Abstract
SIRT1 is an evolutionary conserved NAD+-dependent deacetylase that is at the pinnacle of metabolic control, all the way from yeast to humans. SIRT1 senses changes in intracellular NAD+ levels, which reflect energy level, and uses this information to adapt the cellular energy output, such that the it matches cellular energy requirements. Generally, but not exclusively, the changes induced by SIRT1 activation are transcriptional in nature and are related to an increase in mitochondrial metabolism and antioxidant protection. These attractive features have validated SIRT1 as a therapeutic target in the management of metabolic disease and prompted an intensive search to identify pharmacological SIRT1 activators. In this review we will first give an overview of the SIRT1 biology with a particular focus on its role in metabolic control. We will then analyze the pros and cons of the current strategies used to activate SIRT1 and explore the emerging evidence indicating that modulation of NAD+ levels could provide an effective way to achieve such goals.
INTRODUCTION
During the last decade the mammalian sirtuin family (formed by paralogues SIRT1 to SIRT7) has emerged as a constellation of enzymes with key roles in whole body metabolic homeostasis and an interesting therapeutic potential applicable to multiple pathophysiological states.
The history of sirtuins initiates almost three decades ago with the identification of Sir2 (Silent Information Regulator 2), a protein forming part of a complex that enabled gene silencing at selected regions of the yeast genome (Ivy et al., 1986; Shore et al., 1984). A major turning point in the history of Sir2 came from the discovery that Sir2 was involved in the yeast replicative aging process (Kaeberlein et al., 1999). The accumulation of extrachromosomal rDNA circles (ERCs) as the organisms ages is believed to be a major determinant of yeast replicative lifespan (Sinclair and Guarente, 1997). While the mechanism by which the accumulation of ERCs influence lifespan are not fully understood, different genetic manipulations promote reasonable, despite correlational, evidence that the accumulation of ERCs is negatively correlated with yeast replicative aging (Defossez et al., 1999; Kaeberlein et al., 1999; Sinclair and Guarente, 1997). It was originally thought that the impact of Sir2 on replicative lifespan of yeast was consequent to its silencing activity on ERCs. However, the effects of Sir2 on aging extend further than ERCs silencing, as genetic manipulations of Sir2 orthologues can also affect lifespan of higher eukaryotes, such as the nematode Caenorhabditis elegans (Berdichevsky et al., 2006; Rizki et al., 2011; Tissenbaum and Guarente, 2001; Viswanathan et al., 2005) and insects like Drosophila melanogaster (Bauer et al., 2009; Rogina and Helfand, 2004), where ERCs are not thought to cause ageing. However, there are some caveats on the consistency, amplitude and mammalian translation of the lifespan extension effects of Sir2 orthologs (Burnett et al., 2011; Kaeberlein and Powers, 2007; Lombard et al., 2011; Viswanathan and Guarente, 2011), which suggest that the effects of Sir2 on organismal lifespan might be indirect and/or largely depend on a specific repertoire of third-party modulators.
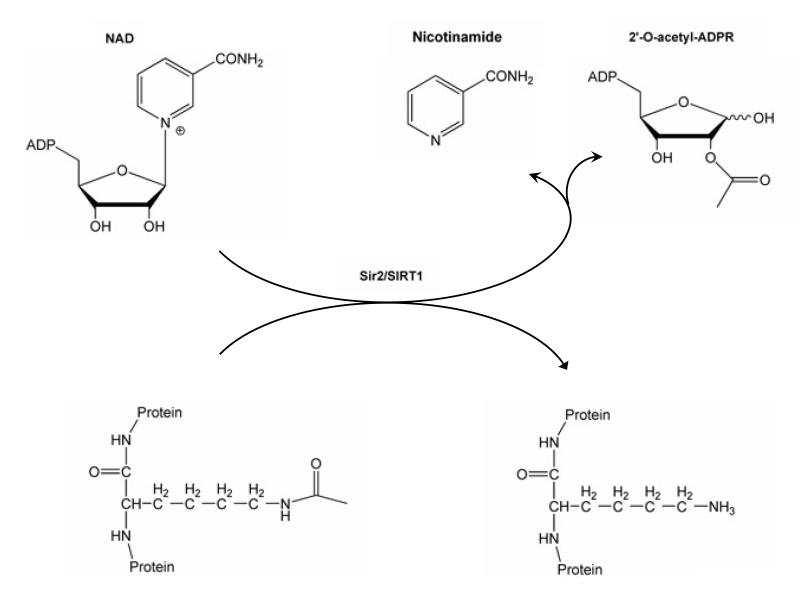
If not acting primarily as lifespan determinants, what is then the exact function of this family of proteins? A first glimpse of the real function of Sir2, or its orthologs, was grasped when the activity of Sir2 as a silencing enzyme was more precisely defined as a NAD+ dependent deacetylase (Imai et al., 2000). In the reaction catalyzed by sirtuins, an acetylated substrate gets deacetylated, using NAD+ as a cosubstrate and yielding the deacetylated substrate, nicotinamide and 2′-O-acetyl-ADP-ribose (Figure 1). The NAD+ dependence and the relatively high Km of the Sir2 enzyme for NAD+ hence immediately suggested a potential link between Sir2 activity and the metabolic state of the cell (Guarente, 2000).
Figure 1. The NAD+-dependent SIRT1 deacetylase reaction.
SIRT1 uses NAD+ as a substrate to remove acetyl groups from a target protein. In addition to the deacetylated substrate, the reaction yields nicotinamide and 2′-O-acetyl-ADP ribose as products.
In mammals there are seven Sir2 orthologs (SIRT1-7), which constitute the sirtuin family of enzymes. All of them are ubiquitously expressed and share a conserved catalytic core comprising 275 amino acids (for a review, see (Dali-Youcef et al., 2007; Michan and Sinclair, 2007)). The different members of the mammalian sirtuin family, however, show distinct features that probably endow them with specialized functions. For example, mammalian sirtuins differ in their subcellular localization. SIRT1, the best characterized family member, resides mainly in the nucleus (Michishita et al., 2005), but can shuttle from the nucleus to the cytosol (Tanno et al., 2007), where several of its targets are found. SIRT2 is mainly localized in the cytoplasm, although it can also regulate gene expression by deacetylation of transcription factors that shuttle from the cytoplasm to the nucleus (Jing et al., 2007), and it contributes to chromatin compaction upon disassembly of the cell nucleus during mitosis (Vaquero et al., 2006). SIRT3, SIRT4 and SIRT5 are generally considered as mitochondrial proteins (Michishita et al., 2005; Onyango et al., 2002; Schwer et al., 2002), whereas SIRT6 and SIRT7 are nuclear proteins. However, while SIRT6 is predominantly located in the heterochromatin, SIRT7 is thought to be mainly enriched in the nucleoli (Michishita et al., 2005).
In addition to their differential cellular location, the sirtuin family members can also be distinguished by their different enzymatic activities. SIRT1 and SIRT5 act as deacetylases (Imai et al., 2000; Vaziri et al., 2001), while SIRT4 seems to be mono-ADP-ribosyl transferase (Haigis et al., 2006). SIRT2, SIRT3 and SIRT6 can display both activities (Liszt et al., 2005; Michishita et al., 2008; North et al., 2003; Shi et al., 2005). The activity of SIRT7 has not been clearly established, even though it has been hypothesized to act as a deacetylase (Vakhrusheva et al., 2008). Of note, SIRT5 was recently described to demalonylate and desuccinylate proteins (Peng et al., 2011; Du et al, 2011). It is tempting to speculate that the spectrum of action of sirtuin is not limited to deacetylation, but would cover a much wider range of –acylation based post-translational modifications. The identification of sirtuin substrates during the last decades has clearly pointed out a prominent role of sirtuins as metabolic regulators. For the purpose of this review, we will mostly focus on the actions of SIRT1. For extensive discussion of the actions of other sirtuin members, we refer the reader to some recent reviews (Dali-Youcef et al., 2007; Finkel et al., 2009; Guarente, 2011; Michan and Sinclair, 2007; Schwer and Verdin, 2008; Yamamoto et al., 2007).
1. SIRT1 IN A NUTSHELL
1.a SIRT1: what and where is it?
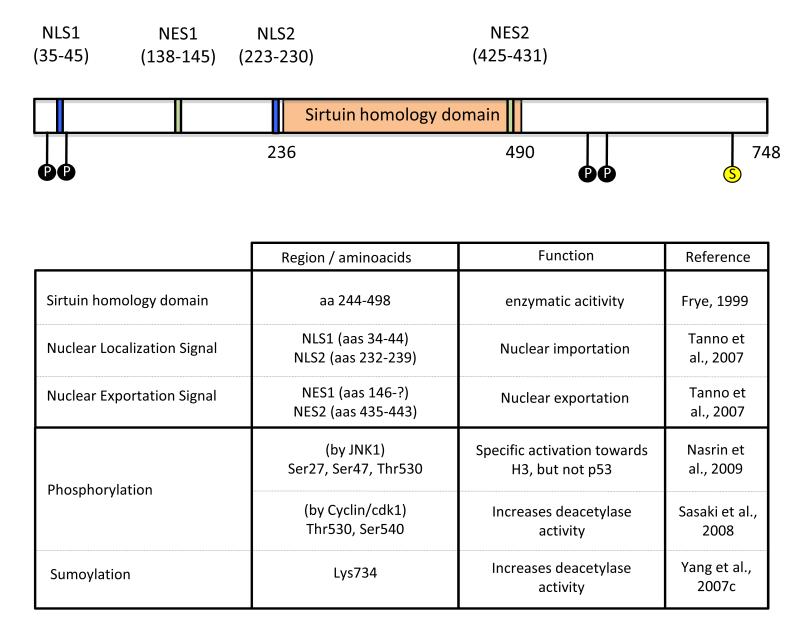
Among all sirtuins, SIRT1 is the best characterized. Human SIRT1 contains the conserved catalytic core of sirtuins and both N- and C-terminal extensions that all span ~240 aminoacids (Figure 2). These extensions serve as platforms for interaction with regulatory proteins and substrates. In total the human SIRT1 spans 747 amino acids. SIRT1 contains 2 nuclear localization signals (NLS) as well as 2 nuclear exportation signals (NES) (Tanno et al., 2007). The balanced functionality of these signals determines the presence of SIRT1 in either the nuclear or the cytoplasmic compartment, and explains that SIRT1 location may differ depending on the cell type or tissue evaluated. For instance, while SIRT1 is mainly found in the nuclear compartment in COS-7 cells (McBurney et al., 2003; Sakamoto et al., 2004), it is abundantly found in the cytosol of rodent β-cells, myotubes and cardiomyocytes (Moynihan et al., 2005; Tanno et al., 2007). While the implications and regulation of SIRT1 shuttling are still largely unknown, some experiments indicate that SIRT1 shuttles from the nuclei to the cytosol upon inhibition of insulin signalling (Tanno et al., 2007). The latter observations suggested a link between SIRT1 activity and the sensing of the metabolic status of the cell, as discussed in the next chapter.
Figure 2. Relevant domains in the human form of the SIRT1 protein.
The figure schematizes the span of the conserved sirtuin homology domain as well as the nuclear localization (NLS) and nuclear exportation signals (NES). The residues subject to phosphorylation, by JNK1 and Cyclin/cdk1, and sumoylation are also indicated.
1.b SIRT1 as an NAD+ sensor
SIRT1 activity is generally increased in situations of energy/nutrient stress. The fact that SIRT1 activity is regulated by NAD+ raised the hypothesis that NAD+ could act as a metabolic sensor in situations of energy stress, where NAD+ levels are generally affected. Some aspects of this hypothesis are still controversial (see (Canto and Auwerx, 2009) for review). First amongst them is whether sirtuin activity can really respond to changes in intracellular NAD+ levels. One premise, at least, must be met in that context, i.e. that the Km of the sirtuins for NAD+ falls into the physiological range of NAD+ bioavailability. Direct experimental evidence supporting this point is, in most cases, preliminary or absent. In great part, this is due to the fact that the true bioavailable NAD+ levels are (still) difficult to evaluate. The estimated total intracellular content of NAD+ is in the 0,2-0,5 mM range (for review see (Houtkooper et al., 2010; Sauve et al., 2006)), which lies within the estimated Km values of SIRT1 for NAD+. This would indicate that NAD+ might actually be rate-limiting in certain circumstances in order to propel SIRT1 to its maximal activity. These levels, however, do not discriminate free and protein-bound NAD+. Similarly, this approximation does not take into account the existence of cellular compartmentalization of NAD+.
Changes in intracellular NAD+ rarely fluctuate more than 2-fold (Canto et al., 2009; Chen et al., 2008; Fulco et al., 2008; Rodgers et al., 2005), which is a likely range to impact on sirtuin activity. In general, NAD+ levels increase in mammalian tissues in response to energy/nutrient stresses like exercise (Canto et al., 2009; Canto et al., 2010; Costford et al., 2010), fasting (Canto et al., 2010; Rodgers et al., 2005) or calorie restriction (Chen et al., 2008). Accordingly, SIRT1 activity is enhanced by all these conditions. Interestingly, it has been recently reported that NAD+ levels fluctuate in a circadian fashion (Nakahata et al., 2009; Ramsey et al., 2009). The influence of SIRT1 on the control of clock-related gene expression (Asher et al., 2008; Nakahata et al., 2008) makes it very attractive to conceive this relation as a way by which feeding/fasting cycles influence the circadian clock. In general, high glycolytic rates in the fed state would bring about higher NAD+ reduction rates, while the reduced glycolytic in the fasted state would enhance mitochondrial oxidative metabolism, derived from fatty acid oxidation, which is generally paired with higher NAD+ levels. This scenario constitutes a beautiful mechanism by which metabolism would be directly coupled to the enzymatic activity of SIRT1 and down-stream pathways.
SIRT1 activity is also controlled by other NAD+-derived metabolites. It was proposed that NADH would compete with NAD+ binding to SIRT1 and inhibit SIRT1 activity (Lin et al., 2004). However, NADH can only competitively inhibit NAD+ binding only at the millimolar range, which is well above its physiological levels (Schmidt et al., 2004). A more prominent and consistent inhibitory effect is achieved with nicotinamide (NAM), which exerts a potent end-product inhibition on SIRT1 activity in a non-competitive fashion with NAD+ (Anderson et al., 2003; Bitterman et al., 2002). Kinetic studies demonstrate that NAM acts at a Km between 30 and 200 μM (Bitterman et al., 2002). The reference values for the intracellular concentration and subcellular compartmentalization of NAM are still far from determined, an issue that is further complicated by the diffusive nature of NAM (van Roermund et al., 1995). Indirect evidence of the large influence of NAM on SIRT1 activity is derived from experiments that manipulate NAM metabolism through changing the activity of nicotinamide phosphorybosyltransferase (Nampt). In the cell NAM is used as a substrate for NAD+ resynthesis through the action of Nampt (Revollo et al., 2004). Inhibition or downregulation of Nampt leads to NAM accumulation and NAD+ depletion, ultimately decreasing SIRT1 activity (Revollo et al., 2004). This highlights how NAM can influence SIRT1 activity through different means: first, as a non-competitive SIRT1 inhibitor, and, second, as an NAD+ precursor. Low levels of NAM might hence be beneficial for SIRT1 activity, as it can act as an NAD+ precursor, but more important accumulation of NAM could be deleterious through the inhibition of SIRT1 (Yang and Sauve, 2006).
1.c SIRT1 actions (I): Nuclear targets
In agreement with its dual cellular localization, SIRT1 targets can be found in both the nuclear and cytosolic compartments. SIRT1 activity in the nucleus articulates dynamic and varied transcriptional responses through the deacetylation of a large spectrum of transcriptional regulators. Therefore, the deacetylation by SIRT1 can lead to the direct activation or inhibition of transcriptional regulators and modify their interaction profiles, depending on the cellular context. From a metabolic perspective, it is exciting to see that many SIRT1 deacetylation targets are key metabolic regulators, further enhancing the notion that SIRT1 is a metabolic stress effector governing transcriptional adaptations aimed to synchronize energy metabolism with nutrient availability. SIRT1 activity and targets, however, expand beyond the realm of metabolism. For example, SIRT1 has marked anti-inflammatory effects in diverse tissues and cell models (Pfluger et al., 2008; Purushotham et al., 2009; Yoshizaki et al., 2009; Yoshizaki et al., 2010), probably through the negative regulation of the NF-kB pathway (Yeung et al., 2004). Also, SIRT1 activity has a strong influence on cell proliferation, apoptosis and cancer. While the data in vitro is controversial, the work on genetically engineered mouse models indicate that enhanced SIRT1 activity would be protective against the development of some types of cancer (Herranz et al., 2010b). Another field where SIRT1 may be of interest is in the central nervous system. It was recently proven that SIRT1 has key roles modulating cognitive function and synaptic plasticity (Gao et al., 2010; Michan et al., 2010). Additionally, there is evidence that enhanced SIRT1 activity could be protective in diseases as neurodegeneration, Alzheimer’s disease and amyotrophic lateral sclerosis (Araki et al., 2004; Chen et al., 2005a; Donmez et al., 2010; Kim et al., 2007a). As we will mainly focus on the metabolic impact of SIRT1, we refer the reader to other recent reviews for discussion of these other fields of action for SIRT1 (Finkel et al., 2009; Guarente, 2011; Herranz and Serrano, 2010).
The identification of p53 as a SIRT1 substrate enlightened the scientific community on the versatility of SIRT1, which was until then largely considered a histone deacetylase. Two different labs simultaneously reported how SIRT1 interacts with and deacetylates p53 (Luo et al., 2001; Vaziri et al., 2001). While p53 can be acetylated in up to 6 residues, SIRT1 seems to preferentially deacetylate Lys379 (human Lys382). The deacetylation of p53 by SIRT1 attenuated its activity on the p21 promoter and inhibited p53-dependent apoptosis (Luo et al., 2001; Vaziri et al., 2001). This link between p53 and SIRT1 activities, led to the premature hypothesis that SIRT1 inhibition could lead to tumor suppression and, the other way round, that SIRT1 activation would promote tumor formation. However, SIRT1 transgenic models challenge this hypothesis and, actually, point out that SIRT1 activation suppresses tumor formation (for review, see (Herranz and Serrano, 2010). This discrepancy might stem from different, yet unresolved, issues. For example, it is not clear whether physiological deacetylation of p53 in situations of higher SIRT1 activity are modulated via direct deacetylation or take place indirectly through changes in other cellular processes, such as affecting its interaction with p300 (Bouras et al., 2005). Also, as will be described later for other transcriptional regulators, p53 activity does not only depend on the modulation of its acetylation levels, but on many other post-translational modifications, which create a “bar-code”-like situation determining specific activity (Murray-Zmijewski et al., 2008). A very nice example about why p53 “activation” should be reworded as “specification” is provided by the actions of p53 on mitochondrial metabolism. In general, p53 activation has been linked to enhanced mitochondrial oxidation, while p53 deletion is associated with defective mitochondrial respiratory rates (Matoba et al., 2006; Saleem et al., 2009; Zhou et al., 2003). However, it has been recently reported how activation of p53 in the context of DNA damage can paradoxically lead to decreased mitochondrial biogenesis (Sahin et al., 2011). This highlights how the activity of transcription factors can be channelled in different ways depending on the biological context of their activation and, likely, on a differential post-translational modification “bar-code”.
The Forkhead-O-box (FOXO) family of transcription factors constitute another example of how SIRT1 channels, rather than activates/inhibits, transcriptional regulators. FOXOs are key regulators of lipid metabolism, stress resistance and apoptosis (Gross et al., 2008) and SIRT1 was shown to interact with and deacetylate the FOXO family of transcription factors (Brunet et al., 2004; Motta et al., 2004). Interestingly, deacetylation of FOXO3 by SIRT1 inhibited its activity on apoptosis-related gene expression, while driving its actions towards the induction of oxidative stress resistance genes (Brunet et al., 2004). It was later also described that SIRT1-mediated FOXO deacetylation also enhances autophagy (Hariharan et al., 2010). This is in line with the hypothesis that activation of SIRT1 allows the cell to adapt to situations of energy stress. Of note, SIRT1 activity, as well as FOXO1 and FOXO3 deacetylation is prompted by situations of oxidative stress, energy stress and fasting (Brunet et al., 2004; Canto et al., 2009; Canto et al., 2010). Also notorious, FOXO and SIRT1 orthologs in lower eukaryotes have both been linked to lifespan extension (Canto and Auwerx, 2009; Greer and Brunet, 2008). Altogether, the correlative activation and effects on lifespan, metabolism and adaptation to energy stress suggest that SIRT1 and FOXO activities might be linked mechanistically through this SIRT1-mediated deacetylation. When the acetylatable residues in FOXO are mutated to mimic a constant acetylated state (K->Q), FOXO becomes more sensitive to Akt-mediated phosphorylation and nuclear exclusion (Qiang et al., 2010). Conversely, when the mutations mimic the deacetylated state (K->R), FOXO is retained in the nucleus (Qiang et al., 2010). These mutants further confirmed that FOXO deacetylation is required for the effects of oxidative stress FOXO nuclear trapping and the induction of stress resistance gene expression (Qiang et al., 2010) Interestingly, the mere coexistence of FOXOs and SIRT1 in the nucleus is insufficient to promote their interaction in the absence of energy or oxidative stresses (Brunet et al., 2004), underscoring the necessity of an additional stress-derived signal to trigger their functional interaction.
A similar case can be made for the transcriptional coactivator peroxisome proliferator-activated receptor (PPAR)γ coactivator 1α (PGC-1α). PGC-1α acts as a master regulator of mitochondrial biogenesis in vertebrates (Puigserver et al., 1998; Rodgers et al., 2005) and orchestrates a constellation of transcription factors, such as the estrogen-related receptors (ERRs), the nuclear respiratory factors (NRFs) 1 and 2 or PPARs, to induce mitochondrial gene expression (Rodgers et al., 2005; Wu et al., 1999). Seminal work by the Puigserver lab illustrated how PGC-1α is acetylated and how deacetylation of PGC-1α by SIRT1 is a key event required for its activation (Lerin et al., 2006; Rodgers et al., 2005). In situations of energy stress or SIRT1 activation, PGC-1α is prominently deacetylated and activated (Canto et al., 2009; Gerhart-Hines et al., 2007; Rodgers et al., 2005). PGC-1α can be acetylated in up to 13 lysine residues (Rodgers et al., 2005), and although there is still not a clear idea on the differential contribution of each residue, mutation of all 13 lysine into arginine (mimicking constant deacetylation), constitutively activates PGC-1α (Rodgers et al., 2005). PGC-1α is a nuclear protein, and, therefore, might coexist with SIRT1 in the nucleus. However, as happened with the FOXOs, only upon energy stress physiological activation of PGC-1α by SIRT1 is prominent, indicating that additional signals are required to prompt SIRT1-mediated PGC-1α deacetylation. The mechanism by which this specification happens has recently been unveiled. Energy or nutrient stress is generally translated in imbalanced AMP/ATP ratios (Hardie, 2007). Whenever there is an increase in the AMP/ATP ratio or ADP/ATP ratio, be it by enhanced ATP consumption or defective ATP synthesis, the enzymatic activity of the AMP-activated protein kinase (AMPK) is enhanced (see (Hardie, 2007) for a mechanistic review). PGC-1α is a substrate for AMPK phosphorylation leading to its activation (Jager et al., 2007), even though how this phosphorylation activates PGC-1α remains still elusive. Our lab recently described how the phosphorylation of PGC-1α by AMPK in situations of energy stress is required to prime it for subsequent deacetylation and activation by SIRT1 (Canto et al., 2009). This AMPK/SIRT1/PGC-1α signalling pathway is furthermore the mechanism by which several hormones enhance mitochondrial metabolism, such as the case for adiponectin (Iwabu et al., 2010), leptin (Li et al., 2010) or FGF21 (Chau et al., 2010). Of note, SIRT1 can deacetylate non-phosphorylated PGC-1α in vitro (Nemoto et al., 2005), indicating that the cellular context poses some constraints for this reaction/interaction to happen. It is likely that AMPK-mediated phosphorylation of PGC-1α modifies the nuclear localization of PGC-1α and/or allows the interaction with third-party proteins that reinforce the stability of the SIRT1 and PGC-1α interaction. It is also tempting to hypothesize that a similar mechanism explains why, despite their coexistence in the nucleus, FOXOs are only interacting with SIRT1 in situations of energy stress. In fact, FOXOs are also phosphorylated in response to energy stress by AMPK (Greer et al., 2007). Further experiments will have to verify whether, as is the case for PGC-1α, this phosphorylation by AMPK converges with SIRT1 deacetylation to target FOXO toward specific gene sets, such as those related to protection against oxidative stress. In all, the AMPK and SIRT1 signaling pathway highlights the interactive nature of different post-translational modifications on transcriptional activators, such as PGC-1α and FOXO, that allow them to select specific downstream pathways.
Both PGC-1α and FOXOs are transcription factors that, upon deacetylation by SIRT1, will enhance lipid catabolism and mitochondrial respiration. However, SIRT1 can also directly block lipid anabolism by interfering with PPARγ and the Liver X Receptor (LXR) signalling. PPARγ is a nuclear receptor that is mainly expressed in white adipose tissue, and plays key roles in adipocyte differentiation, lipid synthesis and storage (Heikkinen et al., 2007). SIRT1 represses PPARγ activity, even though it is not clear whether this is mediated by acetylation-related events in the PPARγ protein (Picard et al., 2004). A recent report indicates that PPARγ can be directly deacetylated by SIRT1, although the relevance of this acetylation for PPARγ activity is not yet known (Han et al., 2010). It is known, however, that the repressive effect of SIRT1 requires the formation of a correpressor complex that also involves the Nuclear receptor Co-Repressor 1 (NCoR1) (Picard et al., 2004). Hence, during fasting, SIRT1 associates with NCoR1 and represses PPARγ function, favoring fat mobilization instead of storage. This work also highlights how many actions of SIRT1 are determined by its interaction with specific protein complexes. Therefore, understanding the dynamics of how SIRT1 merges into different protein complexes will be key to understand how to drive SIRT1 towards specific sets of actions.
While PPARγ is a major controller of lipid anabolism in adipose tissue, other nuclear receptors can also perform similar functions in other tissues. For example, LXRα and β are well-known for their ability to sense oxysterols and regulate genes that decrease total body cholesterol levels (Kalaany and Mangelsdorf, 2006). LXRs, however, also are potent stimulators of lipid anabolism through the induction of the sterol regulatory element binding protein-1c (SREBP-1c) and its downstream targets, stearoyl-CoA deasturase 1 (SCD-1), acyl-CoA carboxylase (ACC) and fatty acid synthase (FAS) (Kalaany and Mangelsdorf, 2006). LXRs are acetylated at Lys432 in LXRα and Lys433 in LXRα (Li et al., 2007). Upon LXR activation, LXRs interacts with SIRT1, which then removes their acetyl groups (Li et al., 2007). Deacetylation of LXR increases its transcriptional activity, even though the deacetylated lysine residue of LXR makes it also more prone to ubiquitination and degradation (Li et al., 2007). The key role of SIRT1 in the modulation of LXRs activity fits with the impaired cholesterol homeostasis and hepatic cholesterol accumulation observed in SIRT1 null mice. The impact of SIRT1 on cholesterol homeostasis was further supported by other studies showing how the absence of SIRT1 reduced the expression of CYP7A1, the rate-limiting enzyme in the bile acid synthesis (Rodgers and Puigserver, 2007), even though whether this phenomenon is strictly LXR-dependent is currently unclear. Given that SIRT1 stimulates both macrophage cholesterol efflux to the liver (Li et al., 2007) and the hepatic conversion of cholesterol into bile acids potentially through LXR (Rodgers and Puigserver, 2007), the SIRT1-LXR pathway seems therefore to be important for reverse cholesterol transport. The effects of LXR and SIRT1 on lipid homeostasis are, however, more conflictive. Theoretically, LXR activation by SIRT1 should increase liver triglyceride accumulation. Liver-specific deletion of SIRT1, however, induces hepatic steatosis, while gain of SIRT1 function is protective (see Section 2). Although the latter observation fits with the higher oxidative metabolism expected after SIRT1 activation, it seems incompatible with LXR activation. A likely explanation is that SIRT1 activation does not only deacetylate LXR, but also its major mediator in the induction of triglyceride synthesis, SREBP-1c (Ponugoti et al., 2010; Walker et al., 2010). In fact, SREBP-1c is stabilized by p300-mediated acetylation (Giandomenico et al., 2003), and the deacetylation of SREBP-1c by SIRT1 at Lys289 and Lys309 makes the protein prone to ubiquitin-mediated degradation (Giandomenico et al., 2003; Ponugoti et al., 2010). The abrogation of SREBP-1c activity would hence allow SIRT1 to promote beneficial effects on cholesterol metabolism through activation of LXR in the absence of detrimental effects on liver lipid accumulation.
While the modulation of all the above transcription factors mainly influence lipid metabolism in white adipose tissue and liver, SIRT1 also modulates carbohydrate metabolism via deacetylation of other transcription factors. The liver maintains blood glucose levels during fasting through gluconeogenesis. A key transcriptional regulator of gluconeogenic gene expression is the cAMP Response Element-Binding (CREB) protein, whose activity is largely controlled by the binding of its coactivator CREB-regulated transcriptional coactivators (CRTCs) (Altarejos and Montminy, 2011). Amongst the 3 members of the CRTC family, CRTC-2 has been reported to be key to properly induce gluconeogenic gene expression (Altarejos and Montminy, 2011). In the fed state, CRTC-2 is hyperphosphorylated, probably by the salt-inducible kinase 2 (SIK2), which sequesters it in the cytosol by avid binding to 14-3-3 platform proteins (Koo et al., 2005; Screaton et al., 2004). Upon exposure to cAMP or calcium signals during fasting, CRTC2 is dephosphorylated by calcineurin, released from 14-3-3 and therefore able to translocate to the nucleus, where it binds and activates CREB on relevant gluconeogenic gene promoters, such as those of the phosphoenolpyruvate carboxykinase (PEPCK) and Glucose 6 Phosphatase (G6P) genes (Screaton et al., 2004). The coactivation of CREB by CRTC2 is, however, only transient during the early stages of fasting (Liu et al., 2008). Upon prolonged fasting, and coinciding with the hepatic increase in NAD+ and SIRT1 activation, CRTC2 activity decreases (Liu et al., 2008; Rodgers et al., 2005). SIRT1 activation in fact deacetylates CRTC2 at Lys628, leading to the COP-1-mediated ubiquitylation and proteasome-dependent degradation of CRTC2 (Liu et al., 2008). Since gluconeogenesis consumes ATP, this action of SIRT1 may attenuate gluconeogenesis in an effort to prevent premature energy depletion upon protracted fasting. Of note, restraining the activity of orthologs of CRTC and CREB in C. elegans prolongs lifespan (Mair et al., 2011), which suggest that decreased levels of CRTCs after deacetylation may be one way by which SIRT1 orthologs may affect lifespan in worms.
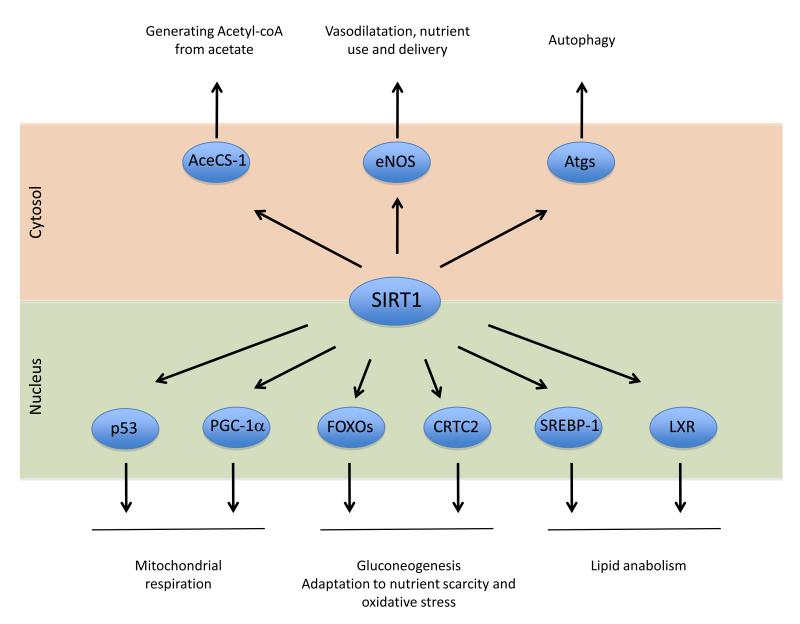
Altogether, the ensemble of metabolic transcriptional regulators directly affected by SIRT1 enables it to orchestrate cellular and whole body metabolism to extract energy from non-carbohydrate sources, especially by mitochondrial respiration-based routes (Figure 3). This is in line with the observation that SIRT1 is activated in situations of nutrient deprivation and energy stress.
Figure 3. SIRT1 metabolic targets.
SIRT1 deacetylates a large arrav of protein targets involved in metabolic regulation. The bottom part of the figure highlights nuclear targets implicated in transcriptional metabolic adaptations. SIRT1’s cytosolic targets are illustrated in the top part. The full names for the abbreviations can be found on the main text.
1.d SIRT1 actions (II): cytosolic targets
As mentioned before, SIRT1 is also present in the cytosol in many cell types, especially when insulin signals are lacking (Tanno et al., 2007). This suggests that SIRT1 also modifies the activity of cytosolic enzymes through direct deacetylation (Figure 3).
Initial evidence for the existence cytosolic SIRT1 targets came from the discovery that the cytosolic Acetyl-CoA Synthetase 1 (AceCS-1) enzyme, is deacetylated by SIRT1, but not by other sirtuins (Hallows et al., 2006). AceCS-1 can generate Acetyl-CoA from acetate. While this enzyme has a key role in bacterial energy metabolism, the impact of acetate metabolism and AceCS-1 on mammalian whole-body metabolism is not yet clear. AceCS-1 is acetylated on Lys661 in the catalytic domain (Hallows et al., 2006). The activity of AceCS-1 is almost 50 times lower in its acetylated state (Hallows et al., 2006) and SIRT1 deacetylation, therefore, serves as an activation switch. The dynamic regulation of AceCS-1 acetylation in response to physiological events has, however, not yet been explored.
Another cytoplasmic enzyme deacetylated by SIRT1 is the endothelial nitric oxide synthase (eNOS) (Mattagajasingh et al., 2007). SIRT1 deacetylates Lys496 and Lys506 in the calmodulin binding domain of eNOS, thereby activating it to boost endothelial nitric oxide levels (Mattagajasingh et al., 2007). Endothelial inhibition of SIRT1 leads to inefficient endothelium-dependent vasodilatation (Mattagajasingh et al., 2007), a process key for proper nutrient supply to tissues. The activation of eNOS by SIRT1 could hence be a mechanism by which nutrient scarcity increases energy delivery into tissues. Of note, impaired eNOS function has major consequences on whole body metabolism, as it impacts on peripheral glucose uptake (Kapur et al., 1997; Li et al., 2004) and mitochondrial biogenesis (Le Gouill et al., 2007; Nisoli et al., 2003; Nisoli et al., 2005).
The impact of SIRT1 in metabolic cytosolic processes was further underscored by the discovery that SIRT1 forms molecular complexes with critical components of the autophagy machinery, including Atg5, Atg7 and Atg8 (Lee et al., 2008). SIRT1 deacetylates these proteins in an NAD+-dependent manner, even though the substrate residues and the consequences of this deacetylation have not yet been fully elucidated (Lee et al., 2008). Autophagy during starvation is hence impeded in embryonic fibroblasts of SIRT1−/− mice and leads to the accumulation of damaged organelles, especially mitochondria (Lee et al., 2008). This phenomenon contributes to the fact that impaired SIRT1 activity systematically correlates with deficiencies in energy metabolism and fits with the hypothesis of SIRT1 being a master metabolic switch driving the cell to obtain energy from non-carbohydrate energy sources.
2. SIRT1 AND METABOLIC DISEASE: EVIDENCE FROM MICE MODELS
The attractive effects of SIRT1 orthologs in lower organisms, as well as at the cellular and molecular level in mammalian cells, prompted the generation of mouse models to evaluate the impact of SIRT1 on whole body metabolism. This goal proved more difficult to achieve than expected, as inbred germline SIRT1 deficient mice have high prenatal death rates (McBurney et al., 2003). The very few pups that were born presented severe neurological and cardiac defects, resulting in early postnatal death (McBurney et al., 2003). Outbred mice with the SIRT1 mutation, however, were viable (McBurney et al., 2003). From a metabolic perspective, SIRT1 knock-out mice were metabolically inefficient and showed impaired calorie restriction-induced effects on metabolism and longevity (Boily et al., 2008). The outbred mouse line, however, is not ideal for metabolic studies, and inducible models will be required to evaluate how whole body deletion of the SIRT1 gene will affect global metabolism.
In the meantime, several tissue-specific somatic SIRT1 deficient mouse models, however, already provided ample evidence that most of the in vitro biology of SIRT1 translates into an in vivo context. Most of the metabolic work on SIRT1 has been focussed on muscle cells and hepatocytes. Surprisingly, while most evidence in cultured muscle cells indicates a key role for SIRT1 in the modulation of mitochondrial metabolism (Canto et al., 2009; Gerhart-Hines et al., 2007), initial studies in the muscle-specific SIRT1 knock-out mice indicate that SIRT1 is not required for exercise-induced deacetylation of PGC-1α or mitochondrial biogenesis in skeletal muscle (Philp et al., 2011). Complementary mice models and physiological challenges will be required to help clarify the role of SIRT1 in skeletal muscle. In contrast to the situation in muscle, many different studies have focussed on liver-specific SIRT1 gene deletion (Chen et al., 2008; Purushotham et al., 2009; Wang et al., 2010). The lack of SIRT1 in liver does not induce an overt phenotype on chow diet and these mice respond normally to calorie restriction (Chen et al., 2008). However, diametrically opposite results became apparent when the physiological impact of high-fat diet in two independent liver-specific SIRT1 deficient mice lines was evaluated. Whereas the Guarente group found that the liver-specific SIRT1 null mice gained less weight upon high-fat feeding, maintained better glucose tolerance and were protected against hepatic steatosis (Chen et al., 2008), the study by Purushotham et al reported that liver SIRT1 deletion increased susceptibility to hepatic steatosis and body weight gain upon high-fat feeding (Purushotham et al., 2009). Furthermore, the latter study also reported that the lack of SIRT1 in liver also enhanced hepatic triglyceride accumulation upon fasting (Purushotham et al., 2009). A subsequent study, using yet another hepatocyte SIRT1 knock-out mouse line, reported also prominent liver steatosis in chow fed mice at young ages, which worsened with age (Wang et al., 2010). It must be noted that germline heterozygous SIRT1 deficient mice also show a marked tendency towards liver lipid accumulation (Xu et al., 2010). This conclusion would also be totally in line with the in vitro observations suggesting that SIRT1 enhances fat oxidation and that SIRT1 activity downregulates SREBP-1c, a master controller of fatty acid synthesis (Ponugoti et al., 2010; Rodgers and Puigserver, 2007; Walker et al., 2010).
Other investigators have analyzed the role of SIRT1 in the liver by acutely knocking its expression down through tail vein injection of adenoviruses carrying a SIRT1 shRNAs. Strikingly, no alterations in triglyceride accumulation were observed in livers acutely depleted of SIRT1 (Rodgers and Puigserver, 2007). Instead, glucose homeostasis was severely impaired and gluconeogenic capacity was defective upon SIRT1 reduction (Rodgers and Puigserver, 2007). This role of SIRT1 in glucose homeostasis was, however, not observed in any of the above mentioned hepatic SIRT1 deficient mouse lines. This might indicate that either the adenoviral shRNA delivery is causing additional effects or that the defects in glucose homeostasis upon acutely decreasing SIRT1 levels are somehow compensated in the liver-specific SIRT1 knock-out and the germline heterozygote SIRT1 deficient mice, which have chronic reductions in hepatic SIRT1 expression. A role of SIRT1 in gluconeogenesis is furthermore highly debated. From one side, it is speculated that SIRT1 enhances gluconeogenesis via the deacetylation and activation of PGC-1α, which, in turn would coactivate CREB on the promoters of gluconeogenic genes (Herzig et al., 2001; Yoon et al., 2001). However, while there is no doubt that artifactual overexpression of PGC-1α enhances gluconeogenic gene expression in liver, it must be pointed out that the evidence indicating that physiological modulation of PGC-1α activity is participating in gluconeogenesis is weak (Herzog et al., 2004). On the other hand, a plethora of scenarios have illustrated how SIRT1 activation in liver is not per se associated with enhanced glucose production, but rather to attenuated gluconeogenic rates (summarized in (Canto and Auwerx, 2010)).
The role of SIRT1 in pancreas function has also been characterized in genetically engineered mouse models. Studies in outbred SIRT1 knock-out mice indicated that SIRT1 deficiency blunts pancreatic insulin secretion (Bordone et al., 2006). The etiology of this defect is not entirely clear, even though it was proposed that it might stem from the negative regulation that SIRT1 exerts on UCP2 expression (Bordone et al., 2006). The lack of SIRT1 leads to higher UCP2 levels, which alter the ability of glucose to modulate ADP/ATP ratios in pancreatic β-cells and trigger insulin release (Zhang et al., 2001). The influence of SIRT1 on insulin release was confirmed in mice that specifically overexpressed SIRT1 in pancreatic β-cells, which manifested enhanced glucose-induced insulin secretion (Moynihan et al., 2005). This study further certified how UCP2 is negatively regulated by SIRT1, therefore allowing better coupling and ATP production in response to high glucose in the SIRT1 overexpressing mice (Moynihan et al., 2005). However, SIRT1 also enhanced insulin secretion upon artificial depolarization with KCl, indicating that SIRT1 alters insulin release by additional mechanisms downstream of depolarization and independent of UCP2 (Moynihan et al., 2005). Interestingly, the beneficial effects of β-cell SIRT1 overexpression were restricted to young mice and lost upon aging (Ramsey et al., 2008). While the explanation for this phenomena is not clear yet, it might originate in the NAD+-dependence of SIRT1, and aging is known to decrease NAD+ levels in rodent tissues (Braidy et al., 2011). Therefore, it is likely that the reduction in NAD+ limits SIRT1 activity during aging, attenuating its beneficial effects on insulin secretion and glucose homeostasis.
The functions of SIRT1 in central nervous system control of metabolism have also not escaped attention. The neuronal deletion of SIRT1 does not affect brain development, but reduces body size as a consequence of a specific deficiency in pituitary growth hormone production (Cohen et al., 2009). While mice with the neuronal deletion of SIRT1 showed no major differences in glucose tolerance compared to wild-type littermates on both chow and high-fat diets at young age, defects in glucose homeostasis were exacerbated upon aging (Cohen et al., 2009). The reasons for these particular phenotypes have not been elucidated. Conversely, brain-specific overexpression of SIRT1 did not result in a major phenotypic change in the basal state (Satoh et al., 2010). Based on these reports, no clear picture of how central nervous system SIRT1 activity influences global metabolism has emerged as of yet.
While other tissue specific mouse models are being generated at present, the role of SIRT1 in metabolism has also been studied using whole body gain-of-function SIRT1 mice. The first SIRT1 gain-of-function mouse model displayed several phenotypes that resembled calorie restricted mice as the animals were leaner, metabolically more active and had increased glucose tolerance suggestive of insulin sensitization (Bordone et al., 2007). The other two mice lines that overexpress SIRT1 further explored the impact of diet- and genetically-induced obesity, both concluding that mild SIRT1 overexpression protects against the development of hyperglycemia, metabolic disease and fatty liver (Banks et al., 2008; Pfluger et al., 2008). The above results would be in line with the observations in liver SIRT1 knock-out mice reporting higher hepatic fat accumulation (Purushotham et al., 2009; Wang et al., 2010). Of note, while SIRT1 transgenic mice were protected against the onset of age-related diseases, such as cancer and metabolic diseases, they did not live longer (Herranz et al., 2010a). These data further underscore that so far no firm evidence has been found indicating that SIRT1 influences lifespan in mammals.
Altogether, the information provided by genetically engineered mouse models supports the notion that SIRT1 activation has metabolic benefits. In the next chapter we will describe the efficiency, specificity and results obtained from different strategies aimed to artificially activate SIRT1.
3. PHYSIOLOGICAL AND PHARMACOLOGICAL MODULATION OF SIRT1 ACTIVITY
3.a The modulation of SIRT1 expression
A first line of action when aiming to enhance the biological action of a protein is to increase its expression levels. Indeed, the simple overexpression of SIRT1 in cells and tissues is enough to increase SIRT1 activity (Banks et al., 2008; Rodgers et al., 2005; Rodgers and Puigserver, 2007), indicating that NAD+ might not be limiting for SIRT1 activity in basal conditions. Strikingly, the elucidation of the transcriptional mechanisms controlling SIRT1 expression has only recently begun. We will here focus on the mechanisms related to the metabolic and redox control of SIRT1 expression (Figure 4).
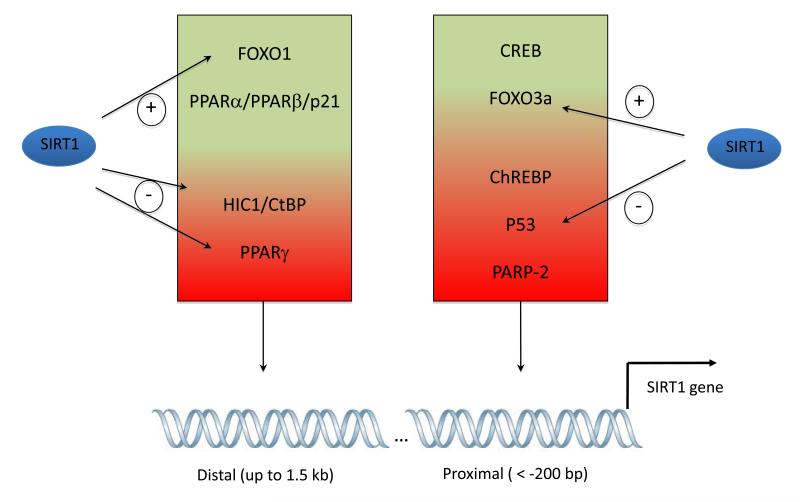
Figure 4. Transcriptional regulation of the SIRT1 gene.
Many transcription factors influence the transcriptional activity through acting on both the proximal and distal regions of the SIRT1 promotor. Transcriptional regulators in the green part of the boxes positively regulate SIRT1 gene expression, while those in the red part of the boxes act as negative regulators. Of note, the SIRT1 protein can create many feed-forward loops by deacetylating and enhancing the activity of some positive regulators (FOXOs) while deacetylating and/or inactivating repressor complexes (p53, PPARγ, HIC1/CtBP). Full names for the abbreviations can be found in the text.
SIRT1 expression is generally higher in situations of low nutrient availability and endurance exercise (Nemoto et al., 2004). In mice and humans it was shown that SIRT1 expression correlates with higher expression of nuclear encoded mitochondrial genes and energy expenditure (Lagouge et al., 2006; Rutanen et al., 2010). The earliest studies on SIRT1 gene expression aimed to understand how SIRT1 mRNA levels increase in response to nutrient deprivation. In these studies, FOXO3a, a member of the FOXO family of transcription factors, was shown to indirectly increase rodent SIRT1 transcription through its proximal promoter. Interestingly, FOXO3a modulates SIRT1 activity via its interaction with p53, and this interaction was shown to be nutrient-dependent (Nemoto et al., 2004). In the absence of this interaction, p53 acts as a repressor of the SIRT1 promoter (Nemoto et al., 2004). Therefore, a model was build where, under normal conditions, p53 represses SIRT1, and, upon nutrient starvation, activated FOXO3a interacts with p53 and relieves the inhibition of SIRT1 transcription, probably by changing the balance of coactivators/correpressors on the SIRT1 promoter. Of note, this interrelation highlights how SIRT1, p53 and FOXO3a activities are interconnected with feedback loops: SIRT1 and p53 negatively regulate each other, via deacetylation (see Section 1.c) and transcriptional events, respectively, generating an homeostatic loop balancing both activities. Conversely, SIRT1-mediated deacetylation of FOXO3a enhances FOXO3a activity, and this will be further amplified by the FOXO3a-mediated induction of SIRT1 expression. Interestingly, the rat SIRT1 promoter contains multiple FOXO1 core binding motifs and a forkhead-like consensus binding site, which enable FOXO1 to directly activate SIRT1 transcription, which is in contrast to the indirect effects of FOXO3a (Xiong et al., 2011). Importantly, as happened with FOXO3a, FOXO1 and SIRT1 would create a feedforward loop in which FOXO1 activation by SIRT1-mediated deacetylation amplifies SIRT1 expression and activity. It will be interesting to elucidate whether these FOXO1 binding sites are evolutionary conserved and how this feedforward loop is integrated with other regulatory mechanisms of SIRT1 transcription. Similarly, it will be key to understand the mechanisms by which FOXOs are driven to the SIRT1 promoter upon glucose deprivation. Importantly, FOXOs are activated by AMPK (Greer et al., 2007), which is known to increase SIRT1 expression (Suwa et al., 2011).
SIRT1 can also promote another negative feedback loops on its own promoter through Hypermethylated in Cancer 1 (HIC1) (Chen et al., 2005b). HIC1 naturally forms a transcriptional correpressor complex with SIRT1, which binds directly to the SIRT1 promoter and downregulates its transcription (Chen et al., 2005). Importantly, the repressive activity of HIC1 on the SIRT1 promoter is regulated by its association with CtBP a sensor of energy/redox stress (Zhang et al., 2007). The binding of CtBP to transcriptional repressors as HIC1 is enhanced by NADH (Zhang et al., 2002). Therefore, in situations of low glycolytic rates, such as seen upon 2-deoxyglucose treatment, NADH will decrease, destabilizing CtBP/HIC1/SIRT1 inhibitory complexes, and therefore allowing the induction of SIRT1 mRNA levels. The increase in SIRT1 levels would hence be a way for metabolic adaptation towards the utilization of non-carbohydrate energy sources.
While SIRT1 content is generally higher upon nutrient deprivation, it is also known that SIRT1 is reduced upon high-fat feeding and in obese individuals (Costa Cdos et al., 2010; Coste et al., 2008). It was therefore interesting to find how PPARγ activation downregulates SIRT1 expression (Han et al., 2010). The distal SIRT1 promoter contains PPAR-response elements (PPREs) (Han et al., 2010; Hayashida et al., 2010), even though a thorough mapping of the sites and their evolutionary conservation is lacking. Upon activation, PPARγ can bind to and repress the SIRT1 promoter and as such provide a mechanism by which SIRT1 expression can be reduced in situations of nutrient overload. Of note, PPREs can bind also other PPARs. Therefore, it would be expected that other PPARs also regulate the SIRT1 promoter. Confirming this speculation, PPARα or PPARβ/δ activation by synthetic ligands enhances SIRT1 expression (Hayashida et al., 2010; Okazaki et al., 2010). While the mechanism through which PPARα regulates the SIRT1 promoter remains unclear, the actions of PPARβ/δ are not mediated by its direct binding to the PPREs, but rather involve binding to p21, which has a conserved binding site in the proximal human SIRT1 promoter (Okazaki et al., 2010). Altogether it is interesting that the PPARs, which act as lipid sensors (Schoonjans et al., 1996) control SIRT1 activity, with PPARγ, related to lipid anabolism, inhibiting SIRT1 expression, while PPARα and PPARβ/δ, both linked to fatty acid oxidation, increase SIRT1 mRNA levels. Therefore, the differential sensing of lipid species might be key to understand how lipid metabolism can influence SIRT1 expression and drive adaptations towards lipid anabolic or catabolic pathways and future work addressing the distinct effects of the different PPARs is warranted.
Another recent interesting finding about the transcriptional regulation of SIRT1 expression came from the studies of a non-sirtuin NAD+ consumer, the poly-ADP-ribose polymerase (PARP)-2 protein. PARP-2 is a member of a large family of PARP proteins (see section 3.e.2.a). While, in general PARP-2 has been mainly considered to be part of the DNA damage repair machinery, it has been lately shown that PARP-2 also acts as a transcriptional modulator (Bai et al., 2007). PARP-2 enhances the transcriptional activity of PPARγ, but not that of PPARα or PPARβ/δ, therefore favoring adipocyte differentiation and fat storage (Bai et al., 2007). In contrast, PARP-2 decrease the activity of the SIRT1 promoter by directly binding to the proximal -91 bp region (Bai et al., 2011a). Consistent with this, decreased PARP-2 levels enhanced SIRT1 gene expression, which translated into higher SIRT1 activity (Bai et al., 2011a). At the whole body level, PARP-2 deletion mimics all the features of SIRT1 activation, such as higher mitochondrial content, enhanced oxidative metabolism and protection against diet-induced obesity and insulin resistance (Bai et al., 2011a). It will be interesting to elucidate how PARP-2 influences SIRT1 transcription and whether this may involve the poly-ADP-ribosylation (PARylation) of other proteins in the vicinity of the SIRT1 promoter. Also, how PARP-2 can both activate and repress transcription requires further study.
While most of the above examples provide a number of candidate transcription factors that influence SIRT1 transcription in vitro, few of them have been clearly linked to the physiological modulation of SIRT1 expression by hormones and the feeding/fasting cycles in vivo. Our lab recently identified how, during feeding, the carbohydrate response element binding protein (ChREBP) directly binds to a composite response element in the proximal SIRT1 promoter and represses its transcription (Noriega et al, 2011). Upon fasting, ChREBP is translocated to the cytosol and its binding site on the promoter is now liberated. This enables CREB, whose activity is enhanced by the cAMP signal generated by the fasting hormones, glucagon and norepinehrine, to bind and enhance SIRT1 gene expression (Noriega et al, 2011). This way the opposite effects of CREB and ChREBP on SIRT1 transcription constitute the first established mechanism for the regulation of SIRT1 expression in response to physiological fasting/feeding cycles. This also highlights that the proximal SIRT1 promoter is a hot spot for its physiological regulation (Figure 4) and further underscores that SIRT1 is a crucial metabolic checkpoint connecting the energetic status with transcriptional programs downstream of SIRT1.
Finally, a interesting mechanism regulating SIRT1 expression is the control by microRNAs (miRNAs), which emerge as key controllers of global gene expression (Neilson and Sharp, 2008). miRNAs bind to the 3′-untranslated region (UTR) of target mRNAs and inhibit their expression by causing mRNA cleavage or inhibition of translation (Neilson and Sharp, 2008). It is assumed that around 30% of all human genes are regulated by miRNAs (Neilson and Sharp, 2008). It has been recently reported that miRNA-34a targets hepatic SIRT1 and negatively correlates with SIRT1 expression (Lee et al., 2010; Yamakuchi et al., 2008). miRNA-34a binds to the 3′ UTR of SIRT1 mRNA in a partial complementary manner and represses its translation (Lee et al., 2010; Yamakuchi et al., 2008). Consistently, high miRNA-34a levels are highly elevated in the livers from diet-induced and genetically obese mice (Lee et al., 2010). Of note, p53, which negatively regulates SIRT1 expression directly, also induces miRNA-34a, providing an additional mechanism to ensure SIRT1 repression upon p53 activation (Yamakuchi and Lowenstein, 2009). Other miRNAs have been reported to also affect SIRT1 expression in different tissues, such as miRNA-132, which downregulates SIRT1 expression in adipose tissue, prompting inflammatory responses (Strum et al., 2009). Therefore, miRNAs are providing a whole new level for the regulation of SIRT1 levels that we are only beginning to grasp.
Together, these mechanisms illustrate the complexity of the regulation of SIRT1 at the level of its expression. Of note, most of the transcriptional regulators described are also substrates for SIRT1, which illustrates the intricate nature of SIRT1 regulation and how it is driven by multiple regulatory loops. However, multiple interactions between proteins and the multi-functionality of the individual proteins involved, makes it difficult to predict how pharmacological targeting of one of the players will affect the others. While the identification of novel players will for sure contribute to our understanding of SIRT1 transcriptional regulation, it is understandable that, at this point, alternative strategies to enhance SIRT1 activity are also of interest. Such strategies will be described in the next sections.
3.b Post-translational modifications (PTMs)
The activity of SIRT1, as that of most enzymes, is also modulated by a number of post-translational modifications. The first report indicating this possibility was the identification of SIRT1 as a nuclear phosphoprotein in a large screening using mass-spectrometry (Beausoleil et al., 2004). While 2 phosphoresidues, Ser27 and Ser47, were identified, their function has not yet been explored. Subsequent efforts identified up to 13 phosphorylable residues, including the two previously found (Sasaki et al., 2008). Dephosphorylated SIRT1 was less active than the phosphorylated form (Sasaki et al., 2008). Amongst these residues, Thr530 and Ser540 were phosphorylated by cyclinB/cdk1, and their mutation resulted in aberrant cell proliferation and cell cycle profiles that could not be explained by changes in SIRT1 stability, but rather resulted from lower SIRT1 activity (Sasaki et al., 2008). Another report indicated that JNK1 can also phosphorylate SIRT1 in three residues, Ser27, Ser47 and Thr530, in response to oxidative stress promoted by H2O2 or anisomycin (Nasrin et al., 2009). In agreement with the previous report, phosphorylation of these sites by JNK1 increased nuclear localization of SIRT1 and its activity (Nasrin et al., 2009). Surprisingly, it also seemed that JNK1 phosphorylation oriented SIRT1 activity towards specific substrates, as it triggered the deacetylation of histone H3, but not p53 (Nasrin et al., 2009). Paradoxically, JNK1 activity is induced in situations of obesity and metabolic disease (Hirosumi et al., 2002), where SIRT1 activity is decreased (Coste et al., 2008). A constellation of other kinases, including casein kinase II (CKII) (Kang et al., 2009; Zschoernig and Mahlknecht, 2009), the dual specificity tyrosine phosphorylation-regulated kinases (DYRKs) (Guo et al., 2010) and the mammalian sterile 20-like kinase 1 (MST-1) (Yuan et al., 2011a), have also been suggested to phosphorylate SIRT1. While these data indicates that the activity of SIRT1 might be modulated through the phosphorylation of the above mentioned residues by multiple kinases, it is discouraging that most of the phosphorylable residues identified or their flanking sequences, are very poorly conserved across species, making it difficult to argue that these residues have been key throughout evolution and participate in the most conserved metabolic functions of SIRT1 orthologs across species. Furthermore, the metabolic roles of these phosphorylation events, as well as their interaction with other post-translational modifications needs to be addressed in vivo to fully understand their true biological function.
A second type of post-translational modification that can impact on SIRT1 activity is sumoylation. SIRT1 is sumoylated at Lys734 upon UV irradiation or H2O2 treatment (Yang et al., 2007c). The sumoylation of SIRT1 increased its intrinsic deacetylase activity (Yang et al., 2007c). Conversely, mutation of the residue or forced desumoylation by the SUMO1/sentrin specific peptidase 1 (SENP1) enzyme, rendered SIRT1 less enzymatically active and cells more prone to apoptosis (Yang et al., 2007c). Although, it was speculated that SIRT1 sumoylation acts as a switch between cell survival and cell death, further work is required to define the mechanisms by which cellular stress enhances the interaction of SIRT1 with sumoylation enzymes and/or decreases the association with SENP. As with phosphorylation, an additional caveat is the poor conservation of the sumoylation residue and its flanking regions. For example, mouse SIRT1 cannot be sumoylated, as the human Lys734 is an Arg residue in mice. While this does not rule out a potential effect of sumoylation on SIRT1 activity in many species, including human, it is unlikely that this will contribute to the activity of SIRT1 in other species, unless sumoylation takes place at other residues.
It is often overlooked that SIRT1 activity can affect many different targets. It is unlikely then, that, upon its activation, SIRT1 unselectively deacetylates all of its targets, which could often lead to opposite physiological effects. Therefore, some specification must exist. A clear hint that this is the case was already mentioned by the work on JNK1 (Nasrin et al., 2009), which illustrated how, despite being intrinsically more active, SIRT1 only deacetylated specific substrates. Consequently, PTMs might not only affect SIRT1 activity at the level of its intrinsic activity, but they might also channel SIRT1 towards specific subsets of targets. Importantly, we have illustrated how phosphorylation of PGC-1α by AMPK is key for SIRT1-mediated deacetylation (Canto et al., 2009). Given the large number of SIRT1 substrates it is very likely that substrate accessibility is also controlled by PTMs. Understanding these specification mechanisms will be key to design future strategies aimed to selectively impact on certain functions of SIRT1, but not others.
3.c Protein interactions
SIRT1 activity is not only be controlled intrinsically at the level of the SIRT1 protein, but also through its association with different protein complexes, which may affect its activation or inhibition, as well as its target specificity. In Section 1c, we already specified how the presence of SIRT1 in a complex with NCoR1 actually inhibits PPARγ action in adipose tissue. In this section we will discuss two additional and physiologically relevant mammalian SIRT1 “non-substrate” interactors.
Two simultaneous reports in 2007 indicated that a nuclear protein Deleted in Breast Cancer-1 (DBC1) forms a stable complex with the catalytic domain of SIRT1 and inhibits SIRT1 activity both in vivo and in vitro (Kim et al., 2008; Zhao et al., 2008). As a consequence, the artificial reduction of DBC1 in cell-based experiments stimulated SIRT1 activity, diminishing the acetylation levels of p53, and inhibiting p53-depending apoptosis (Kim et al., 2008; Zhao et al., 2008). The physiological pathways influencing the dynamic interaction of SIRT1 and DBC1 are, however, still unexplored. The in vitro evidence indicating that DBC1 is a SIRT1 inhibitor was further supported by observations in mouse liver indicating that DBC1 and SIRT1 colocalize within the nucleus and coimmunoprecipitate in nuclear extracts (Escande et al., 2010). Mice with a germline deletion of the DBC1 gene showed a 2- to 4-fold increase in endogenous SIRT1 activity in a wide range of tissues, rendering p53 hypoacetylated (Escande et al., 2010). Importantly, the dynamics of the DBC1/SIRT1 interaction have been evaluated in vivo, demonstrating that, under normal feeding conditions, at least 50% of total liver SIRT1 is associated with DBC1, and that this interaction was nearly absent after starvation, contributing to the increase in SIRT1 activity observed in fasting livers (Escande et al., 2010). Importantly, the increase in SIRT1 activity during fasting was blunted in DBC1 knock-out mice (Escande et al., 2010). In contrast to the effects observed during fasting, high fat feeding stabilized the association of DBC1 with SIRT1 (Escande et al., 2010). Understanding the molecular determinants influencing the association between DBC1 and SIRT1 will be an interesting field for future investigation. The overall phenotype of the DBC1 knock-out mice did not differ from wild-type littermates on chow diet. These mice were, however, protected against the development of hepatic steatosis and liver damage induced by high-fat feeding (Escande et al., 2010), in line with the data obtained in the liver-specific SIRT1 knock-out models (Purushotham et al., 2009; Wang et al., 2010).
Around the same time that DBC1 was reported as a sirtuin inhibitor, another report identified a possible activator of SIRT1 activity, the Active Regulator of SIRT1 (AROS) (Kim et al., 2007b). The association of AROS with SIRT1 in, presumably, its catalytic domain enhances SIRT1 activity 2-fold, resulting in p53 inhibition through deacetylation (Kim et al., 2007). Conversely, the artificial reduction in AROS levels, sensitized cells to p53-induced apoptosis (Kim et al., 2007). Interestingly, AROS interacts specifically with SIRT1, but not with other sirtuins (Kim et al., 2007). The exact nature of the actions of AROS on SIRT1 and how it impacts on metabolism will require future study.
These data on NCoR1, DBC1, and AROS illustrate how SIRT1 activity is influenced by its association with specific protein partners. It will be important to elucidate whether these partnerships also influence SIRT1 substrate selectivity. It is reasonable to think that the known, and yet to be identified, SIRT1 interactors also have an impact on the deacetylation level of SIRT1 substrates other than p53, such as PGC-1α, FOXO1, which could have far-reaching implications for metabolic regulation. It will also be interesting for future studies to understand to what extent the deacetylase activity of SIRT1 influences the activity of corepressor or coactivator complexes and to identify the possible roles of SIRT1 as an adaptor protein. It goes without saying that modifying the interaction of SIRT1 with such interactors could constitute a promising avenue to modulate SIRT1 activity.
3.d SIRT1 activating compounds (STACs)
An obvious strategy to artificially enhance SIRT1 would be through chemical compounds that directly bind and activate SIRT1. In 2003, the Sinclair lab, using a screening strategy with a fluorescently-labelled substrate, identified resveratrol and a few other polyphenols, including quercetin and piceatannol, as natural compounds that could enhance the deacetylating activity of SIRT1 (Howitz et al., 2003). A number of subsequent studies, mainly using resveratrol, showed that it activated SIRT1 in diverse species (see (Baur, 2010) for review). Resveratrol treatment mimics numerous aspects of calorie restriction in all eukaryotes tested to date (Barger et al., 2008; Baur et al., 2006; Howitz et al., 2003; Lagouge et al., 2006; Pearson et al., 2008; Valenzano et al., 2006; Wood et al., 2004), and, in most of them, this effect seems to depend on SIRT1 (Howitz et al., 2003; Lagouge et al., 2006; Wood et al., 2004). Not surprisingly, in several (Baur et al., 2006; Howitz et al., 2003; Valenzano et al., 2006; Wood et al., 2004), albeit not all (Pearson et al., 2008), models tested resveratrol increased lifespan. In mice, resveratrol promoted SIRT1 activation and energy expenditure (Baur et al., 2006; Lagouge et al., 2006). Upon high fat feeding, resveratrol prominently prevented the onset of diet-induced obesity and metabolic disease, which ended up protecting the treated mice against the lifespan curbing associated with high caloric diets (Baur et al., 2006; Lagouge et al., 2006). At the molecular level, resveratrol boosted mitochondrial content, as a result of the activation of the SIRT1/PGC-1α axis (Lagouge et al., 2006). Interestingly, resveratrol also improved mitochondrial function and fatty acid oxidation in humans, as demonstrated in a recent study, but at much lower concentrations that those used in mice (Timmers et al, 2011). Altogether, resveratrol proved to be an effective way to activate SIRT1 in vivo and promote beneficial health effects, most of which resemble the effects observed upon the overexpression of SIRT1.
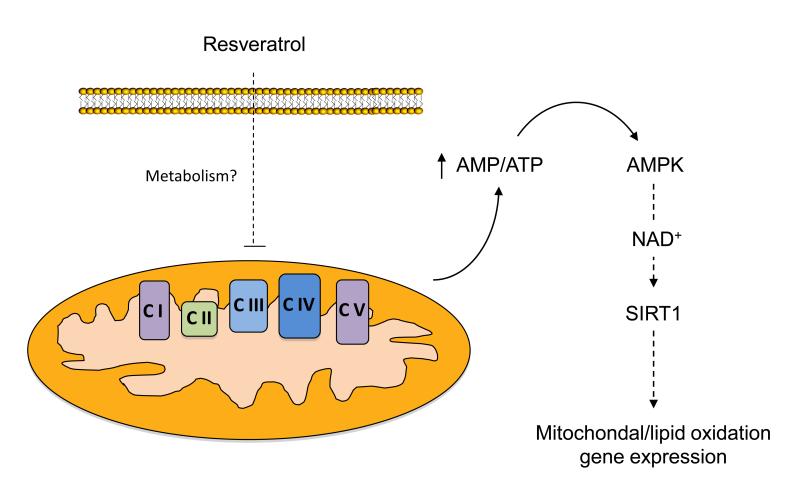
However, the ability of resveratrol to directly activate SIRT1 was seriously questioned by results demonstrating that the non-physiological fluorescent “Fluor de Lys” substrate used for SIRT1 activity assays can lead to artefactual results (Borra et al., 2005; Kaeberlein et al., 2005). Blinded by the beneficial effects in line with SIRT1 activation, resveratrol actions on other possible molecules/pathways have been largely neglected. In fact, resveratrol was much earlier reported as a polyphenol that interfered with the mitochondrial respiratory chain (Zini et al., 1999). It is furthermore relevant that many reports have recently demonstrated that resveratrol can also activate AMPK (Baur et al., 2006; Dasgupta and Milbrandt, 2007; Feige et al., 2008; Park et al., 2007; Zang et al., 2006), which is consistent with its possible effect on the mitochondrial respiratory chain. Elegant studies, using isogenic cell lines stably expressing AMPK complexes containing AMP-insensitive γ2 subunit variants (R531G), convincingly demonstrated that AMPK activation in response to resveratrol derives from and AMP/ATP imbalance as a consequence from interference with mitochondrial respiration (Hawley et al., 2010). AMPK activation by resveratrol is very fast and already prominent after a couple of minutes, while the activation of SIRT1 becomes only detectable after a few hours (Cantó and Auwerx, Unpublished observations), clearly indicating that AMPK activation is an earlier event upon resveratrol treatment. While it has been argued that resveratrol action on AMPK is SIRT1-dependent (Suchankova et al., 2009), the use of MEF cells from SIRT1 knock-out mice unequivocally demonstrated that SIRT1 is dispensable for resveratrol-induced AMPK-activation (Dasgupta and Milbrandt, 2007; Um et al., 2010). Conversely, several approaches have convincingly proven that resveratrol cannot activate SIRT1 in the absence of functional AMPK (Canto et al., 2010; Um et al., 2010). A final picture of the actions of resveratrol on AMPK and SIRT1 activation was finally drawn by the demonstration that SIRT1 is the downstream mediator of AMPK actions. AMPK activation initially leads to a gradual increase in NAD+ levels, as a consequence to the activation of fatty acid oxidation, subsequently activating SIRT1 (Canto et al., 2009). This initial raise in NAD+ is sustained by enhanced Nampt expression which favors the synthesis of NAD+ from NAM through the NAD+ salvage pathway (Fulco et al., 2008). The requirement for NAD+ accumulation also explains why the activation of SIRT1 is not immediate upon resveratrol treatment. Therefore, SIRT1 may be essential for resveratrol action but as a downstream consequence of AMPK activation, rather than as a direct molecular target of resveratrol (Figure 5).
Figure 5. Resveratrol promotes mitochondrial biogenesis and lipid oxidation gene expression through indirect AMPK and SIRT1 activation.
While still a matter of debate, most data currently indicate that the metabolic actions of resveratrol or its metabolites stem from its ability to act as a mild mitochondrial poison, impairing ATP synthesis. The energy stress induced by resveratrol activates AMPK, subsequently stimulating SIRT1 by enhancing NAD+ levels. Then, SIRT1 activates key downstream targets through deacetylation (see Section 1.c), ultimately leading to an adaptative potentiation of mitochondrial biogenesis and lipid oxidation pathways. CI-V represent mitochondrial respiratory complexes I-V. The full names for other abbreviations can be found in the main text.
A more recent screening for other possible small molecular SIRT1 activators provided a second batch of compounds, among which SRT1720 has been best characterized (Milne et al., 2007). SRT1720 was in vitro a much more potent and efficient activator of SIRT1 than resveratrol (Milne et al., 2007). Treatment of rodents with SRT1720 prevented diet-induced obesity and ameliorated the diabetic phenotype of genetically obese mice (Feige et al., 2008; Milne et al., 2007). Similar to what was observed with resveratrol, SRT1720 enhanced SIRT1 activity, oxidative metabolism and mitochondrial biogenesis in mouse tissues (Feige et al., 2008). Recently, however, a new study also indicated that direct activation of SIRT1 by SRT1720 observed in the in vitro setting suffered from the same artifacts derived from the use of the “Fluor de Lys” moiety and that were already described for resveratrol (Pacholec et al., 2010). Therefore, one needs to consider that SIRT1 activation in tissues from mice fed with SRT1720 might be an indirect event, potentially again involving activation of AMPK, which was evident upon long-term treatment of SRT1720 in vivo (Feige et al., 2008). In addition to that, while the in vitro assays indicated that SRT1720 was a more potent activator of SIRT1 than resveratrol, this was not translated in vivo, indicating that the actions of these compounds in vivo involve indirect activation of SIRT1 (Feige et al., 2008) and/or rather poor bioavailability.
3.e Indirect modulation through affecting NAD+ metabolism
We have described in section 1b how sirtuins in general, and SIRT1 in particular meet all the requirements to act as intracellular NAD+ sensors. Therefore, enhancing NAD+ availability could be used to activate SIRT1 and promote beneficial health effects. We will now review different strategies used to achieve such goal and discuss whether the experimental results support that modulating NAD+ metabolism might be a useful tool for health benefits (Figure 6).
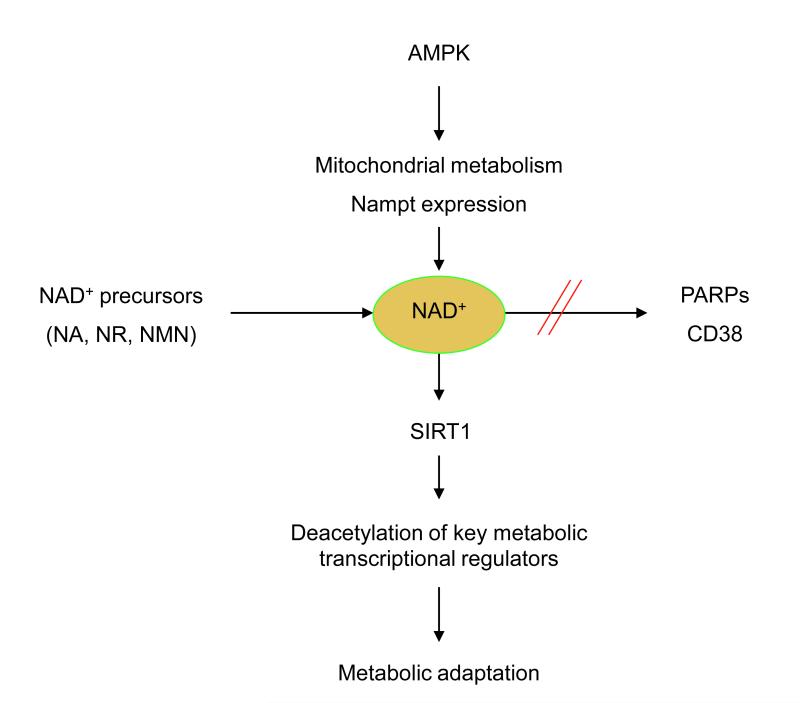
Figure 6. NAD+ as a nodal point for metabolic regulation.
Most evidence to date points out that NAD+ could be rate-limiting for the SIRT1 reaction in diverse conditions. SIRT1 activity would hence be stimulated by interventions which increase NAD+ levels, such as AMPK activation, enhancement of NAD+ biosynthesis through precursor (NA, NR, NMN) supplementation, or through inhibition of alternative NAD+ consuming activities, such as PARPs or CD38. The stimulation of SIRT1 activity by these distinct means improves the capacity of the cells/organism to adapt to external metabolic cues.
3.e.1 Modulation by increasing NAD+ synthesis
The most obvious strategy to test how enhancing NAD+ levels impacts on sirtuin activity consists in boosting NAD+ synthesis by supplementation with NAD+ precursors. Different precursors can be used to promote NAD+ synthesis. The primary de novo synthesis of NAD+ generally initiates from tryptophan (Houtkooper et al., 2010). Nicotinic acid (NA) is another NAD+ precursor that is transformed into NAD+ through the Preiss-Handler pathway, therefore converging with the same NAD+ synthesis pathway used by tryptophan. It is, however, assumed that perhaps the principle source of NAD+ comes from salvage pathways from other adenine nucleotide metabolites (Houtkooper et al., 2010). The main NAD+ precursors that funnel through the salvage pathways are nicotinamide (NAM) and, the more recently described, nicotinamide riboside (NR). NAM generates NAD+ through an independent pathway, in which the rate-limiting enzyme nicotinamide phosphorybosyltransferase (Nampt), transforms NAM into nicotinamide mononucleotide (NMN), which on its turn is converted into NAD+ by NMN adenylyltransferase (NMNAT) (Revollo et al., 2004). NR is phosphorylated upon its entry in the cell by the NR kinases (NRKs), generating NMN, which is then converted to NAD+ by NMNAT (Bieganowski and Brenner, 2004).
Few studies to date have described how supplementation with these precursors influence NAD+ levels and in particular affect sirtuin activity. Both NA and NAM can lead to higher NAD+ levels, even though the effects might be tissue-specific. NAM seems to be a more stable NAD+ precursor in the liver (Collins and Chaykin, 1972), but NA seems to be a more efficient in the kidney (Hara et al., 2007). The metabolism of these NAD+ precursors by the gut flora and intestinal enzymes can also contribute to their absorption and their efficacy to increase NAD+ (Gross and Henderson, 1983). Cell-based experiments also support the existence of cell-specific differences. For example, in NIH3T3 cells concentrations of up to 5 mM NAM were unable to increase intracellular NAD+ levels (Revollo et al., 2004). While NAM also was a relatively unefficient NAD+ precursor in HEK293 cells, requiring 5 mM concentrations to increase NAD+, NA was more efficient, and concentrations as low as 20 μM doubled the NAD+ content (Hara et al., 2007). The cell/tissue specific efficiencies of distinct NAD+ precursors might be consequent to the differential expression of the rate-limiting enzymes in their respective metabolic pathways. NA metabolism into NAD+ is rate-limited by the NA phosphoribosyltransferase (NAPT), which is highly enriched in some tissues, like liver or kidney (Hara et al., 2007). A reduction in NAPT activity in HEK293 cells does not affect basal NAD+ levels, but impairs NA-induced NAD+ accumulation (Hara et al., 2007). To date, however, no clear link between NA supplementation and sirtuin activity has been made.
Both NA and NAM have been used since a long time in a clinical setting. Niacin, basically composed of NA and NAM, has been widely used as an efficient way to overcome situations of dietary tryptophan deficits (Sauve, 2008). Niacin is also used to treat hypercholesterolemia, as it efficiently decreases VLDL synthesis, lowers LDL cholesterol levels, while concurrently increasing HDL cholesterol (Altschul et al., 1955; Karpe and Frayn, 2004). The use of niacin as a lipid-lowering agent results in beneficial effects on coronary artery disease and type 2 diabetes mellitus. It is not clear whether these effects rely on SIRT1 activation. Many of the beneficial actions of niacin in mice and humans - as well as some of the undesired effects, such as spontaneous flushing - have been attributed to the activation of a putative NA-activated G-coupled receptor, GPR109A (Benyo et al., 2005; Tunaru et al., 2003). Whereas the role of GPR109A in mediating niacin-induced flushing is rather well-established (Benyo et al., 2005), the hypothesis that the therapeutic efficacy of niacin is mediated by GPR109A activation needs to be revisited. First, even if GPR109A has a relatively high affinity for NA (EC50 ~100 nM), such levels of NA are rarely found in plasma unless pharmacologically primed (Kirkland, 2009), indicating that the activation of GRP109A by NA is probably fortuitous, but not biologically relevant in the basal state. Second, many of the beneficial effects of niacin, such as the lipid lowering effects (Kamanna and Kashyap, 2008), take place at concentrations higher than those required for GPR109A activation, but which lead to intracellular NAD+ accumulation (Jackson et al., 1995). It is therefore tempting to speculate that some of the effects promoted by niacin might be achieved through an NAD+-induced activation of SIRT1 and the consequent deacetylation of the multiple SIRT1 targets that act as critical regulators of fatty acid, lipid and sterol homeostasis in eukaryotes, such as PGC-1α, FOXOs, LXR or SREBP-1c (described in section 1.c). An additional appealing possibility to explain the effects of niacin involves the fact that it leads to adiponectin release from the white adipose tissue (Vaccari et al., 2007; Westphal et al., 2007). Adiponectin then activates AMP-activated protein kinase (AMPK) in muscle and liver tissues, which would enhance NAD+ content and SIRT1 activity (Canto et al., 2009; Iwabu et al., 2010). Therefore, future work is urgently needed to elucidate the contribution of SIRT1 as a potential mediator of niacin’s beneficial health effects.
The effects of NR, the most recently discovered NAD+ precursor, remain also largely unknown. While NR can increase intracellular NAD+ in mammalian cells (Yang et al., 2007b), it is still not reported whether this is enough to influence sirtuin activity. However, evidence obtained in yeast models suggests that this might in fact be the case. Supplementation of yeast with NR enhanced Sir2-dependent repression of recombination, improved gene silencing and extended replicative lifespan (Belenky et al., 2007). All these actions were completely dependent of NAD+ synthesis (Belenky et al., 2007). These experiments using NR constitute the first solid evidence that enhancing NAD+ bioavailability via NAD+ precursor supplementation can also enhance sirtuin activity and increase lifespan, even though they are limited to yeast models. Furthermore, the discovery that NR is present in milk (Bieganowski and Brenner, 2004) poses an interesting opportunity for food-based preventive or therapeutic interventions in NAD+-dependent metabolism.
In addition to boosting NAD+ levels through administration of NAD+ precursors, NAD+ levels can also be artificially modulated by changing the activity of rate-limiting enzymes in NAD+ biosynthesis. This principle is well-illustrated by the modulation of Nampt activity, which has a clear impact on NAD+ levels in virtually any mammalian cell tested (for examples, see (Fulco et al., 2008; Pittelli et al., 2010; Rongvaux et al., 2008; van der Veer et al., 2005)). In most of these cases, the alterations in NAD+ levels promoted by differential Nampt activity were associated changes in SIRT1 activity (Fulco et al., 2008; Revollo et al., 2007; van der Veer et al., 2007; van der Veer et al., 2005). However, and as explained in section 1.b, a complication from these experiments is that the influence that Nampt exerts on SIRT1 activity may derive not only from altering NAD+ availability, but also from more effective NAM clearance (Bitterman et al., 2002). Nampt inhibitors are currently being developed to deplete NAD+ levels and as such induce tumor cell apoptosis (Hasmann and Schemainda, 2003). Also inhibitors of kynurenine 3-monooxygenase (KMO), involved in de novo NAD+ biosynthesis pathways, have already been identified (Zwilling et al., 2011). Despite the facility to develop enzyme inhibitors, it will be more challenging to discover and develop compounds that pharmacologically activate the NAD+ salvage and/or de novo synthesis pathways. Potentially, such activators could enhance NAD+ levels and activate SIRT1 to promote metabolic fitness.
3.e.2 Modulation by decreasing NAD+ consumption
Another attractive way to modulate NAD+ levels and favour sirtuin activity resides in the modulation of the activity of non-sirtuin NAD+-consuming enzymes. There are two major families of alternative NAD+ consumers: the poly(ADP-ribosyl) polymerases (PARPs) and the cADP-ribose synthases (CD38 and CD157) (Houtkooper et al., 2010) (Figure 6).
3.e.2.a PARPs
PARPs use NAD+ as a substrate for a cellular process in which the ADP-ribose moiety is not transferred to an acetyl group, as happens with sirtuins, but to build ADP-ribosyl polymers onto acceptor proteins (Chambon et al., 1963; Krishnakumar and Kraus, 2010). PARP activity is robustly enhanced upon DNA damage and oxidative stress. Most PARP activity upon oxidative damage is driven by PARP-1, except for a residual 5-10%, which is accounted by PARP-2 (Ame et al., 1999; Shieh et al., 1998). Overactivation of PARP-1 upon oxidative damage rapidly depletes intracellular NAD+ levels (Goodwin et al., 1978; Skidmore et al., 1979). In line with the hypothesis that NAD+ might be rate-limiting for SIRT1 action, SIRT1 activity is downregulated in situations of PARP-1 activation (Bai et al., 2011b; Pillai et al., 2005). Significantly, SIRT1 does not seem to be directly regulated through PARylation events (Bai et al., 2011a; Bai et al., 2011b), indicating that PARP-1 and SIRT1 activity are connected through competition for a limited NAD+ pool. Importantly, PARP-1 has a very low Km (~20 μM) and a relatively high Vmax for NAD+ (Mendoza-Alvarez and Alvarez-Gonzalez, 1993), indicating that PARP-1 might limit SIRT1 action, but not the other way round.
We have recently demonstrated how downregulation of PARP-1 activity favours SIRT1 activation (Bai et al., 2011b). Genetic and pharmacological approaches that abrogate PARP-1 activity induced NAD+ levels. While PARP activity is generally accepted to be rather low in the basal state, recent evidence indicates that it naturally fluctuates in a circadian fashion (Asher et al., 2010). In line with the existence of basal PARP activity, inhibition of its activity gradually leads to a build up of NAD+ levels in cultured cells (Bai et al., 2011b). Similarly, tissues from PARP-1 knock-out mice have an increased NAD+ content (Bai et al., 2011b), roughly 2-fold higher than wild type littermates. The increase in NAD+ levels promoted by PARP inhibition leads, both in vivo and in vitro, to higher SIRT1 activity (Bai et al., 2011b). This way, PARP inhibition associates with the induction of the expression of genes involved in mitochondrial and lipid oxidation in a SIRT1-dependent manner. From a physiological perspective the better metabolic fitness derived from SIRT1 activation offers the PARP-1 null mice protection against the onset of metabolic disease in the context of diet-induced obesity (Bai et al., 2011b). More acute inhibition of PARP activity, using pharmacological PARP inhibitors, was also enough to enhance mitochondrial gene expression in vivo and in vitro (Bai et al., 2011b). Importantly, there are currently 9 different PARP inhibitors at different stages of clinical development, and at least 3 highly selective PARP inhibitors in late preclinical development against diverse types of cancer (see (Yuan et al., 2011b) for review). Interestingly, most cancer cells rely on anaerobic glycolysis to sustain their high proliferation rates, even if oxygen is plentiful, an event that is known as “the Warburg effect”, after Otto Warburg who first described it, and which is believed to be key for malignant transformation. Apart from their undisputed effect on DNA repair (Krishnakumar and Kraus, 2010), PARP inhibitors might also contribute to inhibit cancer progression by promoting oxidative metabolism and acting as anti-Warburg agents. As most PARP inhibitors target both PARP-1 and PARP-2, they would furthermore potentiate SIRT1 function both at the enzymatic level - through PARP-1 inhibition and raising NAD+ levels - and at the transcriptional level - through PARP-2 inhibition and induction of SIRT1 expression (see above section 3a). Furthermore, the results mentioned above also indicate that PARP inhibitors may be retooled to boost oxidative metabolism in metabolic disease where mitochondrial function is impaired, such as inherited and acquired mitochondrial diseases.
The interaction between PARP-1 and SIRT1 activities also open the perspective of a possible role of PARPs in ageing, as SIRT1 activity has been postulated as a mediator of the beneficial effects of calorie restriction on health- and life-span (Canto and Auwerx, 2011). Hence, PARP inhibition could be a nice strategy to activate SIRT1 and mimic the calorie-restricted state. In line with this hypothesis, it was recently reported that PARP activity is higher in aged tissues, leading to decreased SIRT1 activity (Braidy et al., 2011) (Canto C, Houtkooper R, Mouchiroud L and Auwerx J, unpublished observations). Confirming the hypothesis that higher PARP-1 activity might be detrimental for SIRT1 function and global metabolism was the fact that mice expressing and additional copy of the human PARP-1, have reduced median lifespan, impaired glucose homeostasis and higher susceptibility to age-related diseases (Mangerich et al., 2010). Further studies should evaluate the possible influence of PARP activity on aging.
Interestingly, the NAD+ boosting effects of PARP inhibition enhances the activity of SIRT1, but not that of SIRT2 or SIRT3 (Bai et al., 2011b). The major difference between these three sirtuins is their subcellular localization, as, amongst them, only SIRT1 is a nuclear sirtuin. This suggests the existence of compartment-specific NAD+ pools in the cell. Supporting this possibility, elegant studies by the Sinclair lab showed the existence of independently regulated NAD+ pools (Yang et al., 2007a).
3.e.2.b CD38
Another small family of NAD+ consumers are the cADP-Ribose Synthases, CD38 and the less characterized CD157. CD38 and CD157 are multifunctional enzymes that use NAD+ as substrate to generate second messengers, mainly cADP-ribose, which, in turn, regulates calcium mobilization (Lee, 2006). Most studies on how cADP-ribose synthases impact on NAD+ levels and SIRT1 activity have been done with CD38. As true for PARP-1, CD38 displays a very low Km for NAD+ (15-25 μM) (Cakir-Kiefer et al., 2001). Importantly, the stoichiometry of the reaction catalyzed by CD38 involves massive amounts of NAD+, around 100 molecules, to yield a single cADP-ribose (Dousa et al., 1996). Therefore, even low levels of CD38 activity might have a dramatic influence on intracellular NAD+ metabolism. Remarkably, the enzymatic activity of CD38 is located outside the plasma membrane (De Flora et al., 1997), which makes it challenging to envision how NAD+ can access this enzyme, as NAD+ is only present in minute quantities outside the cell (O’Reilly and Niven, 2003). It was recently, however, suggested that CD38 might also be present in the nuclear compartment, which would then set the stage for a role of CD38 as a main intracellular NAD+ consumer (Aksoy et al., 2006). The function of CD38 as an intracellular NADase was subsequently proven right when mice lacking CD38 displayed a 30-fold increase in intracellular NAD+ levels (Aksoy et al., 2006b). This increase in NAD+ levels is far superior compared to the ~2-fold increases generally observed in most genetic (PARP-1 deletion), pharmacological (NAD+ precursors) or physiological interventions (fasting, calorie restriction) that enhance NAD+ content. The increase in intracellular NAD+ elicited by CD38 deletion significantly activated SIRT1 and prompted a similar clinical phenotypes to those expected for SIRT1 activation, including protection against diet-induced obesity and a robust deacetylation of SIRT1 targets (Aksoy et al., 2006). The fact that the 30-fold increase in NAD+ triggered by CD38 gene deletion has a similar biological effect than the 2-fold effect observed in PARP-1 knockout mice suggests that the effective NAD+-sensing range of SIRT1 might be largely exceeded in the CD38 knockout mice. Given the nuclear and plasma membrane localization of CD38, it will be interesting to evaluate whether the increase in NAD+ is homogeneous between different cellular compartments and whether other sirtuins are similarly affected. As for the PARPs, specific CD38 inhibitors are being developed (Dong et al., 2011; Sauve and Schramm, 2002), and it will be relevant to evaluate how they influence SIRT1 activity in situations where oxidative metabolism is impaired (e.g. acquired and inherited mitochondrial defects).
4. CONCLUSIONS AND FUTURE DIRECTIONS
The wealth of new data on SIRT1, generated in species beyond yeast, has recently spontaneously refocused the attention of the field from a potential - but so far not clearly proven - role in increasing lifespan towards its ability to modulate whole body metabolism. Probably evolved as a mediator of the metabolic and transcriptional adaptations to situations of energy stress and nutrient deprivation, SIRT1 activation enhances the ability of organisms to enhance fat consumption and use mitochondrial respiration as a way optimize energy harvesting. Metabolic disease has been strongly linked to impaired energy homeostasis and mitochondrial function. Therefore, manipulations aimed to enhance SIRT1 activity might turn out to be attractive for the prevention and treatment of metabolic disease. Studies in genetic engineered mouse models support the notion that higher SIRT1 activity is protective against metabolic disease without necessarily influencing lifespan. In contrast, defective SIRT1 activity in mouse models is generally linked to metabolic inefficiency. While the quest for direct SIRT1 activators has delivered interesting compounds, such as resveratrol and SRT1720, most of them have been proven to activate SIRT1 in very indirect and unspecific fashion in vivo. Therefore, novel approaches to activate SIRT1 have emerged. Amongst them, strategies aimed to modify the intracellular NAD+ content have provided exquisite correlative evidence that higher NAD+ bioavailability is matched by higher SIRT1 activity. Importantly, the activation of SIRT1 seems to be a key convergent downstream effect, common between the many different approaches used to modulate NAD+ levels, such as increasing NAD+ synthesis rates or decreasing NAD+ consumption rates. This way, the modulation of NAD+ levels appears as one of the most promising strategies to control SIRT1 activity in order to achieve health benefits. One of the advantages of raising NAD+ levels is that that it can be achieved from many different angles and both strategies that boost NAD+ levels and inhibit NAD+ consumption are being pursued for nutraceutical (e.g. NA precursors) and pharmaceutical (e.g. PARP and CD38 inhibitors) development. It should furthermore not be forgotten that despite the fact that raising NAD+ levels might allow SIRT1 to reach higher enzymatic rates, other factors, ranging from transcriptional, translational to post-translational (e.g post-translational modifications strictu sensu and protein complex formation) mechanisms, also profoundly affect SIRT1 activity. In our review, we have highlighted how different post-translational modification in either SIRT1 on in its substrates can strongly drive SIRT1 activity towards specific cellular processes. This indicates that the activity of sirtuins on certain substrates might not only be coupled to NAD+ availability, but also to the accessibility of the target, which is determined by another post-translational modification. This case is perfectly illustrated by PGC-1α, whose deacetylation is much more efficient when phosphorylated. Similarly, it will be very relevant to understand how the ability of SIRT1 to interact with specific substrates is determined by the cohabitation with other molecular coactivator (e.g. PGC-1α) or corepressor (e.g. NCoR1) complexes. The complexity of SIRT1 physiology, which is largely conserved throughout evolution, makes unravelling the intricacy of the SIRT1 signalling and putting it to fruition in drug development, a challenging task that will occupy scientists for years to come. Therefore, those who are waiting for a SIRT1 activating entity that will grace them with eternal youth, will have to wait a little longer.
ACKNOWLEDGEMENTS
The work in the laboratory of the authors was supported by grants of the Ecole Polytechnique Fédérale de Lausanne, Swiss National Science Foundation, the European Research Council Ideas programme (Sirtuins; ERC-2008-AdG23118), and the Velux foundation. The authors thank all the members of the Auwerx lab for inspiring discussions and Beatrice Desvergne for critical reading the manuscript.
Footnotes
The authors declare no conflict of interests.
REFERENCES
- Aksoy P, Escande C, White TA, Thompson M, Soares S, Benech JC, Chini EN. Regulation of SIRT 1 mediated NAD dependent deacetylation: a novel role for the multifunctional enzyme CD38. Biochem Biophys Res Commun. 2006;349(1):353–9. doi: 10.1016/j.bbrc.2006.08.066. [DOI] [PubMed] [Google Scholar]
- Altarejos JY, Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol. 2011;12(3):141–51. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul R, Hoffer A, Stephen JD. Influence of nicotinic acid on serum cholesterol in man. Arch Biochem Biophys. 1955;54(2):558–9. doi: 10.1016/0003-9861(55)90070-9. [DOI] [PubMed] [Google Scholar]
- Ame JC, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, Muller S, Hoger T, Menissier-de Murcia J, de Murcia G. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J Biol Chem. 1999;274(25):17860–8. doi: 10.1074/jbc.274.25.17860. [DOI] [PubMed] [Google Scholar]
- Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423(6936):181–5. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305(5686):1010–3. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134(2):317–28. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- Asher G, Reinke H, Altmeyer M, Gutierrez-Arcelus M, Hottiger MO, Schibler U. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell. 2010;142(6):943–53. doi: 10.1016/j.cell.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Bai P, Canto C, Brunyanszki A, Huber A, Szanto M, Cen Y, Yamamoto H, Houten SM, Kiss B, Oudart H, Gergely P, Menissier-de Murcia J, Schreiber V, Sauve AA, Auwerx J. PARP-2 regulates SIRT1 expression and whole-body energy expenditure. Cell Metab. 2011a;13(4):450–60. doi: 10.1016/j.cmet.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai P, Canto C, Oudart H, Brunyanszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, Schoonjans K, Schreiber V, Sauve AA, Menissier-de Murcia J, Auwerx J. PARP-1 Inhibition Increases Mitochondrial Metabolism through SIRT1 Activation. Cell Metab. 2011b;13(4):461–8. doi: 10.1016/j.cmet.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai P, Houten SM, Huber A, Schreiber V, Watanabe M, Kiss B, de Murcia G, Auwerx J, Menissier-de Murcia J. Poly(ADP-ribose) polymerase-2 [corrected] controls adipocyte differentiation and adipose tissue function through the regulation of the activity of the retinoid X receptor/peroxisome proliferator-activated receptor-gamma [corrected] heterodimer. J Biol Chem. 2007;282(52):37738–46. doi: 10.1074/jbc.M701021200. [DOI] [PubMed] [Google Scholar]
- Banks AS, Kon N, Knight C, Matsumoto M, Gutierrez-Juarez R, Rossetti L, Gu W, Accili D. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 2008;8(4):333–41. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barger JL, Kayo T, Vann JM, Arias EB, Wang J, Hacker TA, Wang Y, Raederstorff D, Morrow JD, Leeuwenburgh C, Allison DB, Saupe KW, Cartee GD, Weindruch R, Prolla TA. A low dose of dietary resveratrol partially mimics caloric restriction and retards aging parameters in mice. PLoS ONE. 2008;3(6):e2264. doi: 10.1371/journal.pone.0002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer JH, Morris SN, Chang C, Flatt T, Wood JG, Helfand SL. dSir2 and Dmp53 interact to mediate aspects of CR-dependent lifespan extension in D. melanogaster. Aging (Albany NY) 2009;1(1):38–48. doi: 10.18632/aging.100001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur JA. Resveratrol, sirtuins, and the promise of a DR mimetic. Mech Ageing Dev. 2010;131(4):261–9. doi: 10.1016/j.mad.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444(7117):337–42. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci U S A. 2004;101(33):12130–5. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenky P, Racette FG, Bogan KL, McClure JM, Smith JS, Brenner C. Nicotinamide riboside promotes Sir2 silencing and extends lifespan via Nrk and Urh1/Pnp1/Meu1 pathways to NAD+ Cell. 2007;129(3):473–84. doi: 10.1016/j.cell.2007.03.024. [DOI] [PubMed] [Google Scholar]
- Benyo Z, Gille A, Kero J, Csiky M, Suchankova MC, Nusing RM, Moers A, Pfeffer K, Offermanns S. GPR109A (PUMA-G/HM74A) mediates nicotinic acid-induced flushing. J Clin Invest. 2005;115(12):3634–40. doi: 10.1172/JCI23626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdichevsky A, Viswanathan M, Horvitz HR, Guarente L. C. elegans SIR-2.1 interacts with 14-3-3 proteins to activate DAF-16 and extend life span. Cell. 2006;125(6):1165–77. doi: 10.1016/j.cell.2006.04.036. [DOI] [PubMed] [Google Scholar]
- Bieganowski P, Brenner C. Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell. 2004;117(4):495–502. doi: 10.1016/s0092-8674(04)00416-7. [DOI] [PubMed] [Google Scholar]
- Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277(47):45099–107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- Boily G, Seifert EL, Bevilacqua L, He XH, Sabourin G, Estey C, Moffat C, Crawford S, Saliba S, Jardine K, Xuan J, Evans M, Harper ME, McBurney MW. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS One. 2008;3(3):e1759. doi: 10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordone L, Cohen D, Robinson A, Motta MC, van Veen E, Czopik A, Steele AD, Crowe H, Marmor S, Luo J, Gu W, Guarente L. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007;6(6):759–67. doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- Bordone L, Motta MC, Picard F, Robinson A, Jhala US, Apfeld J, McDonagh T, Lemieux M, McBurney M, Szilvasi A, Easlon EJ, Lin SJ, Guarente L. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 2006;4(2):e31. doi: 10.1371/journal.pbio.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borra MT, Smith BC, Denu JM. Mechanism of human SIRT1 activation by resveratrol. J Biol Chem. 2005;280(17):17187–95. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- Bouras T, Fu M, Sauve AA, Wang F, Quong AA, Perkins ND, Hay RT, Gu W, Pestell RG. SIRT1 deacetylation and repression of p300 involves lysine residues 1020/1024 within the cell cycle regulatory domain 1. J Biol Chem. 2005;280(11):10264–76. doi: 10.1074/jbc.M408748200. [DOI] [PubMed] [Google Scholar]
- Braidy N, Guillemin GJ, Mansour H, Chan-Ling T, Poljak A, Grant R. Age related changes in NAD+ metabolism oxidative stress and sirt1 activity in wistar rats. PLoS One. 2011;6(4):e19194. doi: 10.1371/journal.pone.0019194. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303(5666):2011–5. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Burnett C, Valentini S, Cabreiro F, Goss M, Somogyvari M, Piper MD, Hoddinott M, Sutphin GL, Leko V, McElwee JJ, Vazquez-Manrique RP, Orfila AM, Ackerman D, Au C, Vinti G, Riesen M, Howard K, Neri C, Bedalov A, Kaeberlein M, Soti C, Partridge L, Gems D. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477(7365):482–5. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakir-Kiefer C, Muller-Steffner H, Oppenheimer N, Schuber F. Kinetic competence of the cADP-ribose-CD38 complex as an intermediate in the CD38/NAD+ glycohydrolase-catalysed reactions: implication for CD38 signalling. Biochem J. 2001;358(Pt 2):399–406. doi: 10.1042/0264-6021:3580399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Auwerx J. Caloric restriction, SIRT1 and longevity. Trends Endocrinol Metab. 2009;20(7):325–31. doi: 10.1016/j.tem.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Auwerx J. AMP-activated protein kinase and its downstream transcriptional pathways. Cell Mol Life Sci. 2010;67(20):3407–23. doi: 10.1007/s00018-010-0454-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Auwerx J. Interference between PARPs and SIRT1: a novel approach to healthy ageing? Aging (Albany NY) 2011 doi: 10.18632/aging.100326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458(7241):1056–60. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Jiang LQ, Deshmukh AS, Mataki C, Coste A, Lagouge M, Zierath JR, Auwerx J. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab. 2010;11(3):213–9. doi: 10.1016/j.cmet.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen DE, Supinski AM, Bonkowski MS, Donmez G, Guarente LP. Neuronal SIRT1 regulates endocrine and behavioral responses to calorie restriction. Genes Dev. 2009;23(24):2812–7. doi: 10.1101/gad.1839209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins PB, Chaykin S. The management of nicotinamide and nicotinic acid in the mouse. J Biol Chem. 1972;247(3):778–83. [PubMed] [Google Scholar]
- Costa Cdos S, Hammes TO, Rohden F, Margis R, Bortolotto JW, Padoin AV, Mottin CC, Guaragna RM. SIRT1 transcription is decreased in visceral adipose tissue of morbidly obese patients with severe hepatic steatosis. Obes Surg. 2010;20(5):633–9. doi: 10.1007/s11695-009-0052-z. [DOI] [PubMed] [Google Scholar]
- Coste A, Louet JF, Lagouge M, Lerin C, Antal MC, Meziane H, Schoonjans K, Puigserver P, O’Malley BW, Auwerx J. The genetic ablation of SRC-3 protects against obesity and improves insulin sensitivity by reducing the acetylation of PGC-1{alpha} Proc Natl Acad Sci U S A. 2008;105(44):17187–92. doi: 10.1073/pnas.0808207105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costford SR, Bajpeyi S, Pasarica M, Albarado DC, Thomas SC, Xie H, Church TS, Jubrias SA, Conley KE, Smith SR. Skeletal muscle NAMPT is induced by exercise in humans. Am J Physiol Endocrinol Metab. 2010;298(1):E117–26. doi: 10.1152/ajpendo.00318.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambon P, Weill JD, Mandel P. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem Biophys Res Commun. 1963;11:39–43. doi: 10.1016/0006-291x(63)90024-x. [DOI] [PubMed] [Google Scholar]
- Chau MD, Gao J, Yang Q, Wu Z, Gromada J. Fibroblast growth factor 21 regulates energy metabolism by activating the AMPK-SIRT1-PGC-1alpha pathway. Proc Natl Acad Sci U S A. 2010;107(28):12553–8. doi: 10.1073/pnas.1006962107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Bruno J, Easlon E, Lin SJ, Cheng HL, Alt FW, Guarente L. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008;22(13):1753–7. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhou Y, Mueller-Steiner S, Chen LF, Kwon H, Yi S, Mucke L, Gan L. SIRT1 protects against microglia-dependent amyloid-beta toxicity through inhibiting NF-kappaB signaling. J Biol Chem. 2005a;280(48):40364–74. doi: 10.1074/jbc.M509329200. [DOI] [PubMed] [Google Scholar]
- Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005b;123(3):437–48. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Dali-Youcef N, Lagouge M, Froelich S, Koehl C, Schoonjans K, Auwerx J. Sirtuins: the ‘magnificent seven’, function, metabolism and longevity. Ann Med. 2007;39(5):335–45. doi: 10.1080/07853890701408194. [DOI] [PubMed] [Google Scholar]
- Dasgupta B, Milbrandt J. Resveratrol stimulates AMP kinase activity in neurons. Proc Natl Acad Sci U S A. 2007;104(17):7217–22. doi: 10.1073/pnas.0610068104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Flora A, Guida L, Franco L, Zocchi E. The CD38/cyclic ADP-ribose system: a topological paradox. Int J Biochem Cell Biol. 1997;29(10):1149–66. doi: 10.1016/s1357-2725(97)00062-9. [DOI] [PubMed] [Google Scholar]
- Defossez PA, Prusty R, Kaeberlein M, Lin SJ, Ferrigno P, Silver PA, Keil RL, Guarente L. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol Cell. 1999;3(4):447–55. doi: 10.1016/s1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- Dong M, Si YQ, Sun SY, Pu XP, Yang ZJ, Zhang LR, Zhang LH, Leung FP, Lam CM, Kwong AK, Yue J, Zhou Y, Kriksunov IA, Hao Q, Lee HC. Design, synthesis and biological characterization of novel inhibitors of CD38. Org Biomol Chem. 2011;9(9):3246–57. doi: 10.1039/c0ob00768d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donmez G, Wang D, Cohen DE, Guarente L. SIRT1 suppresses beta-amyloid production by activating the alpha-secretase gene ADAM10. Cell. 2010;142(2):320–32. doi: 10.1016/j.cell.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dousa TP, Chini EN, Beers KW. Adenine nucleotide diphosphates: emerging second messengers acting via intracellular Ca2+ release. Am J Physiol. 1996;271(4 Pt 1):C1007–24. doi: 10.1152/ajpcell.1996.271.4.C1007. [DOI] [PubMed] [Google Scholar]
- Du J, Xhou Y, Su X, Yu J, Khan S, Jian H, Kim J, Woo J, Kim JH, Choi BH, He B, Chen W, Zhang S, Cerione RA, Auwerx J, Hao Q, Lin H. Sirt5 is an NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011 doi: 10.1126/science.1207861. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escande C, Chini CC, Nin V, Dykhouse KM, Novak CM, Levine J, van Deursen J, Gores GJ, Chen J, Lou Z, Chini EN. Deleted in breast cancer-1 regulates SIRT1 activity and contributes to high-fat diet-induced liver steatosis in mice. J Clin Invest. 2010;120(2):545–58. doi: 10.1172/JCI39319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige JN, Lagouge M, Canto C, Strehle A, Houten SM, Milne JC, Lambert PD, Mataki C, Elliott PJ, Auwerx J. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8(5):347–58. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460(7255):587–91. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA, Sartorelli V. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell. 2008;14(5):661–73. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Wang WY, Mao YW, Graff J, Guan JS, Pan L, Mak G, Kim D, Su SC, Tsai LH. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466(7310):1105–9. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26(7):1913–23. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giandomenico V, Simonsson M, Gronroos E, Ericsson J. Coactivator-dependent acetylation stabilizes members of the SREBP family of transcription factors. Mol Cell Biol. 2003;23(7):2587–99. doi: 10.1128/MCB.23.7.2587-2599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin PM, Lewis PJ, Davies MI, Skidmore CJ, Shall S. The effect of gamma radiation and neocarzinostatin on NAD and ATP levels in mouse leukaemia cells. Biochim Biophys Acta. 1978;543(4):576–82. doi: 10.1016/0304-4165(78)90312-4. [DOI] [PubMed] [Google Scholar]
- Greer EL, Brunet A. FOXO transcription factors in ageing and cancer. Acta Physiol (Oxf) 2008;192(1):19–28. doi: 10.1111/j.1748-1716.2007.01780.x. [DOI] [PubMed] [Google Scholar]
- Greer EL, Oskoui PR, Banko MR, Maniar JM, Gygi MP, Gygi SP, Brunet A. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem. 2007;282(41):30107–19. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- Gross CJ, Henderson LM. Digestion and absorption of NAD by the small intestine of the rat. J Nutr. 1983;113(2):412–20. doi: 10.1093/jn/113.2.412. [DOI] [PubMed] [Google Scholar]
- Gross DN, van den Heuvel AP, Birnbaum MJ. The role of FoxO in the regulation of metabolism. Oncogene. 2008;27(16):2320–36. doi: 10.1038/onc.2008.25. [DOI] [PubMed] [Google Scholar]
- Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 2000;14(9):1021–6. [PubMed] [Google Scholar]
- Guarente L. Franklin H. Epstein Lecture: Sirtuins, aging, and medicine. N Engl J Med. 2011;364(23):2235–44. doi: 10.1056/NEJMra1100831. [DOI] [PubMed] [Google Scholar]
- Guo X, Williams JG, Schug TT, Li X. DYRK1A and DYRK3 promote cell survival through phosphorylation and activation of SIRT1. J Biol Chem. 2010;285(17):13223–32. doi: 10.1074/jbc.M110.102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126(5):941–54. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A. 2006;103(27):10230–5. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Zhou R, Niu J, McNutt MA, Wang P, Tong T. SIRT1 is regulated by a PPAR{gamma}-SIRT1 negative feedback loop associated with senescence. Nucleic Acids Res. 2010;38(21):7458–71. doi: 10.1093/nar/gkq609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara N, Yamada K, Shibata T, Osago H, Hashimoto T, Tsuchiya M. Elevation of cellular NAD levels by nicotinic acid and involvement of nicotinic acid phosphoribosyltransferase in human cells. J Biol Chem. 2007;282(34):24574–82. doi: 10.1074/jbc.M610357200. [DOI] [PubMed] [Google Scholar]
- Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8(10):774–85. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- Hariharan N, Maejima Y, Nakae J, Paik J, Depinho RA, Sadoshima J. Deacetylation of FoxO by Sirt1 Plays an Essential Role in Mediating Starvation-Induced Autophagy in Cardiac Myocytes. Circ Res. 2010;107(12):1470–82. doi: 10.1161/CIRCRESAHA.110.227371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasmann M, Schemainda I. FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res. 2003;63(21):7436–42. [PubMed] [Google Scholar]
- Hawley SA, Ross FA, Chevtzoff C, Green KA, Evans A, Fogarty S, Towler MC, Brown LJ, Ogunbayo OA, Evans AM, Hardie DG. Use of cells expressing gamma subunit variants to identify diverse mechanisms of AMPK activation. Cell Metab. 2010;11(6):554–65. doi: 10.1016/j.cmet.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashida S, Arimoto A, Kuramoto Y, Kozako T, Honda S, Shimeno H, Soeda S. Fasting promotes the expression of SIRT1, an NAD+ -dependent protein deacetylase, via activation of PPARalpha in mice. Mol Cell Biochem. 2010;339(1-2):285–92. doi: 10.1007/s11010-010-0391-z. [DOI] [PubMed] [Google Scholar]
- Heikkinen S, Auwerx J, Argmann CA. PPARgamma in human and mouse physiology. Biochim Biophys Acta. 2007;1771(8):999–1013. doi: 10.1016/j.bbalip.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz D, Munoz-Martin M, Canamero M, Mulero F, Martinez-Pastor B, Fernandez-Capetillo O, Serrano M. Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nat Commun. 2010a;1:3. doi: 10.1038/ncomms1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz D, Munoz-Martin M, Canamero M, Mulero F, Martinez-Pastor B, Fernandez-Capetillo O, Serrano M. Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nat Commun. 2010b;1(1):1–8. doi: 10.1038/ncomms1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz D, Serrano M. SIRT1: recent lessons from mouse models. Nat Rev Cancer. 2010;10(12):819–23. doi: 10.1038/nrc2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, Spiegelman B, Montminy M. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413(6852):179–83. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- Herzog B, Hall RK, Wang XL, Waltner-Law M, Granner DK. Peroxisome proliferator-activated receptor gamma coactivator-1alpha, as a transcription amplifier, is not essential for basal and hormone-induced phosphoenolpyruvate carboxykinase gene expression. Mol Endocrinol. 2004;18(4):807–19. doi: 10.1210/me.2003-0384. [DOI] [PubMed] [Google Scholar]
- Hirosumi J, Tuncman G, Chang L, Gorgun CZ, Uysal KT, Maeda K, Karin M, Hotamisligil GS. A central role for JNK in obesity and insulin resistance. Nature. 2002;420(6913):333–6. doi: 10.1038/nature01137. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Canto C, Wanders RJ, Auwerx J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev. 2010;31(2):194–223. doi: 10.1210/er.2009-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425(6954):191–6. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403(6771):795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Ivy JM, Klar AJ, Hicks JB. Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol Cell Biol. 1986;6(2):688–702. doi: 10.1128/mcb.6.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabu M, Yamauchi T, Okada-Iwabu M, Sato K, Nakagawa T, Funata M, Yamaguchi M, Namiki S, Nakayama R, Tabata M, Ogata H, Kubota N, Takamoto I, Hayashi YK, Yamauchi N, Waki H, Fukayama M, Nishino I, Tokuyama K, Ueki K, Oike Y, Ishii S, Hirose K, Shimizu T, Touhara K, Kadowaki T. Adiponectin and AdipoR1 regulate PGC-1alpha and mitochondria by Ca(2+) and AMPK/SIRT1. Nature. 2010;464(7293):1313–9. doi: 10.1038/nature08991. [DOI] [PubMed] [Google Scholar]
- Jackson TM, Rawling JM, Roebuck BD, Kirkland JB. Large supplements of nicotinic acid and nicotinamide increase tissue NAD+ and poly(ADP-ribose) levels but do not affect diethylnitrosamine-induced altered hepatic foci in Fischer-344 rats. J Nutr. 1995;125(6):1455–61. doi: 10.1093/jn/125.6.1455. [DOI] [PubMed] [Google Scholar]
- Jager S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci U S A. 2007;104(29):12017–22. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing E, Gesta S, Kahn CR. SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metab. 2007;6(2):105–14. doi: 10.1016/j.cmet.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, McDonagh T, Heltweg B, Hixon J, Westman EA, Caldwell SD, Napper A, Curtis R, DiStefano PS, Fields S, Bedalov A, Kennedy BK. Substrate-specific activation of sirtuins by resveratrol. J Biol Chem. 2005;280(17):17038–45. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13(19):2570–80. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW., 3rd Sir2 and calorie restriction in yeast: a skeptical perspective. Ageing Res Rev. 2007;6(2):128–40. doi: 10.1016/j.arr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Kalaany NY, Mangelsdorf DJ. LXRS and FXR: the yin and yang of cholesterol and fat metabolism. Annu Rev Physiol. 2006;68:159–91. doi: 10.1146/annurev.physiol.68.033104.152158. [DOI] [PubMed] [Google Scholar]
- Kamanna VS, Kashyap ML. Mechanism of action of niacin. Am J Cardiol. 2008;101(8A):20B–26B. doi: 10.1016/j.amjcard.2008.02.029. [DOI] [PubMed] [Google Scholar]
- Kang H, Jung JW, Kim MK, Chung JH. CK2 is the regulator of SIRT1 substrate-binding affinity, deacetylase activity and cellular response to DNA-damage. PLoS One. 2009;4(8):e6611. doi: 10.1371/journal.pone.0006611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur S, Bedard S, Marcotte B, Cote CH, Marette A. Expression of nitric oxide synthase in skeletal muscle: a novel role for nitric oxide as a modulator of insulin action. Diabetes. 1997;46(11):1691–700. doi: 10.2337/diab.46.11.1691. [DOI] [PubMed] [Google Scholar]
- Karpe F, Frayn KN. The nicotinic acid receptor--a new mechanism for an old drug. Lancet. 2004;363(9424):1892–4. doi: 10.1016/S0140-6736(04)16359-9. [DOI] [PubMed] [Google Scholar]
- Kim D, Nguyen MD, Dobbin MM, Fischer A, Sananbenesi F, Rodgers JT, Delalle I, Baur JA, Sui G, Armour SM, Puigserver P, Sinclair DA, Tsai LH. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer’s disease and amyotrophic lateral sclerosis. EMBO J. 2007a;26(13):3169–79. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EJ, Kho JH, Kang MR, Um SJ. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol Cell. 2007b;28(2):277–90. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451(7178):583–6. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- Kirkland JB. Niacin status, NAD distribution and ADP-ribose metabolism. Curr Pharm Des. 2009;15(1):3–11. doi: 10.2174/138161209787185823. [DOI] [PubMed] [Google Scholar]
- Koo SH, Flechner L, Qi L, Zhang X, Screaton RA, Jeffries S, Hedrick S, Xu W, Boussouar F, Brindle P, Takemori H, Montminy M. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature. 2005;437(7062):1109–11. doi: 10.1038/nature03967. [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39(1):8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127(6):1109–22. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Le Gouill E, Jimenez M, Binnert C, Jayet PY, Thalmann S, Nicod P, Scherrer U, Vollenweider P. Endothelial nitric oxide synthase (eNOS) knockout mice have defective mitochondrial beta-oxidation. Diabetes. 2007;56(11):2690–6. doi: 10.2337/db06-1228. [DOI] [PubMed] [Google Scholar]
- Lee HC. Structure and enzymatic functions of human CD38. Mol Med. 2006;12(11-12):317–23. doi: 10.2119/2006-00086.Lee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008;105(9):3374–9. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Padhye A, Sharma A, Song G, Miao J, Mo YY, Wang L, Kemper JK. A pathway involving farnesoid X receptor and small heterodimer partner positively regulates hepatic sirtuin 1 levels via microRNA-34a inhibition. J Biol Chem. 2010;285(17):12604–11. doi: 10.1074/jbc.M109.094524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell Metab. 2006;3(6):429–38. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Li J, Hu X, Selvakumar P, Russell RR, 3rd, Cushman SW, Holman GD, Young LH. Role of the nitric oxide pathway in AMPK-mediated glucose uptake and GLUT4 translocation in heart muscle. Am J Physiol Endocrinol Metab. 2004;287(5):E834–41. doi: 10.1152/ajpendo.00234.2004. [DOI] [PubMed] [Google Scholar]
- Li L, Pan R, Li R, Niemann B, Aurich AC, Chen Y, Rohrbach S. Mitochondrial biogenesis and PGC-1{alpha} deacetylation by physical activity: intact adipocytokine-signaling is required. Diabetes. 2010 doi: 10.2337/db10-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang S, Blander G, Tse JG, Krieger M, Guarente L. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol Cell. 2007;28(1):91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18(1):12–6. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;280(22):21313–20. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, Milne J, Meyers DJ, Cole P, Yates J, 3rd, Olefsky J, Guarente L, Montminy M. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456(7219):269–73. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard DB, Pletcher SD, Canto C, Auwerx J. Ageing: longevity hits a roadblock. Nature. 2011;477(7365):410–1. doi: 10.1038/477410a. [DOI] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107(2):137–48. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Mair W, Morantte I, Rodrigues AP, Manning G, Montminy M, Shaw RJ, Dillin A. Lifespan extension induced by AMPK and calcineurin is mediated by CRTC-1 and CREB. Nature. 2011;470(7334):404–8. doi: 10.1038/nature09706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangerich A, Herbach N, Hanf B, Fischbach A, Popp O, Moreno-Villanueva M, Bruns OT, Burkle A. Inflammatory and age-related pathologies in mice with ectopic expression of human PARP-1. Mech Ageing Dev. 2010;131(6):389–404. doi: 10.1016/j.mad.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. p53 regulates mitochondrial respiration. Science. 2006;312(5780):1650–3. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- Mattagajasingh I, Kim CS, Naqvi A, Yamamori T, Hoffman TA, Jung SB, DeRicco J, Kasuno K, Irani K. SIRT1 promotes endothelium-dependent vascular relaxation by activating endothelial nitric oxide synthase. Proc Natl Acad Sci U S A. 2007;104(37):14855–60. doi: 10.1073/pnas.0704329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBurney MW, Yang X, Jardine K, Hixon M, Boekelheide K, Webb JR, Lansdorp PM, Lemieux M. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol. 2003;23(1):38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Alvarez H, Alvarez-Gonzalez R. Poly(ADP-ribose) polymerase is a catalytic dimer and the automodification reaction is intermolecular. J Biol Chem. 1993;268(30):22575–80. [PubMed] [Google Scholar]
- Michan S, Li Y, Chou MM, Parrella E, Ge H, Long JM, Allard JS, Lewis K, Miller M, Xu W, Mervis RF, Chen J, Guerin KI, Smith LE, McBurney MW, Sinclair DA, Baudry M, de Cabo R, Longo VD. SIRT1 is essential for normal cognitive function and synaptic plasticity. J Neurosci. 2010;30(29):9695–707. doi: 10.1523/JNEUROSCI.0027-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404(1):1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452(7186):492–6. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16(10):4623–35. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Jin L, Boss O, Perni RB, Vu CB, Bemis JE, Xie R, Disch JS, Ng PY, Nunes JJ, Lynch AV, Yang H, Galonek H, Israelian K, Choy W, Iffland A, Lavu S, Medvedik O, Sinclair DA, Olefsky JM, Jirousek MR, Elliott PJ, Westphal CH. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450(7170):712–6. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116(4):551–63. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- Moynihan KA, Grimm AA, Plueger MM, Bernal-Mizrachi E, Ford E, Cras-Meneur C, Permutt MA, Imai S. Increased dosage of mammalian Sir2 in pancreatic beta cells enhances glucose-stimulated insulin secretion in mice. Cell Metab. 2005;2(2):105–17. doi: 10.1016/j.cmet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Murray-Zmijewski F, Slee EA, Lu X. A complex barcode underlies the heterogeneous response of p53 to stress. Nat Rev Mol Cell Biol. 2008;9(9):702–12. doi: 10.1038/nrm2451. [DOI] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134(2):329–40. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324(5927):654–7. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrin N, Kaushik VK, Fortier E, Wall D, Pearson KJ, de Cabo R, Bordone L. JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PLoS One. 2009;4(12):e8414. doi: 10.1371/journal.pone.0008414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilson JR, Sharp PA. Small RNA regulators of gene expression. Cell. 2008;134(6):899–902. doi: 10.1016/j.cell.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306(5704):2105–8. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} J Biol Chem. 2005;280(16):16456–60. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- Nisoli E, Clementi E, Paolucci C, Cozzi V, Tonello C, Sciorati C, Bracale R, Valerio A, Francolini M, Moncada S, Carruba MO. Mitochondrial biogenesis in mammals: the role of endogenous nitric oxide. Science. 2003;299(5608):896–9. doi: 10.1126/science.1079368. [DOI] [PubMed] [Google Scholar]
- Nisoli E, Tonello C, Cardile A, Cozzi V, Bracale R, Tedesco L, Falcone S, Valerio A, Cantoni O, Clementi E, Moncada S, Carruba MO. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310(5746):314–7. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11(2):437–44. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- O’Reilly T, Niven DF. Levels of nicotinamide adenine dinucleotide in extracellular body fluids of pigs may be growth-limiting for Actinobacillus pleuropneumoniae and Haemophilus parasuis. Can J Vet Res. 2003;67(3):229–31. [PMC free article] [PubMed] [Google Scholar]
- Okazaki M, Iwasaki Y, Nishiyama M, Taguchi T, Tsugita M, Nakayama S, Kambayashi M, Hashimoto K, Terada Y. PPARbeta/delta regulates the human SIRT1 gene transcription via Sp1. Endocr J. 2010;57(5):403–13. doi: 10.1507/endocrj.k10e-004. [DOI] [PubMed] [Google Scholar]
- Onyango P, Celic I, McCaffery JM, Boeke JD, Feinberg AP. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc Natl Acad Sci U S A. 2002;99(21):13653–8. doi: 10.1073/pnas.222538099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacholec M, Bleasdale JE, Chrunyk B, Cunningham D, Flynn D, Garofalo RS, Griffith D, Griffor M, Loulakis P, Pabst B, Qiu X, Stockman B, Thanabal V, Varghese A, Ward J, Withka J, Ahn K. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J Biol Chem. 2010;285(11):8340–51. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CE, Kim MJ, Lee JH, Min BI, Bae H, Choe W, Kim SS, Ha J. Resveratrol stimulates glucose transport in C2C12 myotubes by activating AMP-activated protein kinase. Exp Mol Med. 2007;39(2):222–9. doi: 10.1038/emm.2007.25. [DOI] [PubMed] [Google Scholar]
- Pearson KJ, Baur JA, Lewis KN, Peshkin L, Price NL, Labinskyy N, Swindell WR, Kamara D, Minor RK, Perez E, Jamieson HA, Zhang Y, Dunn SR, Sharma K, Pleshko N, Woollett LA, Csiszar A, Ikeno Y, Le Couteur D, Elliott PJ, Becker KG, Navas P, Ingram DK, Wolf NS, Ungvari Z, Sinclair DA, de Cabo R. Resveratrol delays age-related deterioration and mimics transcriptional aspects of dietary restriction without extending life span. Cell Metab. 2008;8(2):157–68. doi: 10.1016/j.cmet.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, Luo H, Zhang Y, He W, Yang K, Zwaans BM, Tishkoff D, Ho L, Lombard D, He TC, Dai J, Verdin E, Ye Y, Zhao Y. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteomics. 2011 doi: 10.1074/mcp.M111.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci U S A. 2008;105(28):9793–8. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philp A, Chen A, Lan D, Meyer GA, Murphy AN, Knapp AE, Olfert IM, McCurdy CE, Marcotte GR, Hogan MC, Baar K, Schenk S. Sirtuin 1 (SIRT1) deacetylase activity is not required for mitochondrial biogenesis or peroxisome proliferator activated receptor-{gamma} coactivator-1{alpha} (PGC-1{alpha}) deacetylation following endurance exercise. J Biol Chem. 2011 doi: 10.1074/jbc.M111.261685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429(6993):771–6. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai JB, Isbatan A, Imai S, Gupta MP. Poly(ADP-ribose) polymerase-1-dependent cardiac myocyte cell death during heart failure is mediated by NAD+ depletion and reduced Sir2alpha deacetylase activity. J Biol Chem. 2005;280(52):43121–30. doi: 10.1074/jbc.M506162200. [DOI] [PubMed] [Google Scholar]
- Pittelli M, Formentini L, Faraco G, Lapucci A, Rapizzi E, Cialdai F, Romano G, Moneti G, Moroni F, Chiarugi A. Inhibition of nicotinamide phosphoribosyltransferase: cellular bioenergetics reveals a mitochondrial insensitive NAD pool. J Biol Chem. 2010;285(44):34106–14. doi: 10.1074/jbc.M110.136739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponugoti B, Kim DH, Xiao Z, Smith Z, Miao J, Zang M, Wu SY, Chiang CM, Veenstra TD, Kemper JK. SIRT1 deacetylates and inhibits SREBP-1C activity in regulation of hepatic lipid metabolism. J Biol Chem. 2010;285(44):33959–70. doi: 10.1074/jbc.M110.122978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92(6):829–39. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9(4):327–38. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang L, Banks AS, Accili D. Uncoupling of acetylation from phosphorylation regulates FoxO1 function independent of its subcellular localization. J Biol Chem. 2010;285(35):27396–401. doi: 10.1074/jbc.M110.140228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey KM, Mills KF, Satoh A, Imai S. Age-associated loss of Sirt1-mediated enhancement of glucose-stimulated insulin secretion in beta cell-specific Sirt1-overexpressing (BESTO) mice. Aging Cell. 2008;7(1):78–88. doi: 10.1111/j.1474-9726.2007.00355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, Takahashi JS, Imai S, Bass J. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324(5927):651–4. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 2004;279(49):50754–63. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- Revollo JR, Korner A, Mills KF, Satoh A, Wang T, Garten A, Dasgupta B, Sasaki Y, Wolberger C, Townsend RR, Milbrandt J, Kiess W, Imai S. Nampt/PBEF/Visfatin regulates insulin secretion in beta cells as a systemic NAD biosynthetic enzyme. Cell Metab. 2007;6(5):363–75. doi: 10.1016/j.cmet.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizki G, Iwata TN, Li J, Riedel CG, Picard CL, Jan M, Murphy CT, Lee SS. The Evolutionarily Conserved Longevity Determinants HCF-1 and SIR-2.1/SIRT1 Collaborate to Regulate DAF-16/FOXO. PLoS Genet. 2011;7(9):e1002235. doi: 10.1371/journal.pgen.1002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434(7029):113–8. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci U S A. 2007;104(31):12861–6. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004;101(45):15998–6003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongvaux A, Galli M, Denanglaire S, Van Gool F, Dreze PL, Szpirer C, Bureau F, Andris F, Leo O. Nicotinamide phosphoribosyl transferase/pre-B cell colony-enhancing factor/visfatin is required for lymphocyte development and cellular resistance to genotoxic stress. J Immunol. 2008;181(7):4685–95. doi: 10.4049/jimmunol.181.7.4685. [DOI] [PubMed] [Google Scholar]
- Rutanen J, Yaluri N, Modi S, Pihlajamaki J, Vanttinen M, Itkonen P, Kainulainen S, Yamamoto H, Lagouge M, Sinclair DA, Elliott P, Westphal C, Auwerx J, Laakso M. SIRT1 mRNA expression may be associated with energy expenditure and insulin sensitivity. Diabetes. 2010;59(4):829–35. doi: 10.2337/db09-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin E, Colla S, Liesa M, Moslehi J, Muller FL, Guo M, Cooper M, Kotton D, Fabian AJ, Walkey C, Maser RS, Tonon G, Foerster F, Xiong R, Wang YA, Shukla SA, Jaskelioff M, Martin ES, Heffernan TP, Protopopov A, Ivanova E, Mahoney JE, Kost-Alimova M, Perry SR, Bronson R, Liao R, Mulligan R, Shirihai OS, Chin L, DePinho RA. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470(7334):359–65. doi: 10.1038/nature09787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto J, Miura T, Shimamoto K, Horio Y. Predominant expression of Sir2alpha, an NAD-dependent histone deacetylase, in the embryonic mouse heart and brain. FEBS Lett. 2004;556(1-3):281–6. doi: 10.1016/s0014-5793(03)01444-3. [DOI] [PubMed] [Google Scholar]
- Saleem A, Adhihetty PJ, Hood DA. Role of p53 in mitochondrial biogenesis and apoptosis in skeletal muscle. Physiol Genomics. 2009;37(1):58–66. doi: 10.1152/physiolgenomics.90346.2008. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Maier B, Koclega KD, Chruszcz M, Gluba W, Stukenberg PT, Minor W, Scrable H. Phosphorylation regulates SIRT1 function. PLoS One. 2008;3(12):e4020. doi: 10.1371/journal.pone.0004020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh A, Brace CS, Ben-Josef G, West T, Wozniak DF, Holtzman DM, Herzog ED, Imai S. SIRT1 promotes the central adaptive response to diet restriction through activation of the dorsomedial and lateral nuclei of the hypothalamus. J Neurosci. 2010;30(30):10220–32. doi: 10.1523/JNEUROSCI.1385-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauve AA. NAD+ and vitamin B3: from metabolism to therapies. J Pharmacol Exp Ther. 2008;324(3):883–93. doi: 10.1124/jpet.107.120758. [DOI] [PubMed] [Google Scholar]
- Sauve AA, Schramm VL. Mechanism-based inhibitors of CD38: a mammalian cyclic ADP-ribose synthetase. Biochemistry. 2002;41(26):8455–63. doi: 10.1021/bi0258795. [DOI] [PubMed] [Google Scholar]
- Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Annu Rev Biochem. 2006;75:435–65. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- Screaton RA, Conkright MD, Katoh Y, Best JL, Canettieri G, Jeffries S, Guzman E, Niessen S, Yates JR, 3rd, Takemori H, Okamoto M, Montminy M. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119(1):61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Schmidt MT, Smith BC, Jackson MD, Denu JM. Coenzyme specificity of Sir2 protein deacetylases: implications for physiological regulation. J Biol Chem. 2004;279(38):40122–9. doi: 10.1074/jbc.M407484200. [DOI] [PubMed] [Google Scholar]
- Schoonjans K, Staels B, Auwerx J. Role of the peroxisome proliferator-activated receptor (PPAR) in mediating the effects of fibrates and fatty acids on gene expression. J Lipid Res. 1996;37(5):907–25. [PubMed] [Google Scholar]
- Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol. 2002;158(4):647–57. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metab. 2008;7(2):104–12. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Shi T, Wang F, Stieren E, Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem. 2005;280(14):13560–7. doi: 10.1074/jbc.M414670200. [DOI] [PubMed] [Google Scholar]
- Shieh WM, Ame JC, Wilson MV, Wang ZQ, Koh DW, Jacobson MK, Jacobson EL. Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J Biol Chem. 1998;273(46):30069–72. doi: 10.1074/jbc.273.46.30069. [DOI] [PubMed] [Google Scholar]
- Shore D, Squire M, Nasmyth KA. Characterization of two genes required for the position-effect control of yeast mating-type genes. EMBO J. 1984;3(12):2817–23. doi: 10.1002/j.1460-2075.1984.tb02214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91(7):1033–42. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Skidmore CJ, Davies MI, Goodwin PM, Halldorsson H, Lewis PJ, Shall S, Zia’ee AA. The involvement of poly(ADP-ribose) polymerase in the degradation of NAD caused by gamma-radiation and N-methyl-N-nitrosourea. Eur J Biochem. 1979;101(1):135–42. doi: 10.1111/j.1432-1033.1979.tb04225.x. [DOI] [PubMed] [Google Scholar]
- Strum JC, Johnson JH, Ward J, Xie H, Feild J, Hester A, Alford A, Waters KM. MicroRNA 132 regulates nutritional stress-induced chemokine production through repression of SirT1. Mol Endocrinol. 2009;23(11):1876–84. doi: 10.1210/me.2009-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchankova G, Nelson LE, Gerhart-Hines Z, Kelly M, Gauthier MS, Saha AK, Ido Y, Puigserver P, Ruderman NB. Concurrent regulation of AMP-activated protein kinase and SIRT1 in mammalian cells. Biochem Biophys Res Commun. 2009;378(4):836–41. doi: 10.1016/j.bbrc.2008.11.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwa M, Nakano H, Radak Z, Kumagai S. Short-term adenosine monophosphate-activated protein kinase activator 5-aminoimidazole-4-carboxamide-1-beta-D-ribofuranoside treatment increases the sirtuin 1 protein expression in skeletal muscle. Metabolism. 2011;60(3):394–403. doi: 10.1016/j.metabol.2010.03.003. [DOI] [PubMed] [Google Scholar]
- Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem. 2007;282(9):6823–32. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- Timmers S, Konings E, Bilet L, Houtkooper RH, Weijer T, Goossens GH, Hoeks J, Krieken S, Ryu D, Kersten S, Moonen-Kornips E, Hesselink MK, Kunz I, Schrauwen-Hinderling VB, Blaak EE, Auwerx J, Schrauwen P. Calorie-restriction-like effects of 30 days of resveratrol (resVidaTM) supplementation on energy metabolism and metabolic profile in obese humans. Cell Metabolism. 2011 doi: 10.1016/j.cmet.2011.10.002. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410(6825):227–30. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Tunaru S, Kero J, Schaub A, Wufka C, Blaukat A, Pfeffer K, Offermanns S. PUMA-G and HM74 are receptors for nicotinic acid and mediate its anti-lipolytic effect. Nat Med. 2003;9(3):352–5. doi: 10.1038/nm824. [DOI] [PubMed] [Google Scholar]
- Um JH, Park SJ, Kang H, Yang S, Foretz M, McBurney MW, Kim MK, Viollet B, Chung JH. AMP-activated protein kinase-deficient mice are resistant to the metabolic effects of resveratrol. Diabetes. 2010;59(3):554–63. doi: 10.2337/db09-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccari CS, Nagamia S, Thoenes M, Oguchi A, Hammoud R, Khan BV. Efficacy of controlled-release niacin in treatment of metabolic syndrome: Correlation to surrogate markers of atherosclerosis, vascular reactivity, and inflammation. J Clin Lipidol. 2007;1(6):605–13. doi: 10.1016/j.jacl.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Vakhrusheva O, Braeuer D, Liu Z, Braun T, Bober E. Sirt7-dependent inhibition of cell growth and proliferation might be instrumental to mediate tissue integrity during aging. J Physiol Pharmacol. 2008;59(Suppl 9):201–12. [PubMed] [Google Scholar]
- Valenzano DR, Terzibasi E, Genade T, Cattaneo A, Domenici L, Cellerino A. Resveratrol prolongs lifespan and retards the onset of age-related markers in a short-lived vertebrate. Curr Biol. 2006;16(3):296–300. doi: 10.1016/j.cub.2005.12.038. [DOI] [PubMed] [Google Scholar]
- van der Veer E, Ho C, O’Neil C, Barbosa N, Scott R, Cregan SP, Pickering JG. Extension of human cell lifespan by nicotinamide phosphoribosyltransferase. J Biol Chem. 2007;282(15):10841–5. doi: 10.1074/jbc.C700018200. [DOI] [PubMed] [Google Scholar]
- van der Veer E, Nong Z, O’Neil C, Urquhart B, Freeman D, Pickering JG. Pre-B-cell colony-enhancing factor regulates NAD+-dependent protein deacetylase activity and promotes vascular smooth muscle cell maturation. Circ Res. 2005;97(1):25–34. doi: 10.1161/01.RES.0000173298.38808.27. [DOI] [PubMed] [Google Scholar]
- van Roermund CW, Elgersma Y, Singh N, Wanders RJ, Tabak HF. The membrane of peroxisomes in Saccharomyces cerevisiae is impermeable to NAD(H) and acetyl-CoA under in vivo conditions. EMBO J. 1995;14(14):3480–6. doi: 10.1002/j.1460-2075.1995.tb07354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, Serrano L, Sternglanz R, Reinberg D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006;20(10):1256–61. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107(2):149–59. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Viswanathan M, Guarente L. Regulation of Caenorhabditis elegans lifespan by sir-2.1 transgenes. Nature. 2011;477(7365):E1–2. doi: 10.1038/nature10440. [DOI] [PubMed] [Google Scholar]
- Viswanathan M, Kim SK, Berdichevsky A, Guarente L. A role for SIR-2.1 regulation of ER stress response genes in determining C. elegans life span. Dev Cell. 2005;9(5):605–15. doi: 10.1016/j.devcel.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Walker AK, Yang F, Jiang K, Ji JY, Watts JL, Purushotham A, Boss O, Hirsch ML, Ribich S, Smith JJ, Israelian K, Westphal CH, Rodgers JT, Shioda T, Elson SL, Mulligan P, Najafi-Shoushtari H, Black JC, Thakur JK, Kadyk LC, Whetstine JR, Mostoslavsky R, Puigserver P, Li X, Dyson NJ, Hart AC, Naar AM. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 2010;24(13):1403–17. doi: 10.1101/gad.1901210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RH, Li C, Deng CX. Liver steatosis and increased ChREBP expression in mice carrying a liver specific SIRT1 null mutation under a normal feeding condition. Int J Biol Sci. 2010;6(7):682–90. doi: 10.7150/ijbs.6.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal S, Borucki K, Taneva E, Makarova R, Luley C. Extended-release niacin raises adiponectin and leptin. Atherosclerosis. 2007;193(2):361–5. doi: 10.1016/j.atherosclerosis.2006.06.028. [DOI] [PubMed] [Google Scholar]
- Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430(7000):686–9. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98(1):115–24. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- Xiong S, Salazar G, Patrushev N, Alexander RW. FoxO1 mediates an autofeedback loop regulating SIRT1 expression. J Biol Chem. 2011;286(7):5289–99. doi: 10.1074/jbc.M110.163667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Gao Z, Zhang J, Rivera CA, Yin J, Weng J, Ye J. Lack of SIRT1 (Mammalian Sirtuin 1) activity leads to liver steatosis in the SIRT1+/− mice: a role of lipid mobilization and inflammation. Endocrinology. 2010;151(6):2504–14. doi: 10.1210/en.2009-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A. 2008;105(36):13421–6. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakuchi M, Lowenstein CJ. MiR-34, SIRT1 and p53: the feedback loop. Cell Cycle. 2009;8(5):712–5. doi: 10.4161/cc.8.5.7753. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Schoonjans K, Auwerx J. Sirtuin functions in health and disease. Mol Endocrinol. 2007;21(8):1745–55. doi: 10.1210/me.2007-0079. [DOI] [PubMed] [Google Scholar]
- Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, de Cabo R, Sauve AA, Sinclair DA. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007a;130(6):1095–107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Chan NY, Sauve AA. Syntheses of nicotinamide riboside and derivatives: effective agents for increasing nicotinamide adenine dinucleotide concentrations in mammalian cells. J Med Chem. 2007b;50(26):6458–61. doi: 10.1021/jm701001c. [DOI] [PubMed] [Google Scholar]
- Yang T, Sauve AA. NAD metabolism and sirtuins: metabolic regulation of protein deacetylation in stress and toxicity. AAPS J. 2006;8(4):E632–43. doi: 10.1208/aapsj080472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Fu W, Chen J, Olashaw N, Zhang X, Nicosia SV, Bhalla K, Bai W. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat Cell Biol. 2007c;9(11):1253–62. doi: 10.1038/ncb1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23(12):2369–80. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, Newgard CB, Spiegelman BM. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413(6852):131–8. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- Yoshizaki T, Milne JC, Imamura T, Schenk S, Sonoda N, Babendure JL, Lu JC, Smith JJ, Jirousek MR, Olefsky JM. SIRT1 exerts anti-inflammatory effects and improves insulin sensitivity in adipocytes. Mol Cell Biol. 2009;29(5):1363–74. doi: 10.1128/MCB.00705-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizaki T, Schenk S, Imamura T, Babendure JL, Sonoda N, Bae EJ, Oh DY, Lu M, Milne JC, Westphal C, Bandyopadhyay G, Olefsky JM. SIRT1 inhibits inflammatory pathways in macrophages and modulates insulin sensitivity. Am J Physiol Endocrinol Metab. 2010;298(3):E419–28. doi: 10.1152/ajpendo.00417.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan F, Xie Q, Wu J, Bai Y, Mao B, Dong Y, Bi W, Ji G, Tao W, Wang Y, Yuan Z. MST1 promotes apoptosis through regulating Sirt1-dependent p53 deacetylation. J Biol Chem. 2011a;286(9):6940–5. doi: 10.1074/jbc.M110.182543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Liao YM, Hsueh CT, Mirshahidi HR. Novel targeted therapeutics: inhibitors of MDM2, ALK and PARP. J Hematol Oncol. 2011b;4:16. doi: 10.1186/1756-8722-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang M, Xu S, Maitland-Toolan KA, Zuccollo A, Hou X, Jiang B, Wierzbicki M, Verbeuren TJ, Cohen RA. Polyphenols stimulate AMP-activated protein kinase, lower lipids, and inhibit accelerated atherosclerosis in diabetic LDL receptor-deficient mice. Diabetes. 2006;55(8):2180–91. doi: 10.2337/db05-1188. [DOI] [PubMed] [Google Scholar]
- Zhang CY, Baffy G, Perret P, Krauss S, Peroni O, Grujic D, Hagen T, Vidal-Puig AJ, Boss O, Kim YB, Zheng XX, Wheeler MB, Shulman GI, Chan CB, Lowell BB. Uncoupling protein-2 negatively regulates insulin secretion and is a major link between obesity, beta cell dysfunction, and type 2 diabetes. Cell. 2001;105(6):745–55. doi: 10.1016/s0092-8674(01)00378-6. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Piston DW, Goodman RH. Regulation of corepressor function by nuclear NADH. Science. 2002;295(5561):1895–7. doi: 10.1126/science.1069300. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Wang SY, Fleuriel C, Leprince D, Rocheleau JV, Piston DW, Goodman RH. Metabolic regulation of SIRT1 transcription via a HIC1:CtBP corepressor complex. Proc Natl Acad Sci U S A. 2007;104(3):829–33. doi: 10.1073/pnas.0610590104. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451(7178):587–90. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Kachhap S, Singh KK. Mitochondrial impairment in p53-deficient human cancer cells. Mutagenesis. 2003;18(3):287–92. doi: 10.1093/mutage/18.3.287. [DOI] [PubMed] [Google Scholar]
- Zini R, Morin C, Bertelli A, Bertelli AA, Tillement JP. Effects of resveratrol on the rat brain respiratory chain. Drugs Exp Clin Res. 1999;25(2-3):87–97. [PubMed] [Google Scholar]
- Zschoernig B, Mahlknecht U. Carboxy-terminal phosphorylation of SIRT1 by protein kinase CK2. Biochem Biophys Res Commun. 2009;381(3):372–7. doi: 10.1016/j.bbrc.2009.02.085. [DOI] [PubMed] [Google Scholar]
- Zwilling D, Huang SY, Sathyasaikumar KV, Notarangelo FM, Guidetti P, Wu HQ, Lee J, Truong J, Andrews-Zwilling Y, Hsieh EW, Louie JY, Wu T, Scearce-Levie K, Patrick C, Adame A, Giorgini F, Moussaoui S, Laue G, Rassoulpour A, Flik G, Huang Y, Muchowski JM, Masliah E, Schwarcz R, Muchowski PJ. Kynurenine 3-monooxygenase inhibition in blood ameliorates neurodegeneration. Cell. 2011;145(6):863–74. doi: 10.1016/j.cell.2011.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]