Abstract
Background
We characterized 100 USA100 epidemic MRSA from individuals in the community with no known healthcare-associated risk factors.
Findings
Molecular epidemiology demonstrated 21 pulsed-field types and six spa types. SCCmec typing demonstrated that all of the strains possess the type II cassette. The staphylococcal enterotoxin D virulence gene was also present.
Conclusion
Characterization of USA100 MRSA in the community illustrated the importance of nasal carriage, and the genetic diversity of the USA100 clone.
Keywords: Staphylococcus, PFGE, Spa, MRSA, USA100
Remarkably in a mere 40 year evolutionary time period, Staphylococcus aureus has produced multiple highly successful antibiotic-resistant epidemic clones. Among the methicillin-resistant S. aureus (MRSA), are the extensively researched USA300 and the less well characterized USA100 MRSA. Historically, USA300 was described as a community-associated MRSA (CA-MRSA) causing disease in healthy individuals (Chambers 2005; McDougal et al. 2003). Recently the USA300 CA-MRSA designation has been challenged as this pathogen is repeatedly identified as the cause of hospital-acquired MRSA (HA-MRSA) infection. Regardless of the environment from which the strains are isolated, the complement of virulence factors demonstrated in USA300 strains is identical. In contrast, USA100 is considered an HA-MRSA and a cause of invasive infection primarily among persons with healthcare-associated risk factors (Klevens et al. 2007; Limbago et al. 2009). Although detected in nasal swabs from non-institutionalized individuals (Tenover et al. 2008), USA100 MRSA have not been well described in the community environment, the goal of the present study. Herein we characterize 100 USA100 MRSA identified within a collection of isolates from individuals with no known healthcare contact by pulsed-field type (PFT), spa type (ST), SCCmec type, and PCR identification of virulence factors. Our results are compared to those reported for USA100 healthcare-associated isolates.
A total of 100 CA-MRSA from a previously described report were used in this study (Roberts et al. 2006). The control, USA100 (NRS382), was obtained from the Network on Antimicrobial Resistance in S. aureus (NARSA).
PFGE was performed as previously described (Roberts et al. 2006). Genomic DNA from 100 isolates was extracted using the MagNA Pure® and the MagNA Pure LC DNA Isolation Kit III (Roche Diagnostics, Indianapolis, IN) according to manufacturer’s instructions. Typing using the polymorphic region of the spa gene and SCCmec characterization were performed as previously described (Oliveira & de Lencastre 2002; Shopsin et al. 1999). PCR detection of lukPV (Lina et al. 1999), seb, tst, sed (Monday & Bohach 1999) and psmα (Wang et al. 2007) genes was performed as previously reported.
Following our report of USA300 among community-associated S. aureus (Roberts et al. 2006) our laboratory continued to characterize MRSA from individuals with no known healthcare-associated risks using PFGE. Although the majority of isolates added to our collection are USA300 MRSA, we have also identified a large number of USA100 MRSA from the same population. Previous studies which have characterized USA100 strains primarily from healthcare environments by PFGE have demonstrated great diversity in the USA100 pulsed-field types (PFTs) (McDougal et al. 2003; Limbago et al. 2009; Tenover et al. 2008). Similarly, our study of community-associated USA100 MRSA using PFGE illustrated substantial diversity among the strain patterns as represented by 21 PFTs for the 100 isolates (Simpson’s discriminatory index = 0.920). Reportedly, USA100 MRSA preferentially classify in three PFGE patterns (40.7% of strains) among mostly hospital-associated isolates (Limbago et al. 2009). In the present study, the majority of community USA100 strains possess two PFTs (44% of strains) (Figure 1).
Figure 1.
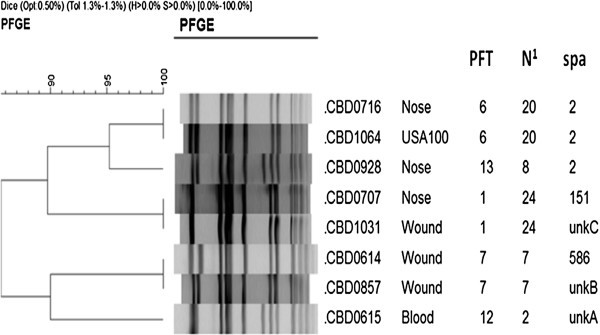
Variation in pulsed-field Patterns of USA100 MRSA. N1 Number of isolates included in the PFT (pulsed-field type) including control strains. Figure 1 Pulsed-field gel electrophoresis classified the majority of the USA100 strains to PFTs 1 (24 isolates including the control strain USA100/NRS382) and PFT 6 (20 isolates). Representative strains for each of the six spa types are shown included three isolates which possessed known repeats but not known repeat patterns and therefore could not be typed (unknown unkA-C). The two typing techniques were not always in agreement as demonstrated by isolates CBD0707 and CBD1031 which possess the same pulsed-field type (Chambers 2005) but different spa types (151 and unkC).
Nasal colonization studies have identified USA100 MRSA as the most common MRSA colonizing non-institutionalized individuals in the United States (Tenover et al. 2008). Characterization of these isolates is important in part because of the established link between S. aureus nasal carriage and staphylococcal disease. Tenover et al. specifically queried nasal strains and found 44.8% of isolates possess USA100 PFTs (Tenover et al. 2008). In the present study we worked in the opposite direction querying first for USA100 PFTs then identifying etiology. Given the results of the aforementioned study, it is consistent that we found 47 isolates (47%) of our strains identified as USA100 were from nasal swabs (Table 1). Other common sources of USA100 MRSA were wound (27%) and blood samples (13%) consistent with the prevalence of USA100 MRSA in invasive infections (Klevens et al. 2007; Limbago et al. 2009).
Table 1.
Summary of results for 100 USA100 Epidemic MRSA
| Molecular typing | Virulence factor PCR | ||||
|---|---|---|---|---|---|
| USA100 | SCCmec II | spa type1 | sed | psmα | |
| Nose | 47 | 47 | 2,151 | 45 (96%) | 47 (100%) |
| Wound | 27 | 27 | 2,586,C | 26 (96%) | 27 (100%) |
| Blood | 13 | 13 | 2,A | 12 (82%) | 13 (100%) |
| Urine | 4 | 4 | 2 | 3 (75%) | 4 (100%) |
| Other | 4 | 4 | 2,151,B | 3 (75%) | 4 (100%) |
| Lung | 3 | 3 | 2 | 3 (100%) | 3 (100%) |
| Unknown | 2 | 2 | 2 | 2 (100%) | 2 (100%) |
| Total | 100 (100%) | 100 (100%) | NA | 94 (94%) | 100 (100%) |
1spa types identified for each disease etiology are listed. A-C are unknown spa types.
Previous studies have demonstrated USA100 MRSA are relatively uniform in SCCmec type and spa genes, typically containing SCCmec II and spa type 2 (TJMBMDMGMK) (McDougal et al. 2003; Limbago et al. 2009; Tenover et al. 2008). In rare cases, USA100 isolates have been identified which contain SCCmec IV (Tenover et al. 2008). We performed SCCmec typing and determined that all of our community USA100 harbor SCCmec type II (Table 1). We then performed spa typing on every other isolate (a total of 50 isolates) identifying six spa types (Table 1). To our knowledge, McDougal et al. (McDougal et al. 2003) is the only published study with spa typing results for a large number of USA100 isolates and the HA-MRSA reported therein were uniformly spa type 2 (McDougal et al. 2003). A total of 90% of our isolates were also spa type 2 (Table 1) and the additional spa types (Table 1) we identified is consistent with data (unpublished) for USA100 strains in the spa database (SpaServer.ridom.de).
Virulence factor characterization for USA100 MRSA has largely centered on the presence of staphylococcal enterotoxin D. The presence of the sed gene in a MRSA strain is in fact considered highly predictive of USA100 (Limbago et al. 2009; Tenover et al. 2008). The percentage of sed positive strains by PCR in our isolates was higher than previously reported (94%) (Limbago et al. 2009; Tenover et al. 2008) (Table 1). Additional virulence genes were selected based on their ability to cause diseases in the community including toxic shock, food poisoning (seb), necrotizing fasciitis and necrotizing pneumonia (psmα and lukPV). All of our isolates were positive for the psmα gene (Table 1), however previous studies have demonstrated that USA100 strains lack PSM protein production despite the presence of the gene (Wang et al. 2007). Finally, although toxic shock toxin, Panton-Valentine leukocidin, and staphylococcal enterotoxin B have rarely been reported in USA100 MRSA (Limbago et al. 2009), we failed to detect any of these virulence factors among our community USA100 MRSA (data not shown).
In conclusion, our study has demonstrated that USA100 MRSA are present in the community among individuals with no known healthcare contact. Comparison to existing studies has illustrated that the pathogen is essentially identical with respect to diversity of pulsed-field types, anatomical location, SCCmec type and virulence factor complement irrespective of the environment from which it was isolated. Perhaps the most significant finding of our study is that when USA100 is found in the community it is most often in the nose. It is tempting therefore to hypothesize that the success of this epidemic clone is partially related to its ability to silently colonize a host. Considering USA100 is the leading cause of invasive MRSA infection, that almost all vancomycin-resistant and intermediate S. aureus strains are USA100, and that antibiotic resistance in USA100 isolates is increasing, the importance of continued surveillance for this pathogen cannot be understated.
This study was supported by U.S. Army Research, Development and Engineering Command, contract W911SR-07-C-0084. Reference strains were provided by the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA).
Footnotes
Competing interests
The author declares that there are no competing interests.
References
- Chambers HF. Community-associated MRSA–resistance and virulence converge. N Engl J Med. 2005;352(14):1485–1487. doi: 10.1056/NEJMe058023. [DOI] [PubMed] [Google Scholar]
- Klevens RM, Morrison MA, Nadle J, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298(15):1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- Limbago B, Fosheim GE, Schoonover V, et al. Characterization of methicillin-resistant Staphylococcus aureus isolates collected in 2005 and 2006 from patients with invasive disease: a population-based analysis. J Clin Microbiol. 2009;47(5):1344–1351. doi: 10.1128/JCM.02264-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lina G, Piemont Y, Godail-Gamot F, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29(5):1128–1132. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol. 2003;41(11):5113–5120. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monday SR, Bohach GA. Use of multiplex PCR to detect classical and newly described pyrogenic toxin genes in staphylococcal isolates. J Clin Microbiol. 1999;37(10):3411–3414. doi: 10.1128/jcm.37.10.3411-3414.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46(7):2155–2161. doi: 10.1128/AAC.46.7.2155-2161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JC, Krueger RL, Peak KK, et al. Community-associated methicillin-resistant Staphylococcus aureus epidemic clone USA300 in isolates from Florida and Washington. J Clin Microbiol. 2006;44(1):225–226. doi: 10.1128/JCM.44.1.225-226.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopsin B, Gomez M, Montgomery SO, et al. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol. 1999;37(11):3556–3563. doi: 10.1128/jcm.37.11.3556-3563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover FC, McAllister S, Fosheim G, et al. Characterization of Staphylococcus aureus isolates from nasal cultures collected from individuals in the United States in 2001 to 2004. J Clin Microbiol. 2008;46(9):2837–2841. doi: 10.1128/JCM.00480-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Braughton KR, Kretschmer D, et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13(12):1510–1514. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]


