Abstract
Increasing evidence supports the contribution of genetic influences on susceptibility/severity in acute lung injury (ALI), a devastating syndrome requiring mechanical ventilation with subsequent risk for ventilator-associated lung injury (VALI). To identify VALI candidate genes, we determined that Brown Norway (BN) and Dahl salt-sensitive (SS) rat strains were differentially sensitive to VALI (tidal volume of 20 ml/kg, 85 breaths/min, 2 h) defined by bronchoalveolar lavage (BAL) protein and leukocytes. We next exploited differential sensitivities and phenotyped both the VALI-sensitive BN and the VALI-resistant SS rat strains by expression profiling coupled to a bioinformatic-intense candidate gene approach (Significance Analysis of Microarrays, i.e., SAM). We identified 106 differentially expressed VALI genes representing gene ontologies such as “transcription” and “chemotaxis/cell motility.” We mapped the chromosomal location of the differentially expressed probe sets and selected consomic SS rats with single BN introgressions of chromosomes 2, 13, and 16 (based on the highest density of probe sets) while also choosing chromosome 20 (low probe sets density). VALI exposure of consomic rats with introgressions of BN chromosomes 13 and 16 resulted in significant increases in both BAL cells and protein (compared to parental SS strain), whereas introgression of BN chromosome 2 displayed a large increase only in BAL protein. Introgression of BN chromosome 20 had a minimal effect. These results suggest that genes residing on BN chromosomes 2, 13, and 16 confer increased sensitivity to high tidal volume ventilation. We speculate that the consomic-microarray-SAM approach is a time- and resource-efficient tool for the genetic dissection of complex diseases including VALI.
Keywords: rodent mechanical ventilation, consomics, bioinformatics, microarrays, candidate gene approach
Mechanical Ventilation is a life-saving intervention in patients with respiratory failure due to acute lung injury (ALI), a devastating clinical syndrome occurring in response to a variety of insults (sepsis, trauma, pneumonia, etc.) resulting in lung inflammation, increased vascular permeability, and protein-rich alveolar edema (36, 46). Unfortunately, although mechanical ventilation is a mainstay of therapy for ALI and the more severe acute respiratory distress syndrome (ARDS), mechanical ventilation potentially contributes directly to de novo lung injury known as ventilator-associated lung injury (VALI) (35, 41, 44), also augmenting pulmonary capillary leakage and acute inflammation (37, 38, 42). A number of studies have demonstrated that rats and mice ventilated with high endinspiratory lung tidal volumes rapidly exhibit lung injury (6, 11, 47) with increased inflammatory gene expression (tumor necrosis factor- , interleukin-6) in pulmonary epithelium (41). In contrast, the landmark ARDS net trial reported a 22% decrease in mortality in ARDS patients when low tidal volume ventilation was applied (compared with traditional, higher tidal volume ventilation) associated with decreases in bronchoalveolar lavage (BAL) leukocytes and inflammatory cytokines (15, 30). However, despite improved understanding of the pathophysiology of ALI and the deleterious effects of ventilation-induced mechanical stress, the underlying mechanisms of the injurious effects of mechanical ventilation remain unclear, and effective pharmacotherapy for ALI/VALI has yet to emerge (1).
Increasing information confirms the considerable heterogeneity in the response to injurious stimuli among the critically ill, suggesting that genetic susceptibility and environmental factors are also key determinants in the development of ALI (2, 4, 12, 14, 15, 24, 27, 28, 30, 34). Unfortunately, there remain significant challenges in the study of ALI genetics and other polygenic diseases. ALI represents a continuum rather than a discrete group of phenotypes, lacking unique, easily measured markers. Furthermore, ALI arises from diverse precipitating factors in a critically ill population and exhibits a variable clinical course (32). Finally, significant difficulty exists in defining the genetic factors that govern susceptibility to or severity of ALI/VALI due to phenotypic variance and likely complex gene-environment interactions (7). The sporadic nature of ALI precludes conventional genetic approaches such as heritability studies or linkage mapping (or “positional cloning”), a strategy potentially effective in other lung disorders, such as asthma, where large families with both affected and unaffected individuals can be examined for loci linked to the trait of interest (13).
We sought to explore genetic influences in the development of VALI and identify potential VALI candidate genes by sequentially integrating differential gene expression profiling with a chromosomal substitution (consomic) approach utilizing inbred rodent strains. We initially determined differential strain susceptibility to ventilator-mediated mechanical stress in both the VALI-sensitive Brown Norway (BN) and the VALI-resistant Dahl salt-sensitive (SS) rat strains followed by a bioinformatic-intensive genomic approach to identify 106 VALI-related candidate genes differentially expressed in response to high tidal volume (HTV) mechanical ventilation. Another 153 genes were differentially regulated at baseline between the two strains, including several well-known VALI-related genes. Combining these pair-wise comparison sets (SS and BN rats at baseline vs. HTV exposure) generated a total of 245 unique genes that were differentially regulated and potentially involved in the development of VALI. We next mapped the differentially expressed probe sets resulting from the microarray analyses to individual chromosomes as one expeditious way to help guide the selection of consomic rats for further evaluation in the VALI model while acknowledging the limits of this method in rigorously distinguishing between absolute chromosomal contributions in VALI. Consomic rats with single chromosome introgression (BN chromosome replacement into SS background) of chromosomes 2, 13, and 16 (SS:BN2, SS:BN13, and SS:BN16) exposed to HTV ventilation protocol exhibited significant increases in either BAL cell counts, BAL protein, or both parameters compared with the resistant SS parent. These results suggest that microarray-defined genes residing on these particular BN chromosomes may confer increased sensitivity to injury by HTV ventilation. BN chromosome 20 introgression into the SS background had minimal effect on the VALI phenotype. We speculate that the sequential microarray-consomic approach is a valuable integrative tool that prioritizes consomic screens seeking to identify candidate genes in complex disease models such as VALI.
MATERIALS AND METHODS
Rats
Male Sprague-Dawley (SD), Dahl SS, BN, and consomic rat strains (with individual BN chromosome introgression into the SS rat background) were obtained from Charles River Laboratories (Boston, MA) and PhysioGenix, Medical College of Wisconsin (http://www.physiogenix.com). All our experiments in animals were approved by the Institutional Animal Care and Use Committee (IACUC) at Johns Hopkins University, and rats weighing 250–350 g were housed in pathogen-free conditions in the Johns Hopkins Asthma and Allergy Center Animal Care Facility where they were cared for in accordance with institutional and National Institutes of Health (NIH) guidelines. During experiments, rats were anesthetized with an intraperitoneal injection of ketamine (75 mg/kg) and acepromazine (1.5 mg/kg) with additional anesthesia administered as necessary. Proper anesthesia was assessed by paw and tail pinching and maintained throughout all experiments with additional doses as needed.
Mechanical ventilation model
All experimental rats underwent tracheostomy and were ventilated with room air at 85 breaths/min, a tidal volume of 20 ml/kg, 0 positive end-expiratory pressure (i.e., PEEP) for 2 h with an Inspira rodent ventilator (Harvard Apparatus, Harvard, MA). A catheter was placed in the carotid artery for arterial blood gas (ABG) analysis and arterial blood pressure monitoring. ABG values were performed, and external dead space was adjusted to maintain an arterial pH of 7.30–7.45 throughout the experiment. Boluses of isotonic phosphate-buffered saline (PBS) were given as needed to maintain a mean arterial pressure greater than 60 mmHg. Airway pressure was monitored continuously and recorded (every 30 min) in all animals as we previously described (17, 31). Control animals were allowed to spontaneously breathe room air. This rodent mechanical ventilation model of VALI including the various levels for pH, breathing rate (breaths/min), and tidal volume has been previously described (7, 21).
BAL fluid and leukocyte counts
At the termination of each experiment, all animals were euthanized by exsanguination under anesthesia in accordance with institutional and NIH guidelines. The pulmonary vasculature was perfused via the pulmonary artery with sterile PBS to remove blood-borne elements and plasma, and the right lung was clamped at the level of the mainstem bronchus, excised, and snap-frozen in liquid nitrogen for subsequent RNA analysis/expression profiling. The left lung was lavaged with 3 ml of PBS, and the BAL fluid (~2 ml) was centrifuged at 2,500 rpm for 20 min at 4°C; then the supernatant removed and frozen at −80°C for subsequent protein analysis. Cell pellets were resuspended in 0.5 ml of red blood cell lysis buffer (ACK Lysing Buffer, BioSource International) for 20 min and then repelleted by centrifugation at 2,500 rpm for 20 min at 4°C. The supernatant was decanted and saved for protein analysis, and the cell pellet was resuspended in 0.2 ml of PBS for cell counting using a standard hemocytometer technique. A total of 300 BAL cells/slide were counted for differential cell using a Diff-Quick-stained kit (Baxter Diagnostics, McGaw Park, IL).
Microvascular permeability to protein
The protein concentration in BAL fluid was determined by a modified Lowry colorimetric assay using a Bio-Rad DC protein assay kit (Bio-Rad Laboratories, Hercules, CA). BAL fluid was prepared according to assay instructions, the absorbances were measured at 750 nm, and protein concentration was determined using standard curves (20).
Tissue Evans blue dye (EBD) deposition
Evans blue dye (EBD, 5 mg/kg) containing 20% bovine serum albumin (BSA, fraction V; Sigma) was administered intravenously via the external jugular vein, 30 before euthanasia and tissue harvesting. The right lung vasculature was perfused with PBS and removed, and its weight was recorded; then it was freeze/thawed and homogenized in 6 ml formamide and incubated at 60°C for 18 h. The extracts were spun (15,000 rpm, for 60 min at 4°C), and the supernatants were collected to quantify lung EBD content in a spectrophotometer (740 nm). Each reading was compared with standard curve of EBD and expressed as milligrams per milliliter per gram body wt (33).
RNA isolation and expression profiling
Right lung mRNA was isolated as we have described previously (26), from three whole lung tissue samples each per strain and condition: SS baseline, SS HTV ventilation, BN baseline, and BN HTV ventilation. Affymetrix Gene-Chip Expression Analysis Manual protocols (Affymetrix, Santa Clara, CA) recommended by the manufacturer were followed, as we have described previously (17). The signal intensity fluorescent images produced during Affymetrix GeneChip Rat Genome 230 2.0 Array hybridizations were read using the Hewlett-Packard GeneArray Scanner G2500A and converted into “GeneChip Cell” files (CEL) by GCOS software (Affymetrix).
Expression data filtering and analysis
Data generated in this study complies with the “minimum information about microarray experiments” (MIAME) standard (5) and may be accessed via the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) with the series accession number GSE7041. CEL files were analyzed by the Robust Multichip Average (RMA) module of Bioconductor package (18, 19). The expression measures of individual probe set were extracted, and background correction, across-array normalization, and summarization were performed. “Summarization” refers to the process designed to obtain a single expression value for each gene and analyzes the signals from all probes (oligos) related to that gene and located on different spots on the array are combined. The global gene expression profile analysis of transformed expression data was conducted using Significance Analysis of Microarrays (SAM) (43). Briefly, SS and BN expression data sets were separated by experimental conditions and strain to corresponding blocks as recommended by the SAM procedure (see the SAM online manual for detailed explanations of the blocking approach, http://www-stat.stanford.edu/ tibs/SAM). Data for each strain was analyzed using a twofold change cutoff and a false discovery rate <5% (with 10,000 permutations). Using this threshold with the SAM approach, we identified candidate genes as genes that were significantly differentially regulated between parental rat strains (BN vs. SS) at baseline and in response to HTV mechanical ventilation for each strain.
To expeditiously guide our consomic selection, we then mapped the HTV-driven differentially expressed probe sets to individual chromosomes to determine chromosomes with the highest probe set density. Though it is important to recognize limitations of this method such as the absence of a qualitative differentiation between probe set-defined transcripts and their roles in VALI, the goal was to devise a time-saving approach for consomic selection that facilitates differentiation between chromosomal contributions in VALI based entirely on a speedy quantitative approach (probe set density). We normalized this observed probe set distribution to the chromosome-specific probe representation on the Affymetrix microarray GeneChip (which generally accounts for the chromosomal size differences for most chromosomes) and generated the ratio of observed over “predicted” chromosomal distribution of the total differentially expressed probe sets (Fig. 4). To allow us to derive this latter parameter, we generated a predicted chromosomal distribution for the 352 differentially regulated HTV-driven probe sets multiplied by the number of chromo-some-specific probes that are fixed on the probe array divided by the total number of probes on the whole microarray platform.
Fig. 4.
Chromosomal distribution of differentially expressed probe set-defined transcripts from rodent lungs: ratio of the observed chromosomal distribution of differentially expressed rodent lung probe sets evoked by HTV mechanical ventilation over that of the predicted distribution (352). Predicted distribution, in this case, equates to an equal distribution of the HTV-driven probe sets (352) between all chromosomes normalized by the individual chromosomal probe representation on the microarray chip. With the goal of accentuating the greatest probe set-dense chromosomes for consomic selection, a chromosome with a ratio greater than 1 (where observed equals predicted) represents a significant trend in surpassing the chromosome's expected probe set density over other chromosomes and, hence, an increase in its potential contribution to the ventilator-associated lung injury (VALI) phenotype. As shown, chromosomes 2, 13, 16, and 17 (as highlighted in black) demonstrated the greatest density of probe sets (over a ratio of 1.2, thick black line), and since the SS:BN17 strain was not available, we studied strains SS:BN13, 16, and 2. We also studied the SS:BN20 strain (shown in striped bar) to study the trend for a chromosome with a low probe set density given the ratio attributed to its chromosome to be lower than 1. Parenthetical annotation reflects the number of unique genes that mapped to the indicated chromosome.
Gene Ontology (GO) terms were dynamically linked to candidate genes using OntoExpress (http://vortex.cs.wayne.edu/Projects.html#Onto-Express) tools in a fashion similar to what has been previously described in MAPPFinder and GenMAPP (9, 10, 17, 26) to examine the expression profiling based on biological pathway activation rather than by single gene expression (22). The relevance of newly identified candidate genes to lung injury was then evaluated using PubMatrix (http://pubmatrix.grc.nia.nih.gov), the automated biomedical literature search engine (3) that measures the co-occurrence of two key words in the National Center for Biotechnology Information literature search service, PubMed. In our analysis, we searched each identified candidate gene against a series of keywords representing ALI, mechanical ventilation, and barrier disruption. The PubMatrix-selected citations of journal articles that referenced “acute lung injury”, “mechanical ventilation”, “coagulation”, “inflammation”, and “permeability” terms in the same context with a given candidate gene were manually evaluated.
Statistical significance
Values are expressed as means ± SE. Intergroup differences (control vs. HTV ventilation) were determined by the Student's unpaired t-test, whereas multiple strain differences were assessed by a simple ANOVA with post hoc analysis using Newman-Keuls multiple comparison test. A value of P < 0.05 was considered statistically significant.
RESULTS
Strain survey and identification of VALI-sensitive and VALI-resistant rodent strains
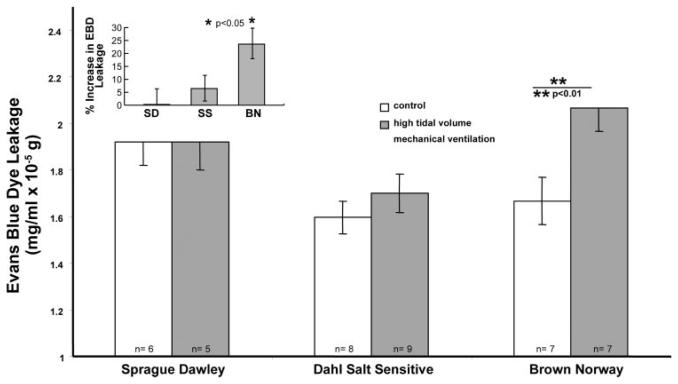
The extent of alveolar injury, inflammation, and barrier disruption (BAL cells, BAL cell differentiation, and BAL protein) and vascular permeability (EBD leakage) were used to assess HTV mechanical ventilation-induced lung injury in adult male SD, Dahl SS, and BN rats. Two hours of HTV mechanical ventilation induced significant alveolar injury and inflammation in the BN strain, with a 103% increase in BAL cell count (2.28 ± 0.53 × 105 vs. 1.12 ± 0.19 × 105 cells/ml in controls, P < 0.01) (Fig. 1A), an observation attributable to an influx of polymorphonuclear cells (PMNs) ( 90% PMNs in BAL). Likewise, exposure to HTV ventilation stimulated a significant inflammatory response in the BN strain, producing a 135% increase in BAL protein (0.67 ± 0.08 vs. 0.28 ± 0.02 mg/ml in controls, P < 0.01) (Fig. 1B). There were no significant differences in either BAL cell counts or BAL protein levels in either the SS or SD rat strains after HTV ventilation challenge compared with baseline values. There were no statistically significant differences in baseline BAL cell count, BAL protein, or tissue EBD between the three strains. Consistent with the observed increase in BAL protein reflecting epithelial permeability, HTV ventilation evoked a significant increase in vascular permeability in the BN strain reflected by increased tissue EBD deposition, whereas there were no significant differences in tissue EBD in either the SS or SD strains after HTV ventilation challenge compared with baseline (Fig. 2). Thus, the BN strain was distinctively sensitive to the effects of HTV mechanical ventilation with evidence of significantly increased alveolar inflammation and vascular/alveolar barrier disruption at 2 h compared with the relatively resistant SS and SD strains.
Fig. 1.
Survey of specific rodent strains for sensitivity to mechanical ventilatory stress: bronchoalveolar lavage (BAL) cell counts and protein levels. BAL inflammatory cells (A) and protein content (B) were used to assess lung injury induced by high tidal volume (HTV) mechanical ventilation in adult male Sprague-Dawley (SD), Dahl salt-sensitive (SS), and Brown Norway (BN) rats exposed to HTV ventilation for 2 h. HTV mechanical ventilation (20 ml/kg, see MATERIALS AND METHODS) induced a significant increase in BAL cells (A) and protein (B) in the BN strain. There were no significant changes in BAL cells or protein in either the SS or SD strains following HTV challenge. Inset: the percent (%) increase in BAL cell counts (A) or protein level (B) by strain compared with controls (i.e., without HTV). **P < 0.01; n = number of animals per condition.
Fig. 2.
Survey of specific rodent strains for sensitivity to mechanical ventilatory stress: Evans blue dye (EBD) vascular leakage. EBD content was quantified to assess albumin leak across the lung vascular bed in rodents injured with HTV mechanical ventilation (20 ml/kg, 2 h). SD, Dahl SS, and BN male rats challenged in this manner revealed significant increases in EBD deposition noted only in the BN rat strain. There were no significant changes in EBD content in either the SS or SD strains. Inset: the percent increase in EBD content by strain compared with controls (i.e., without HTV). *P < 0.05. **P < 0.01.
Gene expression/analysis
We used SAM with a twofold change cutoff and a false discovery rate <5% and found 352 probe sets, from which 106 unique genes were differentially expressed in the two strains in response to injurious HTV mechanical ventilation (Supplemental Tables SII and SIII) (supplemental material for this article is available at the Physiological Genomics web site). Likewise, we found 479 probe sets, from which 153 unique genes were differentially expressed at baseline between the two strains and potentially govern susceptibility to VALI (Supplemental Table SI). Overlap between these two pair-wise comparisons (SS vs. BN at baseline and during HTV exposure) yielded 245 potential VALI-related candidate genes that were differentially regulated. Subsequent GO analysis revealed that the majority of differentially expressed genes (in response to mechanical ventilation) in the HTV-sensitive (BN) strain and the HTV-resistant (SS) strain involved the following gene ontologies: transcription, signal transduction, chemotaxis/cell motility, inflammation, DNA and protein binding, cell proliferation, and cell adhesion (Fig. 3).
Fig. 3.
Gene ontologies (GO) involved in rodent lung responses to mechanical ventilation-induced mechanical stress. We employed OntoExpress, a program that uses a relational database to link genes in a given data set, as another level of filtering of the genomic data derived by microarray analyses. Ontology analysis of the 106 genes in our mechanical ventilation array data set shows biological relationships between genes and gene products, giving experimental gene array expression data a biological context. The “protein binding” ontology was identified as the major mechanical stretch-related acute lung injury (ALI) biological process with the highest percentage of genes per total number of identified genes (106). Significant representation was also identified in the signal transduction, DNA binding, cell proliferation, cell motility/chemotaxis, cell adhesion, and inflammation ontologies. Parenthetical annotation reflects the number of genes that mapped to the indicated gene Ontology. All eight ontologies had were significant at P < 0.001.
To drive subsequent consomic selection, we next evaluated the chromosomal distribution of the HTV-driven differentially regulated probe sets identified by microarray analysis in our model. The lack of an assigned gene name to a probe setdefined transcript does not necessarily reduce its potential functional impact in a phenotype. We, therefore, mapped the 352 differentially regulated HTV-driven probe sets rather than the 106 unique genes (derived from the probe sets) for greater accuracy of representation of the chromosomal involvement in the VALI phenotype. Again, aware of its inherent limits as purely a quantitative indicator, we employed this distribution technique as one method of providing fast and insightful ideas about the potential for varying levels of chromosomal contributions. We normalized the observed data distribution to the chromosome-specific probe representation on the Affymetrix microarray GeneChip (see MATERIALS AND METHODS) and generated the ratio of observed over predicted chromosomal distribution of the total differentially regulated probe sets. This analysis revealed chromosomes 2, 13, 16, and 17 as chromosomes containing the largest number of potential VALI-inducing probe set-defined transcripts and, consequently, genes important in the response to VALI (Fig. 4). Figure 4 also parenthetically depicts the distribution of the 106 candidate genes evoked by HTV ventilation above each individual chromosomal column. The mapping of these candidate genes emphasizes the lack of correlation between the gene density vs. probe set density per chromosome; i.e., chromosome 2 had the greatest number of HTV-related genes (13 genes), whereas chromosome 13 only had 4 genes.
To confirm the contribution of chromosome-specific genes to the susceptibility (or resistance) to injury produced by HTV mechanical ventilation, consomic progeny (including SS:BN2, 13, and 16), with a single BN chromosome substituted into the SS genomic background, were subjected to HTV ventilation according to the VALI protocol (described above) to determine the capacity of introgressed genes to result in ventilator sensitivity in an otherwise resistant strain. Although chromosome 17 had a greater VALI gene representation than 2, 13, and 16, this consomic strain was not immediately available. Furthermore, an available consomic SS:BN20 was studied to determine whether its low level of HTV-driven probe set density was associated with a potentially low likelihood of involvement of chromosome 20 in VALI. Priority was also given to the characteristics that are most relevant for the goal of differentiating the strains for VALI susceptibility: BAL cells > BAL protein. Previous evidence supports the importance of this priority and the prevalence of these individual determinants in the development of an ALI phenotype (25).
Introgression of BN chromosome 2 into the SS background produced an intermediate effect on the VALI phenotype with a significantly higher BAL protein level and, thus, alveolar inflammation but less of a change in BAL cell count, a marker of lung injury. In contrast, introgression of BN chromosomes 13 and 16 into the SS genetic background conferred susceptibility to lung injury due to HTV with significant increases in both BAL cell count and proteins compared with the SS parent (Fig. 5, A-F). Additionally, as would be predicted, the consomic SS:BN20 strain, which contained only a limited number of HTV-driven probe sets by array analysis, displayed minimal susceptibility to VALI with a mild nonsignificant rise in the BAL protein compared with the SS strain and no change in the BAL cell count (Fig. 5, G-H). Table 1 depicts the PubMatrix evaluation of the microarray-derived VALI candidate genes that mapped to these consomics (2, 13, and 16) and details the number of their PubMed citations. This approach revealed several previously identified ALI/VALI candidate genes residing on chromosomes 2, 13, and 16 (Nr4a, Il1b, Btg2, Ifrd1, Jun, Myc, Socs3, Cyr61) (7, 16, 26), as well several potentially novel VALI candidates with few or no PubMed citations.
Fig. 5.
Effect of BN chromosome introgression (chromosomes 2, 13, and 16) into the Dahl SS background on VALI phenotype. SS rats with introgression of BN chromosome 16 into the resistant SS background (SS:BN16) and exposed to HTV mechanical ventilation exhibited increased BAL cell counts (A) and protein level (B). Similar increases were observed with chromosome 13 introgression (SS:BN13) (BAL cell contents, C and D) while chromo-some 2 introgression (SS:BN2) showed divergent responses in BAL cell counts (E) vs. BAL protein level (F) compared with the SS parent. These data suggest that BN chromosomes 13 and 16 conferred increased sensitivity to HTV, and chromosome 2 showed an intermediate phenotype. SS rats with introgression of BN chromosome 20 into the resistant SS background (SS:BN20) and exposed to HTV mechanical ventilation did not exhibit changes in BAL cell counts (G) and a mild nonsignificant increase (P < 0.05) in protein level (H) compared with the SS strain. These data suggest that BN chromosome 20 did not confer increased sensitivity to HTV, unlike chromosomes 2, 13, and 16. *P < 0.05. **P < 0.01.
Table 1.
PubMatrix analysis of previously identified and novel VALI chromosome-specific genes differentially expressed at baseline and after HTV ventilation in the BN and SS rats
| Gene Symbol | Acute Lung Injury | Inflammation | Permeability | Mechanical Ventilation | Coagulation | |
|---|---|---|---|---|---|---|
| Candidate genes on chromosome 2 | ||||||
| Vascular cell adhesion molecule 1 | Vcam1 | 13 | 1,328 | 49 | 10 | 69 |
| Granzyme A | Gzma | 2 | 47 | 9 | 0 | 5 |
| Endothelial cell-specific molecule 1 | Esm1 | 2 | 12 | 2 | 0 | 1 |
| Protein kinase inhibitor, alpha | Pkia | 1 | 113 | 28 | 0 | 5 |
| Chondroitin sulfate proteoglycan 2 | Cspg2 | 1 | 31 | 13 | 1 | 5 |
| S100 calcium binding protein A8 (calgranulin A) | S100a8 | 0 | 77 | 2 | 0 | 0 |
| Carbonic anhydrase 3 | Ca3 | 0 | 16 | 101 | 10 | 14 |
| Interleukin 6 signal transducer | II6 st | 0 | 74 | 1 | 0 | 1 |
| S100 calcium binding protein A9 (calgranulin B) | S100a9 | 0 | 73 | 1 | 1 | 1 |
| Growth hormone receptor | Ghr | 0 | 12 | 0 | 1 | 4 |
| Phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 1 | Pik3r1 | 0 | 9 | 3 | 0 | 1 |
| Natriuretic peptide receptor 3 | Npr3 | 0 | 7 | 10 | 1 | 2 |
| Cysteine-rich protein 61 | Cyr61 | 0 | 7 | 0 | 0 | 1 |
| Cyclin L1 | Ccnl1 | 0 | 4 | 1 | 0 | 0 |
| Guanine nucleotide binding protein, alpha-inhibiting 3 | Gnai3 | 0 | 2 | 2 | 0 | 0 |
| Proteasome (prosome, macropain) subunit, alpha type 5 | Psma5 | 0 | 1 | 0 | 0 | 0 |
| Secreted frizzled-related protein 2 | Sfrp2 | 0 | 1 | 0 | 0 | 0 |
| Calcium/calmodulin-dependent protein kinase II, delta | Camk2d | 0 | 0 | 1 | 0 | 0 |
| Polo-like kinase 2 (Drosophila) | Plk2 | 0 | 0 | 0 | 0 | 0 |
| Forkhead box O1A | Foxo1a | 0 | 0 | 0 | 0 | 0 |
| Muscleblind-like 1 (Drosophila) | Mbnl | 0 | 0 | 0 | 0 | 0 |
| Nexilin | Nexn | 0 | 0 | 0 | 0 | 0 |
| Chaperonin subunit 5 (epsilon) | Cct5 | 0 | 0 | 0 | 0 | 0 |
| PDZ protein Mrt1 | Snx27 | 0 | 0 | 0 | 0 | 0 |
| Protein phosphatase 3, catalytic subunit, alpha isoform | Ppp3ca | 0 | 0 | 0 | 0 | 0 |
| Coagulation factor II (thrombin) receptor-like 2 | F2rl2 | 0 | 0 | 0 | 0 | 0 |
| Candidate genes on chromosome 13 | ||||||
| Coagulation factor 5 | F5 | 129 | 2,022 | 4,051 | 62 | 9,439 |
| Prostaglandin-endoperoxide synthase 2 | Ptgs2 | 20 | 2,007 | 155 | 5 | 53 |
| Troponin T2, cardiac | Tnnt2 | 1 | 54 | 2 | 2 | 12 |
| Laminin, gamma 1 | Lamc1 | 1 | 18 | 4 | 0 | 3 |
| Regulator of G-protein signaling 2 | Rgs2 | 0 | 10 | 6 | 0 | 3 |
| Chemokine (C-X-C motif) receptor 4 | Cxcr4 | 0 | 1 | 0 | 0 | 0 |
| POU domain, class 2, transcription factor 1 | Pou2f1 | 0 | 0 | 0 | 0 | 1 |
| Flavin containing monooxygenase 3 | Fmo3 | 0 | 0 | 0 | 0 | 0 |
| B-cell translocation gene 2, anti-proliferative | Btg2 | 0 | 0 | 0 | 0 | 0 |
| Candidate genes on chromosome 16 | ||||||
| Plasminogen activator, tissue | Plat | 12 | 273 | 53 | 15 | 2,562 |
| Tropomyosin 4 | Tpm4 | 0 | 17 | 6 | 0 | 1 |
| Mitochondrial tumor suppressor 1 | Mtus1 | 0 | 8 | 31 | 0 | 0 |
| Carboxypeptidase E | Cpe | 0 | 2 | 3 | 0 | 1 |
| Deleted in liver cancer 1 | Dlc1 | 0 | 2 | 0 | 0 | 1 |
| ADP-ribosylation factor 4 | Arf4 | 0 | 1 | 3 | 0 | 0 |
| Jun D proto-oncogene | Jund | 0 | 0 | 1 | 0 | 0 |
| Myomesin 2 | Myom2 | 0 | 0 | 0 | 0 | 0 |
| Hook homolog 3 | Hook3 | 0 | 0 | 0 | 0 | 0 |
Five representative terms were chosen for their involvement in ventilator-associated lung injury (VALI) and were used to quantify the PubMed citations of the candidate genes in these categories. Shown are previously identified (located at the top half of the table) and novel (located at the bottom half) VALI chromosome-specific genes differentially expressed at baseline and after high tidal volume (HTV) ventilation in the BN and SS rats.
DISCUSSION
The goal of our study was to utilize a combination of genomic and genetic approaches to help define the contribution of genetic influences to the VALI phenotype. We combined the use of traditional rodent strain analysis with a powerful genomic microarray expression profiling approach to identify candidate genes with potential involvement in the molecular changes which occur in the multigenic VALI syndrome. We examined alterations in lung gene expression in two rat strains exposed to HTV mechanical ventilation: the VALI-sensitive BN strain and the VALI-resistant SS strain. Statistical filtering initially identified 245 potential candidate genes that were significantly differentially expressed between the two strains, either at baseline or in response to HTV mechanical ventilation. GO classification identified significantly involved biological pathways, and PubMatrix literature mining deciphered known and novel genes among those identified in our analysis. Using this microarray analysis as an initial step to study the genetics of VALI allowed us to devise a unique priority schema for consomic selection in an otherwise labor-intensive, and costly, full consomic screen (all 21 rat chromosomes), in which a single chromosome of one parental strain is substituted into the very homogeneous genetic background of the second strain. The partial consomic screen presented here is the first to demonstrate the genetic impact in the development of VALI; introgressions of chromosomes 2, 13, and 16 from the BN into the SS rats confer susceptibility to VALI in an otherwise resistant strain. In addition, this susceptibility of the highly inbred consomic strains including SS:BN2, 13, and 16 to VALI increases the likelihood that candidate genes residing on these chromosomes are involved in the VALI phenotype. Overall, this study evaluates the contributions of selected individual chromosomes in the VALI pathophysiology and provides an important first step in linking phenotypic data to genomic data. In addition to providing initial insights about consomic selection, the integration of microarray technology serves to broaden the scope of candidate genes and pathways that may be relevant to ALI/VALI pathophysiology.
Early animal studies of ALI focused on a single gene (or a few genes) of interest (39, 40, 45). For example, Hsp70 expression was assessed in a rat model of lipopolysaccharide (LPS)-induced ALI (45) and TNF-α expression was evaluated in response to mechanical ventilation in a surfactant depletion model of ALI in rabbits (40). These studies reflect the traditional strategy of preselecting a gene of interest, thus, severely narrowing the scope of study. Because of the likely multigenic etiology of ALI, newer approaches have taken advantage of the availability of genomic techniques/tools to focus on the generation of a robust list of candidate genes that can then be studied individually both in patients and in specific preclinical models of disease. Once a strain-specific phenotypic difference is identified, chromosomal regions encoding genes that regulate a specific phenotype; for example, susceptibility to VALI can then be identified through the comparison of genetically distinct offspring with differing phenotypes. Traditional strain-survey approaches, using carefully selected inbred parental strains with differing sensitivities to a stimulus and subsequent testing of genetically differing offspring, have been successfully used to identify novel candidate genes in a variety of disease. Leikauf et al. (23) investigated the sensitivity of several mouse strains to ALI induced by ozone, ultrafine polytetrafluoroethylene (PTFE), or nickel (NiSO4) exposure, demonstrating that susceptibility to this injury varied between strains, and identified a resistant (C57BL/6J) murine strain whose mean survival time was double that of the sensitive (A/J) strain (with resistance inherited as a dominant trait). Chromosomal analysis of multiple strains of backcrossed mice generated from the two parental strains allowed the identification of quantitative trait loci (QTLs) on murine chromosomes 6, 11, 13, and 17 that contributed to the significant differences in survival time. Subsequent cDNA microarray analysis identified significant differences in the expression of >100 genes involved in cell proliferation, extracellular matrix repair, oxidative stress, hypoxia, and surfactant proteins. Combining the results from traditional linkage studies and QTL mapping as well as microarray analysis, a short list of candidate genes were identified for nickel-induced ALI, as well as several novel genes (23). Such backcross approaches, while highly meritorious, are time-consuming and expensive. More importantly, individual F2 rats from backcrosses add a significant amount of genetic “background noise” due to lack of strain homogeneity and reduce the power to map associated QTLs.
Copland et al. (7) studied in vivo temporal changes in early gene expression in rat lungs with (30 min) HTV ventilation and found that lung injury correlated with increased expression of transcription factors, stress proteins, and inflammatory mediators and decreased expression of metabolic regulatory genes and identified Egr-1, c-Jun, Hsp70, and IL-1β as candidate genes involved in VALI. Our own recent work, using orthologous cross-species expression profiling in multiple animal models of HTV ventilation and in silico bioinformatic analysis, identified both common and novel potential VALI candidate genes including: fibrinogen A, coagulation factor III, plasminogen activator, urokinase receptor, tissue factor, plasminogen activator inhibitor type 1 (PAI1), IL-1β, IL-6 (16), and Egr1 (16, 17, 26). Several of these genes were identified in our current VALI candidate list including those that mapped to chromosomes 2, 13, or 16 in the rodent (calgranulin A, calgranulin B, prostaglandin-endoperoxide synthase 2 or Ptgs2, and cysteine rich protein 61 or Cyr61) (Table 1). Interestingly, calgranulin A was expressed in both the VALI-sensitive BN and VALI-resistant SS strains during HTV exposure; however, these were present at different relative levels possibly corresponding to different states of VALI susceptibility. In addition, Cyr61 was also differentially regulated between the BN and SS strains at baseline, stressing the potential viability of candidate genes within the 153 differentially regulated gene set between our two strains. Although all of these studies possess the standard limitations of microarray approaches, the high frequency of confirmation with these candidate genes in various models of VALI also strengthens their association to VALI and adds appeal for further focused investigation.
In the current study, we expanded the known VALI candidate gene list by identifying potentially 245 candidate genes that participate in differential sensitivities of the BN rats and SS rats to VALI. Similar to our previous orthologous cross-species expression profiling and in silico analysis approaches, as well as the study by Copland et al. (7), blood coagulation and inflammation were two of the highly represented genetic pathways of the candidate genes residing on chromosomes 2, 13, and 16, as evidenced by PubMatrix evaluation. K-mean clustering revealed several well-known ALI/VALI-associated genes (IL6, PAI1, CCL2, COX2) as well as novel ALI genes. In addition to the coagulation pathway, broad ontologies of all candidate genes expressed during HTV exposure included transcription regulation, cell proliferation, and cell motility/chemotaxis. Based on these reports and data generated by our cross-species analysis of ALI, we speculate that mechanical stretch directly regulates cell motility/chemotaxis and cell adhesion in lung tissues (epithelium, endothelium, etc.) accompanied by activation of the coagulation cascade, inflammatory cell recruitment, and the development of ALI, a scenario consistent with clinical reports on the effect of excessive tidal volume ventilation (1).
Acknowledging limitations of microarray technology, it is possible that the regulation of a limited number of genes on parental strains did not occur at the level of steady-state mRNA content and, therefore, could not be identified by the approach used in the present study. In addition, the differential expression of genes on chromosomes 2, 13, and 16 in tissues outside the lungs may have affected the expression of genes on other chromosomes in the lungs. The use of expression microarrays in this study also makes the assumption that genetic variation between the parental strains contributes to disease by directly affecting lung expression of the transcript encoded by that gene. There are, however, additional mechanisms by which genetic polymorphisms could cause or affect disease including a variety of posttranslational modifications, tissue-specific mRNA editing, and amino acids changes that do not change the transcript expression level but rather its protein properties. These possibilities, however, do not negate the importance of deriving a list of differentially regulated genes and their impact to the phenotype via a genomic approach. Advantages to the genomic (microarray technology) approach, as described above, include the discovery of a large number of genes in the development of the VALI phenotype without a priori knowledge of a particular gene or pathway in the disease in contrast to standard linkage approaches (such as the excellent study by Leikauf et al., Ref. 23). Using genome-wide expression analysis, we also used a novel approach to integrate these results to prioritize a consomic screen by mapping differentially regulated probe sets to individual chromosomes. Aware of the limitations of mapping probe sets, we based chromosomal “probe set density” as one easily computable marker for chromosomal contribution in the VALI phenotype.
To systematically approach an association study between the various selected consomic strains and the VALI phenotype, it is important to recognize the difficulty in identifying putative QTLs for complex diseases like VALI. In addition to pleiotropy, incomplete penetrance, and gene-environment interactions, the problem also lies in the complexity and variance of phenotypes with which candidate genes are associated. Complex phenotypes like VALI that are subdivided into intermediate physiological phenotypes can be less complex with oligogenic determination and therefore can exhibit higher heritability. We, thus, focused on distinct determinants of the phenotype: alveolar injury and inflammation measured by BAL cells, and by BAL protein. The differences between these determinants has been previously studied in an ALI model induced by the inoculation of bacterial LPS showing varying responses between acute lung “cell injury” as defined by BAL cell counts and an “inflammatory response” defined by BAL protein (one of multiple inflammatory parameters) (25). We determined that consomic strains SS:BN13 and SS:BN16 conferred full VALI susceptibility, as apparent by significant increases in both BAL cell counts and protein compared with the SS parent. The intermediate effect of SS:BN2 on the VALI phenotype highlights another key advantage to the utility of a consomic approach, that of differentiating a complex disease (like VALI) into individual determinants (in this case, BAL protein but not BAL cell count) that can then be linked to the effect of a single chromosome introgression. Likewise, strain SS:BN20 did not produce either a significant rise in BAL protein or changes to BAL cell counts consistent with the low probe set density distribution (Fig. 4). Ultimately, the continuous spectrum of distribution of consomic strain phenotypes from full VALI susceptibility by SS:BN16 to the minimal VALI susceptibility by SS:BN20 underscores two basic concepts. First, as shown in the study by Leikauf et al. (23), VALI is being controlled by more than one gene and is therefore a polygenic or at least oligogenic syndrome. If VALI was controlled by a single dominant gene, then each consomic strain should demonstrate a single discrete sensitive or resistant phenotype (i.e., all have unidirectional and equal levels of changes in BAL cells and protein). Second, the expected consomic phenotypes also reinforce our probe set density-based chromosomal distribution method that we have employed in driving a consomic selection. Interestingly, the small differences in the probe set ratios between chromosomes correlated with dramatic magnitude differences in phenotypes between the selected consomic strains (i.e., BAL cells in SS:BN16 vs. SS:BN20). Although it is appealing to consider this correlation as an additional strength in the mapping method, this lack of prediction in magnitude can also be interpreted as a limitation given the absence of qualitative differentiation between probe sets in the method that may have accounted for the large phenotypic differences. Despite the possibility that the trend can be attributed to random chance, the general trend of mapped probe densities that rank chromosomal contributions parallel the spectrum of the four consomic phenotypes in our VALI model. We, therefore, present a unique example of a computable systems biology approach to genomic and biomedical research.
Unlike previous studies of VALI genomics and genetics, we chose to employ an inbred consomic rodent strain approach, which minimizes the background noise, as the chromosome (or, in the case of congenics, the portion of a chromosome) of interest represents the only significant difference in the genome between the parental and progeny strains due to selective breeding and rigorous genetic testing. Unlike traditional linkage studies with segregating crosses (as described by Leikauf et al., Ref. 23), the consomic approach provides greater statistical power to detect linkage because of this fixed genetic background. For the same reason, consomic approaches can assign traits like the VALI phenotype to chromosomes with confidence (8). Other advantages to utilizing a consomic approach to study VALI in future genetic studies include the ability to rapidly generate congenic strains that can narrow the region of interest on these particular chromosomes (e.g., chromosome 2, 13, or 16). As the region of interest is narrowed with congenic strains, the list of potential genes is also reduced (8).
An important caveat for consideration is that the consomic approach may include the possibility of gene-gene interactions that may contribute to the VALI phenotype. For example, the difference of chromosome 2 (or for any chromosome introgression) between SS and SS:BN2 can, through extensive interchromosomal gene-gene interactions, result in differential expression of genes on many chromosomes. Collectively, this may contribute to the changes in phenotypes (e.g., an intermediate phenotype) caused by the substitution of chromosome 2, such as a reduction of BAL cell counts but an increase in BAL protein. These gene-gene interactions need to be considered before inferring any strong causal relationships between the particular chromosomes or their residing genes and the VALI phenotype. However, the development of a consomic library by the Medical College of Wisconsin allows the rapid construction of multiple chromosomal substitution models that can then be used to study these gene-gene interactions involved with complex traits (8).
In summary, our results suggest that specific BN chromosomes (2, 13, and 16) containing VALI candidate genes confer increased sensitivity to HTV ventilation (like chromosomes 2, 13, and 16). Thus, the microarray-SAM approach should be considered as a potentially valuable tool for the identification of candidate genes, while also driving a consomic screen in a time- and resource-efficient manner to study the chromosomal contribution in a variety of complex disease models such as VALI. Further analysis of select candidate genes by additional single nucleotide polymorphism discovery and mid- or high-level throughput genotyping and congenic screens of the highlighted chromosomes (2, 13, and 16) from this study will undoubtedly provide important insights into the genetic basis for ALI susceptibility and severity. The explosion in genomic discovery approaches is generating novel mechanistic insights into complex ALI pathobiological processes and may allow us to achieve the ultimate goal of translating this knowledge to the critical care settings.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nicholas Shank for art graphic assistance.
GRANTS
This work was supported by the National Institutes of Health Acute Lung Injury SCCOR Grant HL-073994 (J. G. N. Garcia), National Heart, Lung, and Blood Institute Grant T32-HL-07381 (A. A. Desai), the Lowell Coggeshall Endowment (J. G. N. Garcia), and the Parker B Francis Foundation (J. R. Jacobson).
REFERENCES
- 1.Adhikari N, Burns KE, Meade MO. Pharmacologic treatments for acute respiratory distress syndrome and acute lung injury: systematic review and meta-analysis. Treat Respir Med. 2004;3:307–328. doi: 10.2165/00151829-200403050-00005. [DOI] [PubMed] [Google Scholar]
- 2.Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med. 2001;29:1303–1310. doi: 10.1097/00003246-200107000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Becker KG, Hosack DA, Dennis G, Jr, Lempicki RA, Bright TJ, Cheadle C, Engel J. PubMatrix: a tool for multiplex literature mining. BMC Bioinformatics. 2003;4:61. doi: 10.1186/1471-2105-4-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernard GR, Vincent JL, Laterre PF, LaRosa SP, Dhainaut JF, Lopez-Rodriguez A, Steingrub JS, Garber GE, Helterbrand JD, Ely EW, Fisher CJ., Jr Efficacy and safety of recombinant human activated protein C for severe sepsis. N Engl J Med. 2001;344:699–709. doi: 10.1056/NEJM200103083441001. [DOI] [PubMed] [Google Scholar]
- 5.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 6.Choi WI, Quinn DA, Park KM, Moufarrej RK, Jafari B, Syrkina O, Bonventre JV, Hales CA. Systemic microvascular leak in an in vivo rat model of ventilator-induced lung injury. Am J Respir Crit Care Med. 2003;167:1627–1632. doi: 10.1164/rccm.200210-1216OC. [DOI] [PubMed] [Google Scholar]
- 7.Copland IB, Kavanagh BP, Engelberts D, McKerlie C, Belik J, Post M. Early changes in lung gene expression due to high tidal volume. Am J Respir Crit Care Med. 2003;168:1051–1059. doi: 10.1164/rccm.200208-964OC. [DOI] [PubMed] [Google Scholar]
- 8.Cowley AW., Jr Genomics and homeostasis. Am J Physiol Regul Integr Comp Physiol. 2003;284:R611–R627. doi: 10.1152/ajpregu.00567.2002. [DOI] [PubMed] [Google Scholar]
- 9.Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet. 2002;31:19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- 10.Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dreyfuss D, Basset G, Soler P, Saumon G. Intermittent positive-pressure hyperventilation with high inflation pressures produces pulmonary microvascular injury in rats. Am Rev Respir Dis. 1985;132:880–884. doi: 10.1164/arrd.1985.132.4.880. [DOI] [PubMed] [Google Scholar]
- 12.Gao L, Grant A, Halder I, Brower R, Sevransky J, Maloney JP, Moss M, Shanholtz C, Yates CR, Meduri GU, Shriver MD, Ingersoll R, Scott AF, Beaty TH, Moitra J, Ma SF, Ye SQ, Barnes KC, Garcia JG. Novel polymorphisms in the myosin light chain kinase gene confer risk for acute lung injury. Am J Respir Cell Mol Biol. 2006;34:487–495. doi: 10.1165/rcmb.2005-0404OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia JGN, Moreno Vinasco L. Genomic insights into acute inflammatory lung injury. Am J Physiol Lung Cell Mol Physiol. 2006;291:L1113–L1117. doi: 10.1152/ajplung.00266.2006. [DOI] [PubMed] [Google Scholar]
- 14.Gong MN, Wei Z, Xu LL, Miller DP, Thompson BT, Christiani DC. Polymorphism in the surfactant protein-B gene, gender, and the risk of direct pulmonary injury and ARDS. Chest. 2004;125:203–211. doi: 10.1378/chest.125.1.203. [DOI] [PubMed] [Google Scholar]
- 15.Goodman RB, Pugin J, Lee JS, Matthay MA. Cytokine-mediated inflammation in acute lung injury. Cytokine Growth Factor Rev. 2003;14:523–535. doi: 10.1016/s1359-6101(03)00059-5. [DOI] [PubMed] [Google Scholar]
- 16.Grigoryev DN, Finigan JH, Hassoun P, Garcia JG. Science review: searching for gene candidates in acute lung injury. Crit Care. 2004;8:440–447. doi: 10.1186/cc2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grigoryev DN, Ma SF, Irizarry RA, Ye SQ, Quackenbush J, Garcia JG. Orthologous gene-expression profiling in multi-species models: search for candidate genes. Genome Biol. 2004;5:R34. doi: 10.1186/gb-2004-5-5-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irizarry R, Gautier L, Cope L. An R package for analyses of Affymetrix oligonucleotide arrays. In: Parmigiani G, Garrett ES, Irizarry R, Zeger SL, editors. The Analysis of Gene Expression Data: Methods and Software. Springer; New York: 2003. [Google Scholar]
- 19.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobson JR, Barnard JW, Grigoryev DN, Ma SF, Tuder RM, Garcia JG. Simvastatin attenuates vascular leak and inflammation in murine inflammatory lung injury. Am J Physiol Lung Cell Mol Physiol. 2005;288:L1026–L1032. doi: 10.1152/ajplung.00354.2004. [DOI] [PubMed] [Google Scholar]
- 21.Kamat PP, Slutsky A, Zhang H, Bechara RI, Brown LA, Garcia RC, Joshi PC, Kershaw CD, Guidot DM. Mechanical ventilation exacerbates alveolar macrophage dysfunction in the lungs of ethanol-fed rats. Alcohol Clin Exp Res. 2005;29:1457–1465. doi: 10.1097/01.alc.0000175010.25558.8c. [DOI] [PubMed] [Google Scholar]
- 22.Khatri P, Draghici S, Ostermeier GC, Krawetz SA. Profiling gene expression using onto-express. Genomics. 2002;79:266–270. doi: 10.1006/geno.2002.6698. [DOI] [PubMed] [Google Scholar]
- 23.Leikauf GD, McDowell SA, Wesselkamper SC, Hardie WD, Leikauf JE, Korfhagen TR, Prows DR. Acute lung injury: functional genomics and genetic susceptibility. Chest. 2002;121:70S–75S. doi: 10.1378/chest.121.3_suppl.70s. [DOI] [PubMed] [Google Scholar]
- 24.Lin Z, Pearson C, Chinchilli V, Pietschmann SM, Luo J, Pison U, Floros J. Polymorphisms of human SP-A, SP-B, and SP-D genes: association of SP-B Thr131Ile with ARDS. Clin Genet. 2000;58:181–191. doi: 10.1034/j.1399-0004.2000.580305.x. [DOI] [PubMed] [Google Scholar]
- 25.Lopez A, Yong S. Injury versus inflammatory response in the lungs of rats intratracheally inoculated with bacterial lipopolysaccharide. Am J Vet Res. 1986;47:1287–1292. [PubMed] [Google Scholar]
- 26.Ma SF, Grigoryev DN, Taylor AD, Nonas S, Sammani S, Ye SQ, Garcia JG. Bioinformatic identification of novel early stress response genes in rodent models of lung injury. Am J Physiol Lung Cell Mol Physiol. 2005;289:L468–L477. doi: 10.1152/ajplung.00109.2005. [DOI] [PubMed] [Google Scholar]
- 27.Marshall RP, Webb S, Hill MR, Humphries SE, Laurent GJ. Genetic polymorphisms associated with susceptibility and outcome in ARDS. Chest. 2002;121:68S–69S. doi: 10.1378/chest.121.3_suppl.68s. [DOI] [PubMed] [Google Scholar]
- 28.Meyer N, Garcia JG. Wading in the genomic pool to unravel acute lung injury genetics. Proc Am Thorac Soc. 2007;1:69–76. doi: 10.1513/pats.200609-157JG. [DOI] [PubMed] [Google Scholar]
- 29.Morse D, Choi AM. Heme oxygenase-1: from bench to bedside. Am J Respir Crit Care Med. 2005;172:660–670. doi: 10.1164/rccm.200404-465SO. [DOI] [PubMed] [Google Scholar]
- 30.New England Journal of Medicine Ventilation with lower tidal volumes as compared with traditional tidal volumes for acute lung injury, and the acute respiratory distress syndrome. The Acute Respiratory Distress Syndrome Network. N Engl J Med. 2000;342:1301–1308. doi: 10.1056/NEJM200005043421801. [DOI] [PubMed] [Google Scholar]
- 31.Nonas S, Miller I, Kawkitinarong K, Chatchavalvanich S, Gorshkova I, Bochkov VN, Leitinger N, Natarajan V, Garcia JG, Birukov KG. Oxidized phospholipids reduce vascular leak and inflammation in rat model of acute lung injury. Am J Respir Crit Care Med. 2006;173:1130–1138. doi: 10.1164/rccm.200511-1737OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parsons PE. Mediators and mechanisms of acute lung injury. Clin Chest Med. 2000;21:467–476. doi: 10.1016/s0272-5231(05)70159-3. [DOI] [PubMed] [Google Scholar]
- 33.Patterson CE, Rhoades RA, Garcia JG. Evans blue dye as a marker of albumin clearance in cultured endothelial monolayer and isolated lung. J Appl Physiol. 1992;72:865–873. doi: 10.1152/jappl.1992.72.3.865. [DOI] [PubMed] [Google Scholar]
- 34.Prabhakaran P, Ware LB, White KE, Cross MT, Matthay MA, Olman MA. Elevated levels of plasminogen activator inhibitor-1 in pulmonary edema fluid are associated with mortality in acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2003;285:L20–L28. doi: 10.1152/ajplung.00312.2002. [DOI] [PubMed] [Google Scholar]
- 35.Ranieri VM, Suter PM, Tortorella C, De Tullio R, Dayer JM, Brienza A, Bruno F, Slutsky AS. Effect of mechanical ventilation on inflammatory mediators in patients with acute respiratory distress syndrome: a randomized controlled trial. JAMA. 1999;282:54–61. doi: 10.1001/jama.282.1.54. [DOI] [PubMed] [Google Scholar]
- 36.Rubenfeld GD, Caldwell E, Peabody E, Weaver J, Martin DP, Neff M, Stern EJ, Hudson LD. Incidence and outcomes of acute lung injury. N Engl J Med. 2005;353:1685–1693. doi: 10.1056/NEJMoa050333. [DOI] [PubMed] [Google Scholar]
- 37.Slutsky AS. Lung injury caused by mechanical ventilation. Chest. 1999;116:9S–15S. doi: 10.1378/chest.116.suppl_1.9s-a. [DOI] [PubMed] [Google Scholar]
- 38.Slutsky AS, Tremblay LN. Multiple system organ failure. Is mechanical ventilation a contributing factor? Am J Respir Crit Care Med. 1998;157:1721–1725. doi: 10.1164/ajrccm.157.6.9709092. [DOI] [PubMed] [Google Scholar]
- 39.Takala A, Jousela I, Takkunen O, Kautiainen H, Jansson SE, Orpana A, Karonen SL, Repo H. A prospective study of inflammation markers in patients at risk of indirect acute lung injury. Shock. 2002;17:252–257. doi: 10.1097/00024382-200204000-00002. [DOI] [PubMed] [Google Scholar]
- 40.Takata M, Abe J, Tanaka H, Kitano Y, Doi S, Kohsaka T, Miyasaka K. Intraalveolar expression of tumor necrosis factor-alpha gene during conventional and high-frequency ventilation. Am J Respir Crit Care Med. 1997;156:272–279. doi: 10.1164/ajrccm.156.1.9607072. [DOI] [PubMed] [Google Scholar]
- 41.Tremblay L, Valenza F, Ribeiro SP, Li J, Slutsky AS. Injurious ventilatory strategies increase cytokines and c-fos m-RNA expression in an isolated rat lung model. J Clin Invest. 1997;99:944–952. doi: 10.1172/JCI119259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tremblay LN, Miatto D, Hamid Q, Govindarajan A, Slutsky AS. Injurious ventilation induces widespread pulmonary epithelial expression of tumor necrosis factor-alpha and interleukin-6 messenger RNA. Crit Care Med. 2002;30:1693–1700. doi: 10.1097/00003246-200208000-00003. [DOI] [PubMed] [Google Scholar]
- 43.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Villar J, Flores C, Mendez-Alvarez S. Genetic susceptibility to acute lung injury. Crit Care Med. 2003;31:S272–S275. doi: 10.1097/01.CCM.0000057903.11528.6D. [DOI] [PubMed] [Google Scholar]
- 45.Vreugdenhil HA, Haitsma JJ, Jansen KJ, Zijlstra J, Plotz FB, Van Dijk JE, Lachmann B, Van Vught H, Heijnen CJ. Ventilator-induced heat shock protein 70 and cytokine mRNA expression in a model of lipopolysaccharide-induced lung inflammation. Intensive Care Med. 2003;29:915–922. doi: 10.1007/s00134-003-1747-6. [DOI] [PubMed] [Google Scholar]
- 46.Ware LB, Matthay MA. The acute respiratory distress syndrome. N Engl J Med. 2000;342:1334–1349. doi: 10.1056/NEJM200005043421806. [DOI] [PubMed] [Google Scholar]
- 47.Webb HH, Tierney DF. Experimental pulmonary edema due to intermittent positive pressure ventilation with high inflation pressures. Protection by positive end-expiratory pressure. Am Rev Respir Dis. 1974;110:556–565. doi: 10.1164/arrd.1974.110.5.556. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.