubMed] [Google Scholar]
- Connolly B., White C. I., Haber J. E. Physical monitoring of mating type switching in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jun;8(6):2342–2349. doi: 10.1128/mcb.8.6.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
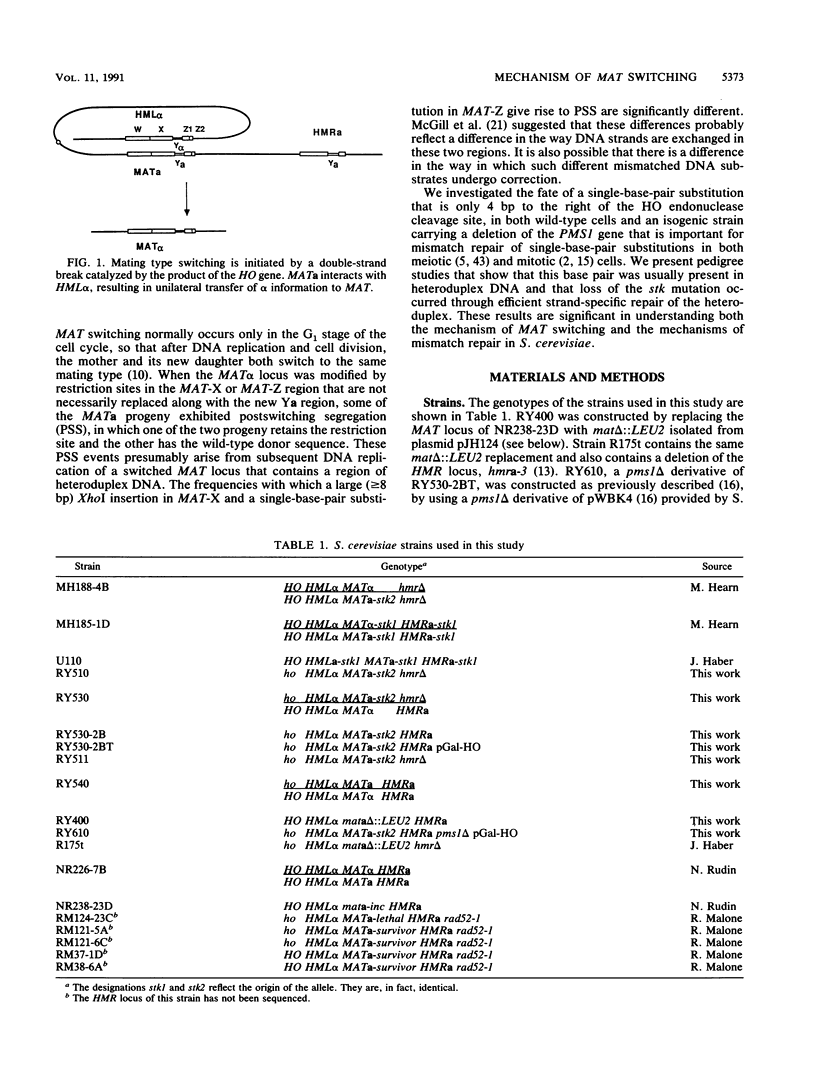
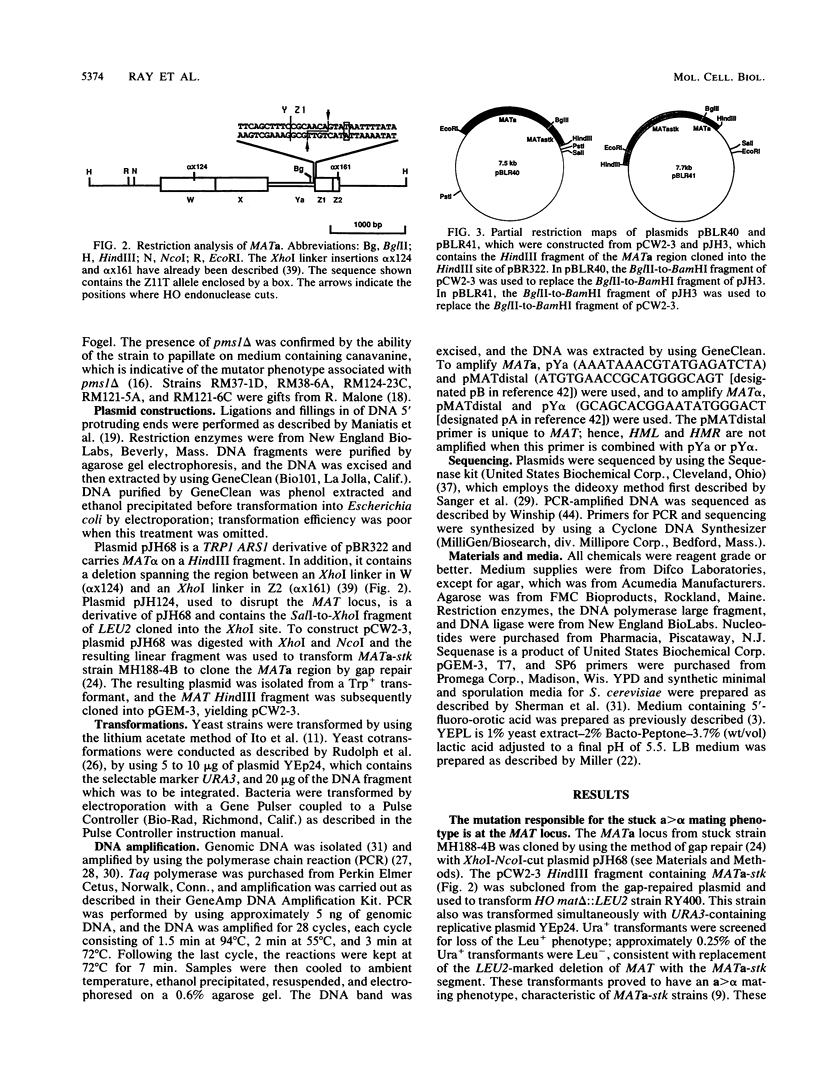
- Haber J. E., Savage W. T., Raposa S. M., Weiffenbach B., Rowe L. B. Mutations preventing transpositions of yeast mating type alleles. Proc Natl Acad Sci U S A. 1980 May;77(5):2824–2828. doi: 10.1073/pnas.77.5.2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks J. B., Herskowitz I. Interconversion of Yeast Mating Types I. Direct Observations of the Action of the Homothallism (HO) Gene. Genetics. 1976 Jun;83(2):245–258. doi: 10.1093/genetics/83.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R. E., Herskowitz I. Directionality and regulation of cassette substitution in yeast. Cold Spring Harb Symp Quant Biol. 1984;49:97–104. doi: 10.1101/sqb.1984.049.01.013. [DOI] [PubMed] [Google Scholar]
- Klar A. J., Hicks J. B., Strathern J. N. Directionality of yeast mating-type interconversion. Cell. 1982 Mar;28(3):551–561. doi: 10.1016/0092-8674(82)90210-0. [DOI] [PubMed] [Google Scholar]
- Klar A. J., Strathern J. N. Resolution of recombination intermediates generated during yeast mating type switching. 1984 Aug 30-Sep 5Nature. 310(5980):744–748. doi: 10.1038/310744a0. [DOI] [PubMed] [Google Scholar]
- Kramer B., Kramer W., Williamson M. S., Fogel S. Heteroduplex DNA correction in Saccharomyces cerevisiae is mismatch specific and requires functional PMS genes. Mol Cell Biol. 1989 Oct;9(10):4432–4440. doi: 10.1128/mcb.9.10.4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer W., Kramer B., Williamson M. S., Fogel S. Cloning and nucleotide sequence of DNA mismatch repair gene PMS1 from Saccharomyces cerevisiae: homology of PMS1 to procaryotic MutL and HexB. J Bacteriol. 1989 Oct;171(10):5339–5346. doi: 10.1128/jb.171.10.5339-5346.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Soni A. Mutagenesis by transient misalignment. J Biol Chem. 1988 Oct 15;263(29):14784–14789. [PubMed] [Google Scholar]
- Mascioli D. W., Haber J. E. A CIS-Acting Mutation within the MATa Locus of SACCHAROMYCES CEREVISIAE That Prevents Efficient Homothallic Mating-Type Switching. Genetics. 1980 Feb;94(2):341–360. doi: 10.1093/genetics/94.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill C., Shafer B., Strathern J. Coconversion of flanking sequences with homothallic switching. Cell. 1989 May 5;57(3):459–467. doi: 10.1016/0092-8674(89)90921-5. [DOI] [PubMed] [Google Scholar]
- Nickoloff J. A., Singer J. D., Heffron F. In vivo analysis of the Saccharomyces cerevisiae HO nuclease recognition site by site-directed mutagenesis. Mol Cell Biol. 1990 Mar;10(3):1174–1179. doi: 10.1128/mcb.10.3.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raveh D., Hughes S. H., Shafer B. K., Strathern J. N. Analysis of the HO-cleaved MAT DNA intermediate generated during the mating type switch in the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1989 Dec;220(1):33–42. [PubMed] [Google Scholar]
- Rudolph H., Koenig-Rauseo I., Hinnen A. One-step gene replacement in yeast by cotransformation. Gene. 1985;36(1-2):87–95. doi: 10.1016/0378-1119(85)90072-1. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf S. J., Horn G. T., Erlich H. A. Direct cloning and sequence analysis of enzymatically amplified genomic sequences. Science. 1986 Sep 5;233(4768):1076–1078. doi: 10.1126/science.3461561. [DOI] [PubMed] [Google Scholar]
- Strathern J. N., Klar A. J., Hicks J. B., Abraham J. A., Ivy J. M., Nasmyth K. A., McGill C. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell. 1982 Nov;31(1):183–192. doi: 10.1016/0092-8674(82)90418-4. [DOI] [PubMed] [Google Scholar]
- Sun H., Treco D., Szostak J. W. Extensive 3'-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell. 1991 Mar 22;64(6):1155–1161. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano I., Kusumi T., Oshima Y. An alpha mating-type allele insensitive to the mutagenic action of the homothallic gene system in Saccharomyces diastaticus. Mol Gen Genet. 1973 Oct 16;126(1):19–28. doi: 10.1007/BF00333478. [DOI] [PubMed] [Google Scholar]
- Tatchell K., Nasmyth K. A., Hall B. D., Astell C., Smith M. In vitro mutation analysis of the mating-type locus in yeast. Cell. 1981 Nov;27(1 Pt 2):25–35. doi: 10.1016/0092-8674(81)90357-3. [DOI] [PubMed] [Google Scholar]
- Weiffenbach B., Haber J. E. Homothallic mating type switching generates lethal chromosome breaks in rad52 strains of Saccharomyces cerevisiae. Mol Cell Biol. 1981 Jun;1(6):522–534. doi: 10.1128/mcb.1.6.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiffenbach B., Rogers D. T., Haber J. E., Zoller M., Russell D. W., Smith M. Deletions and single base pair changes in the yeast mating type locus that prevent homothallic mating type conversions. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3401–3405. doi: 10.1073/pnas.80.11.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White C. I., Haber J. E. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990 Mar;9(3):663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson M. S., Game J. C., Fogel S. Meiotic gene conversion mutants in Saccharomyces cerevisiae. I. Isolation and characterization of pms1-1 and pms1-2. Genetics. 1985 Aug;110(4):609–646. doi: 10.1093/genetics/110.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winship P. R. An improved method for directly sequencing PCR amplified material using dimethyl sulphoxide. Nucleic Acids Res. 1989 Feb 11;17(3):1266–1266. doi: 10.1093/nar/17.3.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]