Dear Editor,
Stem cell niche is a specialized microenvironment crucial for stem cell self-renewal. Drosophila testis contains two types of stem cells: germline stem cells (GSCs) and somatic cyst stem cells (CySCs), providing a good model for studying niche-stem cell interaction1. At the apex of testis tube, non-mitotic somatic cells form a tightly folded structure called the hub. About 10 GSCs, each accompanied by 2 CySCs, make direct contact with the hub cells to form a rose-like structure (Supplementary information, Figure S1A). CySCs rely on the hub cells for niche signals, while GSCs are maintained by niche signals from both the hub cells and CySCs1.
Bone morphogenetic protein (BMP) signaling is critical for GSC maintenance2, while Janus kinase-signal transducer and activator of transcription (JAK-STAT) signaling functions in both CySCs and GSCs. STAT activation in CySCs leads to the expression of zfh-13 and chinmo4, two factors critical for CySC self-renewal. JAK-STAT signaling does not directly regulate the self-renewal of GSCs, but governs GSC-niche adhesion5. Although Zfh-1 is a direct target of activated STAT, eliminating STAT activity only decreased but did not abolish Zfh-1 expression3, suggesting that a paralleled pathway(s) may be involved in the regulation of Zfh-1 expression. In the course of immunostaining adult flies expressing a hh enhancer trap, hh-lacZ6, we observed a strong hh-lacZ staining at the apex of testis (Supplementary information, Figure S1), which is consistent with a previous observation that hh-lacZ was expressed in the hub cells6, suggesting a potential function of Hedgehog (Hh) signaling in the testis.
We examined the expression of several core components of the Hh pathway. Hh is expressed in the hub cells as revealed by hh-lacZ and Hh stainings (Supplementary information, Figure S1), which is consistent with a recent finding from Michel M et al.7. The stainings of Ci-lacZ, Ci, ptc-lacZ and Smo largely coincide with that of Zfh-1 expression (Supplementary information, Figure S1). Immunostainings and real-time PCR analyses strongly suggest that Ci, Ptc and Smo are enriched in cyst cells (Supplementary information, Figure S1 and Data S1). We used different approaches to activate or inactivate Hh signaling and used Ptc expression as a readout of Hh pathway activity, as Ptc itself is a target of Hh signaling. The results strongly argue for the presence of a functional Hh pathway in Drosophila testis (Supplementary information, Figure S2).
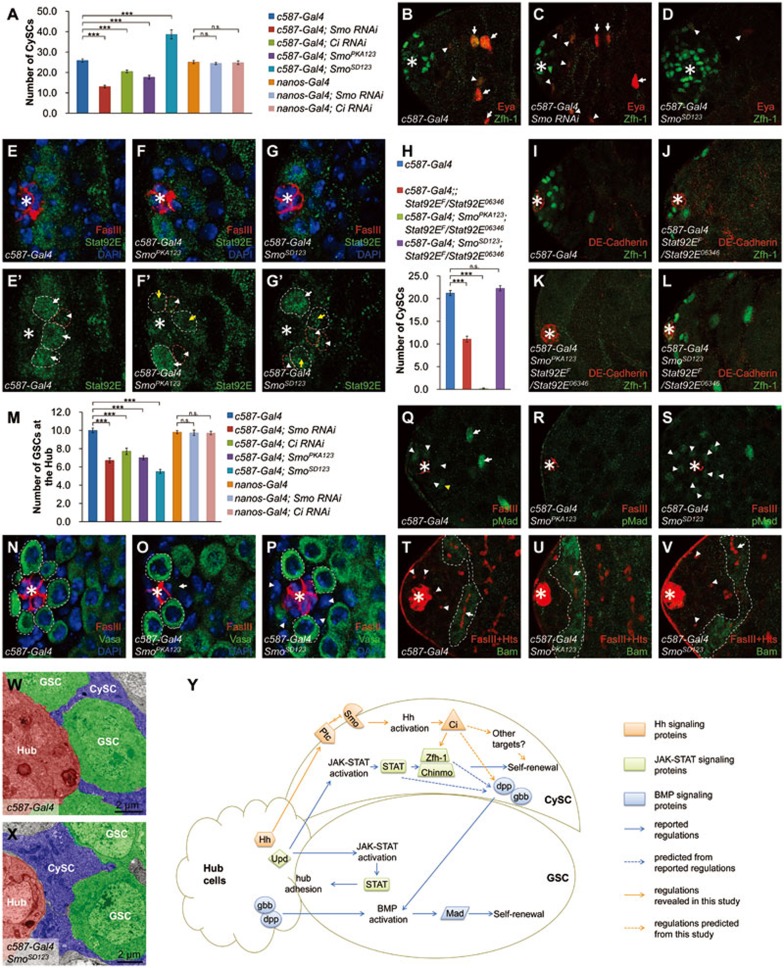
To test whether Hh signaling functions in CySCs, we quantified the number of CySCs by calculating Zfh-1-positive cells under different Hh signaling activities (Supplementary information, Data S1). Smo RNAi, Ci RNAi or the ectopic expression of SmoPKA123 (Smo dominant negative form) driven by cyst cell-specific c587-Gal4 significantly reduced CySC number, whereas the expression of SmoSD123 (Smo active form) in cyst cells significantly increased CySC number (Figure 1A). By contrast, knockdown of Ci or Smo using RNAi driven by germline cell-specific nanos-Gal4, did not significantly affect CySC number (Figure 1A), suggesting that Hh signaling is required in cyst cells but not in germline cells to regulate CySC number. Cyst differentiation marker Eyes absent (Eya) appeared closer to the hub cells in c587-Gal4; Smo RNAi testis and further away from the hub cells in c587-Gal4; SmoSD123 testis (Figure 1B-1D), indicating that Hh signaling inhibits or delays CySC differentiation, and maintains the stemness of CySCs. Similar trends were also observed when Hh signaling activity was manipulated through altering Hh or Ptc expression level (Supplementary information, Figure S3). In addition, Smo3 and PtcIIW null alleles were used to inactivate and activate Hh signaling, respectively, in mosaic analysis with a repressible cell marker (MARCM). The results showed that Smo3 mutant CySCs disappeared rapidly, while PtcIIW mutant CySCs were found in all testes (Supplementary information, Table S1, Figures S4 and S5), which is consistent with the recent findings by others7.
Figure 1.
Dual roles of Hh signaling in the regulation of somatic stem cell self-renewal and germline stem cell maintenance in Drosophila testis. (A) The CySC numbers are counted in the indicated testes. (B-D) Indicated testes are immunostained to show Zfh-1 (green) and Eya (red). Eya-positive cells are marked by arrowheads (weak) and arrows (strong). (E-G') Indicated testes are immunostained to show FasIII (red, to indicate the hub cells), Stat92E (green) and DAPI (blue). GSCs are outlined by white dashed lines and CySCs are outlined by red dashed lines. Arrowheads indicate Stat92E stainings in CySCs. Arrows indicate Stat92E staining in GSCs (white, strong signals; yellow, weak signals). (H) The numbers of CySCs are counted in the indicated testes. (I-L) Representative testes from H are shown with Zfh-1 (green) and DE-Cadherin (red, to indicate the hub cells). (M) The numbers of GSCs at the hub are counted in the indicated testes. (N-P) Indicated testes are immunostained to show Vasa (green), FasIII (red) and DAPI (blue). GSCs are outlined by dashed lines. Arrow indicates an “empty seat” and arrowheads show that GSCs are replaced by non-germline cells. (Q-S) Indicated testes are immunostained to show pMad (green) and FasIII (red). (T-V) Indicated testes are immunostained to show Bam (green) and FasIII plus Hts (red). Round or branched fusomes are marked by arrowheads and arrows, respectively. Bam expression regions are outlined with dashed lines. (W, X) TEM images to show the ultrastructure of testis niche. (Y) A schematic model showing that Hh signaling coordinates with JAK-STAT and BMP signaling to maintain the homeostasis of testis niche. All the apexes of testes are positioned to the left and asterisks indicate the hub cells. Statistics of stem cell number are shown as Mean ± SEM, n ≥10, P-values were obtained by student's t-test between two groups (*** P <0.001, n.s., not significant).
To test the relationship between Hh and JAK-STAT signaling pathways, we examined the level of Stat92E, the only Drosophila STAT protein, under different Hh signaling activities by immunostaining. Stat92E was expressed mainly in GSCs (arrows in Figure 1E') and weakly in CySCs (arrowheads in Figure 1E'). The expression of SmoPKA123 or SmoSD123 driven by c587-Gal4 did not affect the weak staining of Stat92E in CySCs (Figure 1E-1G'), indicating that Hh signaling may not be involved in the regulation of Stat92E expression in CySCs. We then employed a series of genetic interaction experiments (Supplementary information, Data S1) to further clarify whether the regulation of Zfh-1 by Hh signaling depends on Stat92E. Two days after temperature shift, c587-Gal4; Stat92EF/Stat92E06346 flies, which express a temperature-sensitive version of Stat92E, showed a reduced number of CySCs (Figure 1H and 1J). The CySCs in c587-Gal4; SmoPKA123; Stat92EF/Stat92E06346 flies were almost completely lost (Figure 1H and 1K), while in c587-Gal4; SmoSD123; Stat92EF/Stat92E06346 flies, the CySC number was fully rescued (Figure 1H and 1L). On the other hand, Stat92E overexpression driven by c587-Gal4 partially rescued the Smo RNAi-induced CySC loss (Supplementary information, Figure S6). These results demonstrate that Hh signaling acts in parallel with JAK-STAT signaling to promote CySCs self-renewal, and that enhanced Hh signaling activities can compensate for the loss of Stat92E activity to maintain CySCs.
In Drosophila testis niche, GSCs could be regulated by CySCs through CySC-secreted BMP as a niche signal as well as cell competition between GSCs and CySCs3,8. To determine whether Hh signaling affects GSCs through CySCs, we quantified GSC number after altering Hh activities in cyst cells (Supplementary information, Data S1). Smo RNAi, Ci RNAi and overexpression of SmoPKA123 in cyst cells driven by c587-Gal4 reduced the number of GSCs significantly (Figure 1M). Intriguingly, overexpression of SmoSD123 in cyst cells (Figure 1M) or inducing PtcIIW mutant cyst clones (Supplementary information, Figure S4) also significantly reduced the GSC number. In addition, manipulation of Hh expression in the hub cells led to similar results (Supplementary information, Figure S7). Immunostaining of Vasa revealed that GSCs were partially lost around the hub cells in c587-Gal4; SmoPKA123 or c587-Gal4; SmoSD123 testis (Figure 1N-1P). These observations suggest that a proper amount of Hh signaling activity in the CySCs and hub cells is crucial for GSC maintenance. Smo RNAi or Ci RNAi in germline cells driven by nanos-Gal4 did not cause a significant change in GSC numbers (Figure 1M), which is consistent with our previous conclusion that Hh signaling mainly acts within cyst cells.
We next asked whether the BMP signaling activity in GSCs is affected when the Hh signaling in CySCs is perturbed. We measured the level of phospho-Mad (pMad) to determine the BMP signaling activity in GSCs. Figure 1Q showed a representative normal testis, in which pMad signals were mainly detected in GSCs surrounding the hub cells. Some gonialblasts (yellow arrowhead in Figure 1Q) or TA spermatogonia (arrows in Figure 1Q) also showed pMad stainings9. When Hh signaling activity was inhibited in cyst cells by c587-Gal4; SmoPKA123, the pMad staining was reduced throughout the testis (Figure 1R). Overexpression of SmoSD123 in cyst cells increased the pMad staining around the hub cells (Figure 1S). Manipulation of Hh expression in the hub cells also led to similar results of pMad stainings (Supplementary information, Figure S7). Although the pMad staining varied among individual testes, which is consistent with a previous report9, the overall trend indicates that the BMP signaling activity in GSCs could be positively regulated by the Hh signaling activity in CySCs. Furthermore, we examined the endogenous expression of bag of marbles (bam) gene, which is repressed by Mad in GSCs and is expressed only in differentiated spermatogonia10. Compared with normal testis (dashed lines in Figure 1T), Bam-positive cells were observed at a closer distance to the hub cells in c587-Gal4; SmoPKA123 testis (dashed lines in Figure 1U), indicating that the diminished Hh activity in CySCs caused a premature differentiation of GSCs. The Bam stainings in c587-Gal4; SmoSD123 testis appeared a little further away from the hub cells (dashed lines in Figure 1V), which is consistent with the observed increase of pMad stainings (Figure 1S). Analyses of the branched fusomes by Hu-li tai shao (Hts) staining in Bam-positive cells, another marker for differentiation, also led to a similar conclusion (arrows in Figure 1T-1V). These results suggest that the diminished Hh signaling activity in CySCs leads to a decreased BMP signaling activity in GSCs, which results in a reduction of GSC number and premature differentiation of GSCs.
We noticed that in c587-Gal4; SmoPKA123 testis, an “empty seat” judged by the absence of both Vasa and DAPI stainings could be found around the hub cells (arrow in Figure 1O). In contrast, in c587-Gal4; SmoSD123 testis, the seats of lost GSCs were occupied by non-germline cells, which were Vasa-negative and DAPI-positive (arrowheads in Figure 1P). Given the significant increase of Zfh-1-positive cells in c587-Gal4; SmoSD123 testis (Figure 1D), we hypothesized that these non-germline cells could be Zfh-1-positive CySCs. The results from transmission electron microscope (TEM) supported our hypothesis. In wild-type testis, GSCs are attached with the hub cells broadly (Figure 1W). However, in c587-Gal4; SmoSD123 testis, CySCs occupied the space normally belonging to GSCs and made broad contact with the hub cells, expelling GSCs away from the hub cells (Figure 1X). Taken together, these results suggest that cell competition between CySCs and GSCs is likely the cause for the reduced number of GSCs after the hyperactivation of Hh signaling in cyst cells.
Our study identified that Hh signaling activity mainly acts in CySCs and is critical for CySC maintenance in Drosophila testis, which further confirmed an independent finding published recently7. In addition, we revealed three novel aspects of Hh signaling in the testis (Figure 1Y). First, we have demonstrated that Hh pathway acts in parallel with the JAK-STAT pathway in CySCs to regulate Zfh-1 expression. However, it remains to be investigated whether Hh signaling regulates Zfh-1 directly or indirectly. Second, the Hh signaling activity in CySCs also positively regulates BMP signaling activity in GSCs, and the loss of Hh signaling in CySCs leads to precocious differentiation of GSCs. Third, hyperactivated Hh signaling activity promotes the over-production of CySCs, which outcompete GSCs, expelling GSCs away from the hub cells. Thus, proper levels of Hh signaling activity act to keep a balance between GSCs and CySCs in Drosophila testis niche. In summary, our study provides a novel mechanism, by which Hh signaling coordinates with JAK-STAT and BMP signalings to regulate the balance of different stem cell populations in the Drosophila testis.
Acknowledgments
We thank Drs Erika Matunis and Margaret de Cuevas (Johns Hopkins Univ), Ruth Lehmann (New York Univ), Steve Di Nardo (Univ of Pennsylvania), Erika Bach (New York Univ), Zhi-Chun Lai (Pennsylvania State Univ), Dahua Chen (Institute of Zoology, CAS), Xun Huang (Institute of Genetics and Developmental Biology, CAS), Xinhua Lin (Institute of Zoology, CAS), DSHB, VDRC, NIG and Bloomington Stock Center for reagents and fly stocks.This work was supported by grants from the National Basic Research Program of China (973 Program: 2011CB943902, 2010CB912101 and 2012CB945001), the National Natural Science Foundation of China (30971647, 30971646, 31171414 and 31171394), and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA01010405 and XDA01010406) to YZ and LZ, who are scholars of the Hundred Talents Program of the Chinese Academy of Sciences.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Hub cells derived Hh is received by cyst cells in Drosophila testis.
Ptc responds to different Hh signaling activities in testis.
Hh levels in the hub cells regulate Zfh-1 expression in CySCs.
Loss of Smo induces differentiation of CySCs while loss of Ptc promotes CySCs maintenance.
Smo3 mutant CySCs undergo differentiation.
STAT activation partially rescues the CySC loss by diminished Hh activity.
Hh levels in the hub cells influence GSCs maintenance.
Percent of testes containing one or more CySC clones.
Materials and Methods
References
- Issigonis M, Matunis E. Cell. 2011. p. 994. [DOI] [PubMed]
- Kawase E, Wong MD, Ding BC, et al. Development. 2004. pp. 1365–1375. [DOI] [PubMed]
- Leatherman JL, Dinardo S. Cell Stem Cell. 2008. pp. 44–54. [DOI] [PMC free article] [PubMed]
- Flaherty MS, Salis P, Evans CJ, et al. Dev Cell. 2010. pp. 556–568. [DOI] [PMC free article] [PubMed]
- Leatherman JL, Dinardo S. Nat Cell Biol. 2010. pp. 806–811. [DOI] [PMC free article] [PubMed]
- Forbes AJ, Lin H, Ingham PW, et al. Development. 1996. pp. 1125–1135. [DOI] [PubMed]
- Michel M, Kupinski AP, Raabe I, et al. Development. 2012. pp. 2663–2669. [DOI] [PubMed]
- Issigonis M, Tulina N, de Cuevas M, et al. Science. 2009. pp. 153–156. [DOI] [PMC free article] [PubMed]
- Zheng Q, Wang Y, Vargas E, et al. Dev Biol. 2011. pp. 202–210. [DOI] [PMC free article] [PubMed]
- Chen D, Wang Q, Huang H, et al. Development. 2009. pp. 4133–4142. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Hub cells derived Hh is received by cyst cells in Drosophila testis.
Ptc responds to different Hh signaling activities in testis.
Hh levels in the hub cells regulate Zfh-1 expression in CySCs.
Loss of Smo induces differentiation of CySCs while loss of Ptc promotes CySCs maintenance.
Smo3 mutant CySCs undergo differentiation.
STAT activation partially rescues the CySC loss by diminished Hh activity.
Hh levels in the hub cells influence GSCs maintenance.
Percent of testes containing one or more CySC clones.
Materials and Methods