Abstract
Co-option of cis-regulatory modules has been suggested as a mechanism for the evolution of expression sites during development. However, the extent and mechanisms involved in mobilization of cis-regulatory modules remains elusive. To trace the history of non-coding elements, which may represent candidate ancestral cis-regulatory modules affirmed during chordate evolution, we have searched for conserved elements in tunicate and vertebrate (Olfactores) genomes. We identified, for the first time, 183 non-coding sequences that are highly conserved between the two groups. Our results show that all but one element are conserved in non-syntenic regions between vertebrate and tunicate genomes, while being syntenic among vertebrates. Nevertheless, in all the groups, they are significantly associated with transcription factors showing specific functions fundamental to animal development, such as multicellular organism development and sequence-specific DNA binding. The majority of these regions map onto ultraconserved elements and we demonstrate that they can act as functional enhancers within the organism of origin, as well as in cross-transgenesis experiments, and that they are transcribed in extant species of Olfactores. We refer to the elements as ‘Olfactores conserved non-coding elements’.
INTRODUCTION
The sequencing of a large number of vertebrate genomes has enabled the identification of conserved non-coding elements (CNEs) that are constrained during evolution. They were shown to act as tissue-specific enhancers mostly associated with transcription factors that are active during development (1–4). Owing to this role, CNEs are thought to play an important role in gene regulatory networks that specify body plans (5). Genes associated with CNEs require complex spatial and temporal cis-regulation, and indeed key developmental genes contain arrays of CNEs in their intergenic and intronic regions (3,4,6). While CNEs are present in several groups of metazoans such as vertebrates, flies and nematodes and are the most conserved sequences within these groups, they have diverged beyond recognition (if they were originally related) among these groups (6). Such enhancers can work in a modular and autonomous way. They can be active and maintain their specificities regardless of the genetic background, such as associated promoters or genes, and can work in combination with other enhancers from different genomic contexts (7–9). Finally, they can also be functional when transfected from one species to another even if their specificity is not always retained (10–12). Limited evidence also supports the potential activity of enhancers across organisms belonging to different groups. This was shown in the Hox locus for some amphioxus enhancers tested in chicken, mouse and Ciona experiments, as well as Ciona enhancers tested in chicken (13,14).
Most comparative studies to date have focused on mammalian and vertebrate genomes. Two studies (10,15) have been able to find functional CNEs between vertebrates and lancelets (amphioxus), which has been suggested to be the most basal chordate group (16). Recently, an interesting article showed the conservation of both sequence and function of a neural-specific enhancer conserved among human, zebrafish, amphioxus, Saccoglossus, sea urchin and Nematostella (12). However, the sequence of the enhancer is not conserved in tunicate genomes (data not shown; Salvatore D'Aniello, personal communication). Therefore, no CNEs between vertebrates and tunicates have been reported so far, and several studies propose that no such regions exist (2,15). The CNEs discovered in amphioxus act as developmental enhancers and are conserved in syntenic vertebrate regions, which makes the lack of CNEs in tunicates an interesting subject for further studies, given that they are the proposed sister group of vertebrates (17). Regulatory elements may have diversified too much to be recognizable using common comparative genomic strategies, which mainly rely on the identification of collinearly conserved elements found within orthologous loci. It is important to point out that tunicate larvae present the classical chordate body plan but they have greatly diverged at the molecular level leading to the paradox that divergent gene expression programs can lead to similar body plans (18). In addition, in tunicates, many developmental genes and signaling pathways are co-opted differentially as compared with vertebrates [reviewed in (19)], making it difficult to understand where this divergence is embedded.
According to this reasoning, the existence of evolutionary phenomena termed ‘cis-regulatory rewiring’ and ‘enhancer shuffling’ were proposed to account for such differences several times (5,9,20–22). Limited evidence so far relies on individual instances, which have been verified in yeast, insects, sea urchins and tunicates (23–27). Such evidence mostly involve the shift of individual binding sites, a variation that could arise by non-conservation–based mechanisms such as mutations followed by stabilizing selection. Another mechanism for the spreading and shuffling of regulatory regions relies on these regions evolving from transposable elements, which has been observed in mammalian genomes (28). Several studies have previously shown that cis-regulatory elements can shuffle during evolution within the same gene context, i.e. changing location with respect to the gene structure, but maintaining the association to the same gene (26,29,30). A study in plants has also reported an event of promoter shuffling generated by inter-chromosome and subsequent intra-chromosome recombination (31). Kent et al. (32) noticed an unexpected number of small fragments conserved between non-syntenic regions analyzing mammalian genomes, and similarly, in the ENCODE pilot project, the presence of small non-syntenic conserved regions were reported (33). Therefore, non-syntenic rearrangements of conserved (hence potentially functional) sequences did happen during evolution, and they are unlikely to be the mere result of assembly errors, but no further elucidation of their evolution and function has been undertaken so far.
By using an approach that allows the identification of shuffled elements, we have previously demonstrated that the number of functional vertebrate CNEs is significantly higher than reported by using BLAST-like approaches. We identified syntenic rearrangements of regulatory sequences that occurred in vertebrate conserved non-coding regions (29). Our approach has also been successfully adopted in the discovery of elements conserved between vertebrates and the basal chordate amphioxus (10). Now, to evaluate the existence of conserved non-coding sequences between vertebrates and tunicates, we improved and extended our methodology, using progressive alignments, randomizations and a strict false discovery rate (FDR) filtering. We were able to explore the conservation of putative regulatory regions with unprecedented sensitivity and developed a pipeline that led, for the first time, to the discovery of 183 non-coding sequences conserved within Olfactores.
MATERIALS AND METHODS
Data selection
Local installations of the MySQL Ensembl databases version 49 and the relative API (34) were used to extract sequences and annotations from the next Olfactores genomes: Mus musculus, Homo sapiens, Canis familiaris, Danio rerio, Takifugu rubripes, Gasterosteus aculeatus, Oryzias latipes, Ciona intestinalis, Ciona savignyi (see Supplementary Table S1 for information about the used Ensembl core databases and the relative genome builds). Species were divided into three groups according to their phylogenetic classification: mammals, fishes and tunicates. For each group, a representative organism was chosen and its sequences used as the reference genome in VISTA analysis as well as in inter-group analysis. The representative organisms are M. musculus, T. rubripes and C. intestinalis for mammals, fishes and tunicates, respectively. The selection of genes was conducted in each group independently. Basically, using the annotation from Ensembl Compara (35) we selected all the genes containing an homolog classified as ortholog_one2one inside all the species of the group, which led us to collect 14 201, 12 896, 5786 groups of orthologous genes loci for mammals, fishes and tunicates, respectively. For each gene, we extracted the whole genomic repeat-masked sequence containing the transcriptional unit and the flanking sequences up to the preceding and following gene. If there were nested genes present in the locus, they were not taken into consideration to determine the extent of sequence to analyze. The regions were extracted from Ensembl and the 5′–3′ sequence of the locus was stored in a custom database having always the determining gene on the positive strand. The pipeline makes use of custom perl scripts and the Bioperl API (36).
Identification of rCNEs
To define regionally conserved non-coding elements (rCNEs), analyses were conducted independently in each collection of orthologous genes for each group. Global multiple alignments in each group were performed on each collection of homologous genes using MLAGAN (37) with default parameters. The multiple alignments thus obtained were parsed using VISTA (38) and perl scripts with the next parameters according to the following groups:
Mammals and fishes: sliding 20 bp; minimum length 100 bp; minimum identity 80%; minimum length after three species overlap 100 bp.
Tunicate: sliding 20 bp; minimum length 100 bp; minimum identity 60%.
Resulting conserved regions were then filtered stringently to distinguish ‘known genic’ (having evidence of transcription) from ‘non-genic’ (not having evidence of transcription) into Ensembl and to discard redundant sequences. Basically, all the conserved sequences were screened against the annotations of overlapping complementary DNAs (cDNAs), proteins, ESTs and predictions from the Ensembl core, other features and eventually cDNAs databases of the reference organisms. In the case of overlap, the elements were considered ‘genic’ and excluded from the remaining analysis. Finally, in cases in which the upstream region of an analyzed gene coincided with the downstream region of another analyzed gene, rCNEs were counted only once and associated to the locus showing the highest score or the longest transcript in case of equal scores.
Identification of vCNEs
To identify vertebrate conserved non-coding elements (vCNEs), a combined local and multiple alignment strategy was used. This procedure does not look necessarily for collinear elements as the previous one. In the first step, we selected conserved elements between mammals and fish. Each representative mammalian rCNE was aligned against the entire set of representative fish rCNEs by using CHAOS (39) with the next parameters: b = 1, ext = 1, v = 1, co = 10, rsc = 1500, wl = 10, nd = 1 and selecting for segments conserved at least 50 bp sharing an identity of at least 70%. Resulting pairwise alignments were used to extract the corresponding slice from the original rCNE multiple alignments that subsequently were aligned between them using PROLAGAN (37) with default parameters. The cutoff to define the significance of these alignments was determined by randomization analysis. The alignment columns in each rCNE were shuffled so that we maintained the same base composition and identity scores inside the elements but creating not-biologically meaningful sequences. The CHAOS anchoring and PROLAGAN alignment steps were then performed on the randomized rCNEs as reported above and the results used as false positives in the determination of the cutoff for the selection of true vCNEs. The cutoff was calculated on the basis of the overall percentage identity of the multiple alignments to consider significant a percentage of false positives <0.5% (FDR < 0.005) when the same filter is applied to the randomized data.
Identification of oCNEs
The same CHAOS, PROLAGAN, randomization and FDR filtering procedures were used as reported above, aligning the vCNEs with the tunicate rCNEs. The CHAOS analysis was executed between the T. rubripes and C. intestinalis sequences with the next parameters: b = 1, ext = 1, v = 1, co = 10, rsc = 1500, wl = 10, nd = 2 and selecting for segments conserved at least 40 bp sharing an identity of 60% minimum. Finally, the fugu sequences of the resulting 204 mammal/fish/tunicate conserved elements were searched against the repeat-masked sequences of the zebrafish genome by using WU-BLAST with the following parameters: E = 1, W = 5, B = 100, M = 1, N = −1, Q = 2, R = 1, filter = none, hspsepSmax = 10 hspsepQmax = 10, hspmax = 0. All the resulting hits were filtered to present: percentage identity ≥80% and query coverage ≥80%. For each fugu sequence, the top hit was manually chosen, curated and classified as Olfactores conserved non-coding element (oCNE) according to the following criteria in the given order: smaller e-value, presence of the hit on a chromosome, bigger length, higher identity, fugu/zebrafish collinearity, longer contig containing the sequence. We were able to retain 183 of the 204 conserved elements and we focused on this set of conserved regions. It is important to mention that zebrafish was excluded from the initial analysis because it retained many more duplicated loci in respect to other teleosts, and this made the initial 1-to-1 homologous group definition poorly efficient.
Homology analysis
We collected all the Ensembl genes mapped in intervals up to 2 Mb (1 Mb upstream and downstream) around each element in every representative genome per group. For each gene, we collected the evolutionary relationship from Ensembl Compara. We took into consideration the following relationship from the database: ortholog_one2one, ortholog_one2many, ortholog_many2many, between_species_paralog and apparent_ortholog_one2one. For each gene in the interval, we also calculated the number of bystander genes as the number of genes intervening between the gene and the conserved element. We verified within the 1 Mb intervals upstream and downstream of each element, in each pair of species, the presence of evolutionarily related genes as opposed to unrelated genes, taking into account as syntenic conserved oCNEs only those showing a maximum of four bystander genes between the conserved fragment and the closest pair of orthologous genes. For each element, we also measured the number of evolutionarily related pair of genes in the analyzed genomic interval. We also searched for duplicated oCNEs in the genome of M. musculus, D. rerio and C. intestinalis using Blastn with default parameters and selecting only hits showing at least 95% coverage at 95% identity. Results have been manually checked on the Ensembl genome browser in the searched species and in H. sapiens, T. rubripes and C. savignyi.
Aniseed annotation integrations
The next transcript models and annotations were downloaded from the ANISEED database (40): JGI version 1, KYOTOGRAIL2005, KH and ENSEMBL. Data were downloaded from the webpage http://bit.ly/12oO1NL that redirects to the respective archives. Transcript models and annotations were parsed and joined together to form a unique collection, and a MySQL database containing all the information downloaded and generated was used to collect and manage the data using custom perl scripts. Annotations were attached to the data in the pipeline by using the Ensembl transcript ID that represented the ID common to the two sets of data (Ensembl and Aniseed).
oCNEs search in Oikopleura and amphioxus
Oikopleura genome assembly version 3 was downloaded at JGI from http://bit.ly/VPjaD7 with relative annotations from http://bit.ly/TYKrTY and proteome from http://bit.ly/V5Q8yC. Amphioxus genome version 2 was downloaded at JGI from http://bit.ly/12oNxXJ with relative annotations from http://bit.ly/UcbKOg and proteome from http://bit.ly/ZievBL. oCNE multiple alignments containing the sequences of all the analyzed organisms were used to build Hidden Markov Models (HMMs) using the program HMMB from the HMMER tool version 1.8.5 (41). The program HMMFS was then used to search the HMMs against the entire Oikopleura and the amphioxus genomes on both strands. A cutoff score of 20 bits was used to determine whether an oCNE was conserved. This score indicates that a selected match is 220-fold more likely to represent an authentic match than to occur by chance. Putative target genes were considered the genes flanking and overlapping (if any) the regions where HMMs matched the analyzed genomes.
Blast2GO annotation
Protein sequences of the putative target genes in amphioxus and Oikopleura were functionally annotated using the Blast2GO (42) tool with default parameters. We executed the following analysis step: Blastp against NR proteins, Gene Ontology (GO)-mapping, annotation, annotation augmentation, InterProScan. Finally we run the analysis ‘make combined graph’ to count the frequencies and evaluate the scores of the GO classes occurrences in the annotated sequences. We took into consideration the top 10 scoring GO classes with a score higher than 5 at a level higher than 2.
Amphioxus gene pair analysis
We used the proteomes of mouse and sea squirt from Ensembl 49 and the amphioxus proteome version 1 downloaded at JGI from http://bit.ly/ZievBL. We classified all the putative homology relationships between Ciona/amphioxus and mouse/amphioxus proteomes by executing Blastp searches with default parameters but a maximum e-value of 0.001. We took only the best hit (or the best ones in case of equal e-values) showing a minimal coverage of 50% to build a table of putative homologies. We then analyzed the locations of the genes flanking or overlapping oCNEs in mammals and tunicates in the amphioxus genome. The same analysis was repeated 1000 times randomizing the homology associations between Ciona/amphioxus and mouse/amphioxus. Positive elements were considered only those showing at least one pair of associated genes on the same amphioxus scaffold.
Ciona enhancer validations
The Ciona oCNE test fragments were designed cloning the corresponding entire C. intestinalis/C. savignyi conserved block as taken manually from the Ensembl browser (34). Genomic fragments containing the Ciona sequences of the three selected oCNEs were cloned in Gateway constructs (43) upstream of the pFOG basal promoter and the LacZ reporter gene. Each construct has been tested twice and two constructs have been prepared for each element. Ciona electroporated embryos were developed until the early tailbud stage and fixed for Xgal staining. About 100 embryos were inspected for each fragment. The sequences of the primers used are listed in Supplementary Table S2. Each clone was verified by Sanger sequencing.
Zebrafish enhancer validations
The zebrafish oCNE test fragments were designed cloning the corresponding entire zebrafish/mammals conserved block as taken manually from the Vista browser (44). The fragments were amplified from zebrafish genomic DNA and cloned in reporter construct containing zebrafish hsp70 minimal promoter and venus reporter gene (mCherry in transgenesis experiments), using Gateway system (43,45). The Gateway destination vector has been previously modified by introducing medaka Tol2 transposase recognition sequences flanking both sites of the reporter cassette to allow more efficient integration of the transgene into the genome (46). A 570 bp fugu genomic fragment named EK, previously reported (45) to lack enhancer activity, was used as enhancer control. The reporter construct for each element was injected into fertilized zebrafish eggs. The composition of the injection solution was as follows: 15 ng/µl plasmid DNA (reporter construct), 10 ng/µl tol2 in vitro synthesized transposase messenger RNA supplemented with 0.1% Phenol red. Approximately 150–200 (100 in transgenesis experiments) embryos were injected for each reporter construct. The injected zebrafish embryos were analyzed for reporter gene activity between 24 and 28 hpf using Nikon SMZ1500 fluorescent microscope (Olympus ScanR automated microscope in transgenesis experiments). The expression was quantified as percentage of the embryos showing a specific pattern of reporter expression from the total number of normal developing embryos. Oligo sequences used to amplify the zebrafish oCNEs are listed in Supplementary Table S2.
Ultraconserved elements and enhancer browser data overlap
The genomic coordinates of the set of extended ultraconserved elements (UCEs) by Stephen et al. (47) were kindly provided by John S. Mattick. Overlap analyses were performed between the human coordinates of oCNEs and the coordinates of the 5404 vertebrate UCEs. oCNEs overlapping a UCE for at least 50% of their length were considered to be derived from this family of conserved elements. To analyze the overlap of oCNEs with validated UCEs, we downloaded all the elements found in the enhancer browser database (48), together with the functional validation results on 14 November 2012 from http://enhancer.lbl.gov/. The database was composed of 1756 elements. Mouse oCNEs sequences were searched in the downloaded sequences by using Blastn. Fisher exact test was used to test significance for the positive/negative ratio of the complete set of validated conserved sequences and the oCNE overlapping set.
eRNAs overlap
The genomic coordinates related to intergenic transcribed enhancers (eRNAs) were extracted by the supplementary material associated to the article by Kim et al. (49). The dataset contained 5117 single nucleotide positions related to intergenic enhancers of which 2052 were classified as transcribed and 3065 as non-transcribed. oCNEs and vCNEs were considered to overlap eRNAs if their genomic coordinates were overlapping within a 1.5-Kb interval upstream/downstream of the eRNA single-nucleotide position.
Expression analysis
Reverse transcriptase-polymerase chain reaction (RT-PCR) were executed on cDNAs and RNAs extracted by different developmental stages of M. musculus (embryonic day 8.5, 12.5 and adults), D. rerio (dome, shield, 24 hpf and 5 dpf) and C. intestinalis (10 hph). Primers were designed to amplify a fragment of ∼100 bp around each element using Primer3 (50). As positive control, we used primers designed to show the transcription of the following coding transcripts: bActin (D. rerio and M. musculus), otx2 (M. musculus), Ci-ATBF (C. intestinalis). We used the following non-coding transcripts: Ci-Pans (C. intestinalis) (51) and Pans (Mm.221244, the murine homolog of Ci-Pans) (52). All the used controls are known to be expressed at the time of the sampling. As negative control, we used DNAseI-digested RNA that was also used as template for cDNA synthesis. In addition, in C. intestinalis, we also used different combinations of the validated oCNEs forward/reverse primers. The primers used and schemas with the protocols for the reactions can be found in Supplementary Tables S3 and S4.
ENCODE/CSHL Long and Short RNA-seq overlap analysis
Human oCNEs sequences were mapped on the hg19 version of the human genome by using the liftOver tool. A custom perl script was then used to query the public instance of the UCSC MySQL database of the human genome version hg19 at the host genome-mysql.cse.ucsc.edu (53). The script queried all the tables pertaining to the ENCODE/CSHL Long and Short RNA-seq data (54,55) publicly available at the moment of the analysis to test for the overlap of the 183 oCNEs human sequences in every sample. The tables analyzed correspond to 182 samples.
Domain analysis
We collected all the Ensembl genes mapped in intervals up to 2 Mb upstream and downstream around each element in every representative genome per group. For each gene, we collected the domains composition from the Ensembl core annotations and looked for domains common to all the three groups. The same analysis was executed on a set of randomly sampled genomic regions from the three groups of the same dimension as the oCNEs dataset. To avoid methodological biases, the randomly sampled genomic regions were selected in the next way: vCNEs conserved between mammals and fishes were randomly selected and associated to randomly selected Tunicate rCNEs. We considered three different intervals around the conserved elements (500, 1000 and 2000 Kb) and performed the Fisher exact text comparing the proportion of common domains between the real oCNEs set and the random vCNEs/rCNEs associations for each interval and for each domain. P-values were corrected using the Benjamini and Hochberg method.
TFBS analysis
oCNEs sequences from M. musculus, T. rubripes and C. intestinalis were analyzed for their composition in transcription factor binding sites. We used the FAMILY collection of matrices from the 2008 version of the Jaspar4 database (56) together with the TFBS perl API (57). A threshold of 80% was used to map binding sites on sequences. To avoid methodological biases, the same TFBS scanning procedure was performed, for each species, on a set of randomly selected regions of the same length and number of oCNEs extracted from the set of vCNEs (mouse ad fugu) or rCNEs (Ciona) and the results compared using the Fisher exact text. P-values were corrected using the Benjamini and Hochberg method. Only binding sites significantly enriched in all the three organisms tested were kept into consideration.
GO analysis
We considered the association of the conserved elements to the genes determining the genomic regions selected in the data-selection phase. GO functional enrichment analyses were performed on the set of mouse and Ciona genes associated to oCNEs. For the mouse set, we compared the following: (i) functional annotation of the vCNEs with the mammalian rCNEs; and (ii) functional annotation of the vCNEs with the oCNEs by using DAVID (58) and FATIGO (59). The Ciona GO annotations are not present in the above tools nor in the official GO release; therefore, we extracted them from the Aniseed database (40). We then compared the set of genes retaining tunicate rCNEs with the set of genes retaining oCNEs. For each class, we used the Fisher exact text and the P-values were corrected using the Benjamini and Hochberg method.
RESULTS
Discovery of mammalian, fish and tunicate conserved non-coding elements (Olfactores CNEs)
CNEs between vertebrates and tunicates have never been reported so far (15). Therefore, we developed a novel highly sensitive, highly stringent, progressive pipeline to be able to identify the presence of vertebrate CNEs in Ciona (see ‘Material and Methods’ section and Figure 1A). The pipeline used the following combinations of three groups of genomes: (i) Mammals: M. musculus, H. sapiens, C. familiaris; (ii) Fishes: T. rubripes, G. aculeatus, O. latipes; and (iii) Tunicates: C. intestinalis, C. savignyi. We took into consideration all genes for which there were predicted 1-to-1 orthologs within the Ensembl database (34) in all the genomes within each group, which led to the analysis of 14 201, 12 896 and 5786 groups of orthologous loci for mammals, fishes and tunicates, respectively. We began by selecting elements conserved in a collinear manner within each single group using MLAGAN and VISTA (38,60). The results were filtered stringently to discard ‘known genic’ regions (potentially transcribed regions overlapping annotated genes, proteins, EST and predictions) and eliminating redundancies from ‘non-genic’ regions. The analysis produced 92 435, 27 145 and 4525 non-redundant, non-genic, collinearly conserved elements in mammals, fish and tunicates, respectively. We will refer to these initial dataset as rCNEs because they are conserved collinearly within each group. We then proceeded to use these data in a multi-step local and multiple alignment strategy to progressively search for conserved elements among groups, allowing non-collinear conservation and using randomization steps to exclusively select significantly conserved regions with a FDR <0.5% (see ‘Material and Methods’ section) (61). To get sequences conserved in vertebrates, we aligned mammalian rCNEs against fish rCNEs, generating what we will refer to as vertebrate CNEs (vCNEs). We obtained 900 vCNEs, the majority of which overlap the set of CNEs by Woolfe et al. (3). Then, to select sequences significantly conserved across Olfactores, we aligned tunicate rCNEs against the 900 vCNEs, performing a further step of randomizations and FDR filtering (see ‘Material and Methods’ section). The analysis resulted in a final set of 183 oCNEs associated to 91, 93 and 121 genes in mammals, fishes and tunicates, respectively. Table 1 can be used to get a clearer understanding of all the abbreviations most commonly used to refer to conserved elements in this and other articles.
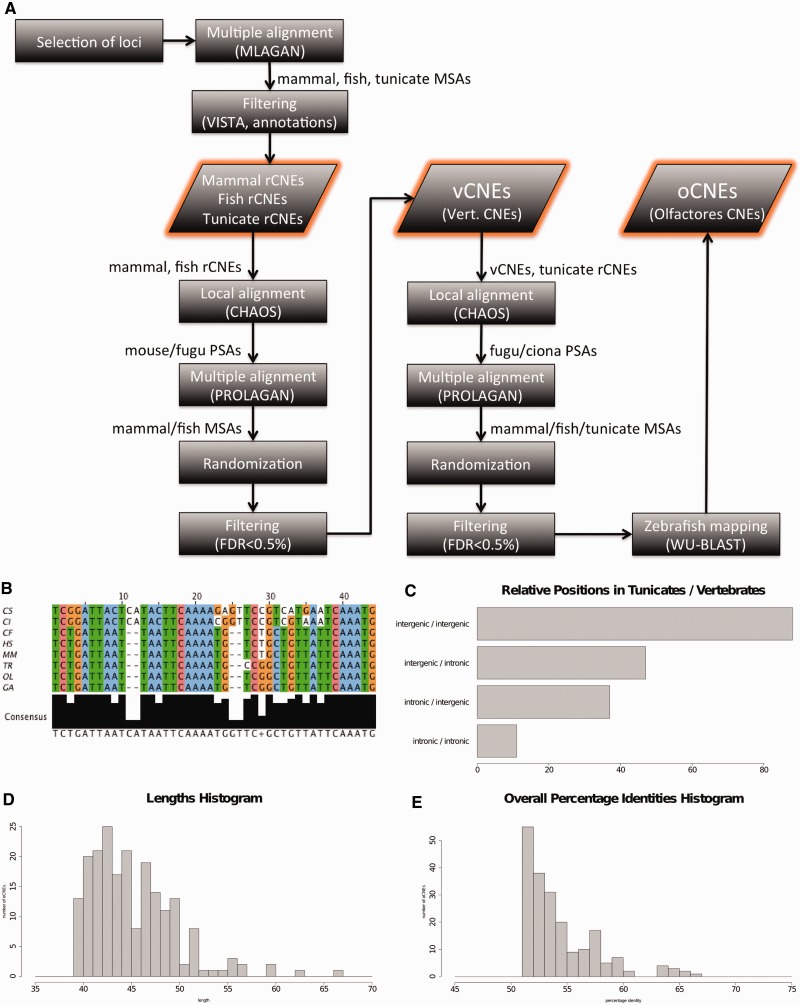
Figure 1.
Description of oCNEs workflow and data: Panel A shows the schema representing the workflow of the pipeline herein presented. In the boxes are indicated the different steps of the procedure, out of the boxes the input and/or output of each step. MSA: multiple sequences alignment. PSA: pairwise sequences alignment. In B is shown an example of the conserved element (oCNE) discovered. Panel C indicates the number of oCNEs classified accordingly to their genomic locations relatively to the associated gene structure in tunicates and vertebrates. The majority of elements are conserved in intergenic regions in both organism groups. Finally, D and E plot the distributions of the length and of the overall percentage identity of the 183 oCNEs.
Table 1.
Abbreviations commonly used for conserved elements
| Abbreviation | Full name | Organismal group | Reference |
|---|---|---|---|
| CNG | Conserved non-genic elements | Mammals | Dermitzakis et al., Nature 2002 |
| UCE | Ultra conserved elements | Mammals, vertebrates | Bejerano et al., Science 2004 |
| CNE | Conserved non-coding elements | vertebrates | Woolfe et al., Plos Biology 2005 |
| SCE | Shuffled conserved elements | Vertebrates | Sanges et al., Genome Biology 2006 |
| PCNE | Phylogenetically conserved non-coding elements | Vertebrates, vertebrates + amphioxus | Hufton et al., Genome Research 2009 |
| rCNE | Regionally conserved non-coding elements | Mammals, fishes, tunicates | This work |
| vCNE | Vertebrate conserved non-coding elements | Vertebrates | This work |
| oCNE | Olfactores conserved non-coding elements | Vertebrates + tunicates | This work |
The table indicates the abbreviations most commonly used to refer to conserved elements in this and other articles. For each acronym, we reported the full text, the group of organisms to which the elements are referring and the first article using it.
The pipeline herein presented progressively joins together groups of species to extract group-specific conserved elements. We focused on the 183 elements conserved among all the Olfactores species considered (see Figure 1B–E for an exemplar element and descriptive charts). Their overall percentage identity (number of identical columns divided by the length of the alignment) spans from 52 to 67% (average 55%). The average length of the elements is 45 bp, and the majority of them are found in intergenic regions in all the analyzed species. Supplementary Table S5 contains all the information about the elements discovered and their sequences.
oCNEs are non-syntenic between vertebrates and tunicates
The 183 oCNEs identified in this study are syntenic among vertebrate loci, but are found in non-syntenic locations in tunicates (i.e. surrounding genes for which the orthologous genes are not found in the corresponding vertebrate locus). It is well known that conserved enhancers can be functional over long distances and that bystander genes can be found between enhancers and their target genes (62,63). To check if oCNEs could be located far from their target genes, we searched for orthologous and/or paralogous genes in regions up to 2 Mb in mouse/fugu for vertebrates and mouse/sea squirt for Olfactores. We verified within 1 Mb intervals upstream and downstream of species, the presence of evolutionarily related genes as opposed to unrelated genes, taking into account as syntenic conserved oCNEs only those showing a maximum of four unrelated genes between the conserved fragment and the orthologous genes. In vertebrates, the majority of the oCNEs are found directly flanking or overlapping orthologous genes and they are also found in prevalence in large syntenic blocks. Only seven elements in mouse show the presence of one unrelated gene and three are separated by two unrelated genes from their putative target gene. In fugu, three elements show one unrelated gene. Overall, >85% of the elements analyzed contain >1 pair of evolutionarily related genes in the analyzed interval, >50% of the elements contain >5 pairs and ∼20% contain >10 pairs. We performed the same check comparing the mouse and the Ciona genomes, and no element could be classified as syntenically conserved. To further verify this finding, we manually screened the genes flanking and overlapping (if any) oCNEs after having integrated the automatically verified Ensembl Ciona annotations with the ones downloaded from the Aniseed database (40). The curated annotations can be found in Supplementary Table S6. We noticed that, using the integrated annotations, a single oCNEs from the whole dataset can be considered as syntenic between vertebrates and ascidians. The element is found in an intron of the FoxP1 gene in both groups. The homology relationship between the two genes was not present in the version of the Ensembl database used in the analyses; therefore, it was classified as non-syntenic. We could not find any other missing relationship.
We also looked for specific duplication of oCNEs elements to check if duplicated elements could be found close to missing orthologous genes. This analysis allowed us to identify 14 duplicated elements of which three are present in all the vertebrate genomes analyzed, two are only found in mammals, two only in fishes, four exclusively in zebrafish (due to additional duplications of loci containing them) and three only in tunicates (see Supplementary Table S7). Elements specifically duplicated in vertebrates, mammals or fishes are associated to paralogous genes, which demonstrate that they were retained after local or whole genomic duplications. In C. intestinalis, on the other hand, the three duplicated elements are found in multiple copies within the same genomic region associated to the same genes. The result in Ciona could be due in some cases to assembly problems, and in fact in two cases the C. savignyi genome contains only a single copy of the same element. One element, however, is present in multiple copies in both genomes.
These results suggest either that oCNEs were eventually shuffled in tunicate genomes or that they were retained after genomic rearrangements and co-opted by different genes. Given the asymmetric design of the starting gene set, for which we used different numbers of genes from each group, we analyzed the number of genes with annotated orthologs, to verify whether the results obtained and the lack of synteny between oCNE containing loci could be due to a lack of inter-group ortholog annotation. Among the total set of 5786 analyzed Ciona gene loci, 3957 (68%) have an annotated ortholog in the set of fish analyzed loci (4107 in mammals), while of the 121 loci containing oCNEs, 61 (50%) have an annotated ortholog in fish (63 in mammals), but no conservation outside the coding exons could be detected between syntenic vertebrate and tunicate orthologous loci. The difference between the proportions is significant (P = 2.1e-05, Fisher exact test) suggesting that our results are not a random sampling of the starting dataset. These results indicate that oCNEs are found in regions showing significantly less annotated orthologous genes.
oCNEs can act as tissue-specific enhancers
CNEs have the ability to act as tissue-specific enhancers. Therefore, to test if oCNEs may carry enhancer activity, we have tested them in sea squirt and in zebrafish. Three genomic DNA fragment containing oCNEs (see Supplementary Table S5 for corresponding id) were chosen to be tested for specific enhancer activity within developing sea squirt and zebrafish embryos. We selected the genomic locus in Ciona containing the highest number of oCNEs. This region is a long intergenic region containing 10 oCNEs, which is found between the ci0100140718 gene (a gene with no annotated homologs in vertebrates, which appears to be a reductase based on protein domain annotation) and a gene named Ci-ATBF. Ci-ATBF is a homeobox transcription factor representing the Ciona homolog of the mammalian ATBF1 gene. It is involved in neuronal differentiation in vertebrates as well as invertebrates (64,65). In Ciona, Ci-ATBF is expressed in mesenchyme, tail epidermis, endoderm, visceral ganglion and nerve cord during development (66). ATBF1 was previously shown to be associated with a cluster of group-specific conserved elements both in vertebrates and in worms (6). The top three most conserved oCNEs from this cluster of 10 were chosen to be validated (Figure 2A).
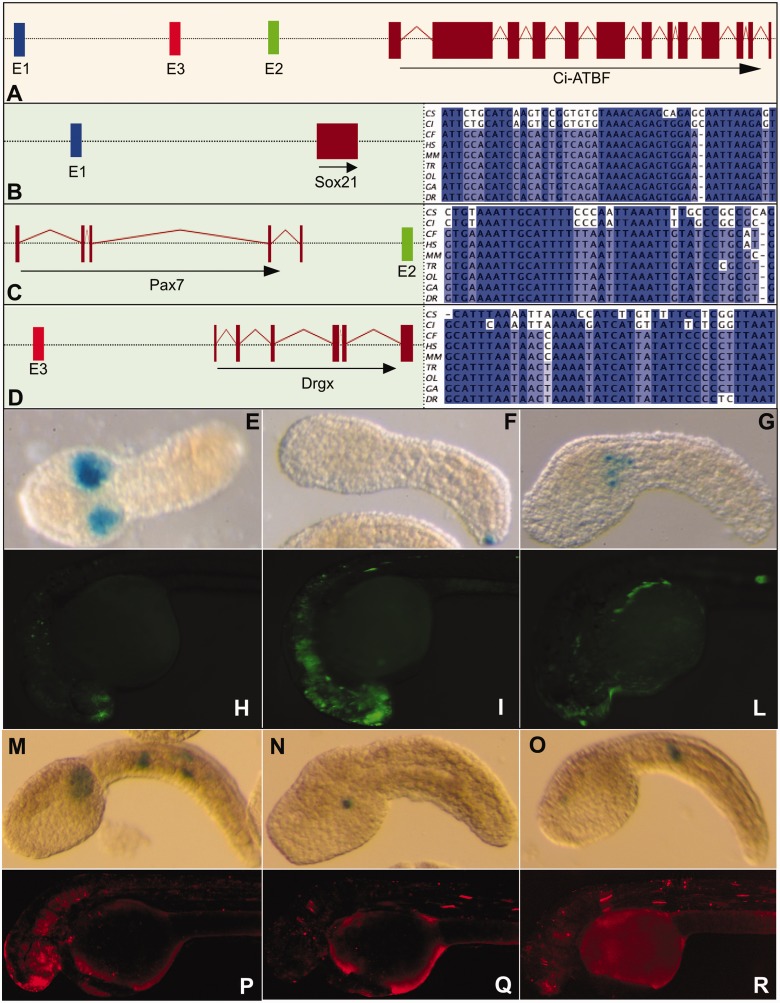
Figure 2.
Functional validation of oCNEs enhancer function: three oCNEs were selected to be validated (E1, E2 and E3). Schemas represent the genomic intervals containing the selected oCNEs and the reciprocal positions of the elements and the associated genes. For clarity purposes the schemas are not respecting a specific scale. Panel A indicates that the three chosen elements are present in the same intergenic region in tunicates associated to the Ci-ATBF gene. In B–D are reported the three distinct vertebrate intervals containing the selected conserved sequences E1, E2, E3 and the respective oCNE alignments in all the analyzed species. Pictures E–G report the most representative expression pattern driven by the elements E1, E2 and E3, respectively, in C. intestinalis. Pictures H, I, L report the most representative expression pattern driven by the elements E1, E2 and E3, respectively, in D. rerio. M–O report the most representative expression pattern driven by the zebrafish elements E1, E2 and E3, respectively, injected in C. intestinalis. Finally P–R report the most representative expression pattern driven by the Ciona elements E1, E2 and E3, respectively, injected in zebrafish embryos.
The first element (E1, id 1351907, 64% overall percentage identity), in mammals, is contained within a known UCE (enhancer browser id 189). This UCE was tested in transgenic mice by Pennacchio et al. (48) and the results in the enhancer browser indicate strong and restricted enhancer function in the neural tube at day 11.5. In mammals and fishes the element is localized in a gene desert upstream of the Sox21 gene known to promote the progression of vertebrate neurogenesis (Figure 2B) (67). The second tested element (E2, id 1353058, 66% overall percentage identity) shows the highest conservation score in vertebrates and is found downstream of the Pax7 gene, which plays a role in neural crest development (Figure 2C) (68). The third element (E3, id 1352705, 60% overall percentage identity) maps upstream of Prrxl1 (Drgx) a transcription factor involved in neuron migration, axonogenesis and nervous system development in vertebrates (Figure 2D). This element is associated with a UCE, which has been tested in mouse and resulted negative at 11.5 days (enhancer browser id 318) (48). Therefore, while these three elements are found inside the same gene desert in Ciona, they are present in three different regions among vertebrates. On the other hand, their genomic organization and the functions of the flanking genes are highly similar, as all elements are localized in intergenic regions flanking a transcription factor gene expressed in the developing neural system. These genes are Sox21, Pax7 and Prrxl1 in vertebrates and Ci-ATBF in tunicates.
Genomic fragments containing the Ciona sequences of the three selected oCNEs were cloned in Gateway constructs (43) upstream of the pFOG basal promoter and the LacZ reporter gene and electroporated in sea squirt embryos (see Figure 2E–G, ‘Material and Methods’ section and Supplementary Table S2). To verify if the elements could be categorized as positive enhancers, we calculated the total percentage of embryos expressing the reporter and used a minimum cutoff of 25% positive embryos as done in similar studies (69). From this analysis, the 600-bp fragment containing the E1 element showed strong enhancer activity exclusively in the mesenchyme (Figure 2E; 61% positive embryos). The 900-bp fragment containing the E2 element also resulted to be a functional enhancer, albeit weaker and in variable tissues (epidermis, muscle, mesenchyme and notochord), where the most representative and specific staining was in two cells at the tip of the tail (Figure 2F; 28% positive embryos). Finally, the 300-bp fragment containing the E3 gave weak mesenchyme staining in a lower number of embryos (20%) and was considered negative. Interestingly, the patterns of expression driven by the E1 and E2 constructs are in good agreement with the expression pattern of Ci-ATBF at the tailbud stage (66).
To evaluate the regulatory potential of these sequences in a vertebrate context, genomic fragments containing the zebrafish sequences of the same oCNEs were inserted in a construct containing a zebrafish hsp70 minimal promoter and the venus reporter gene and microinjected into zebrafish embryos (see ‘Material and Methods’ section and Supplementary Table S2). Subsequently, fluorescence reporter activity was detected to assess their enhancer function (Figure 2H–L). Results from the 500-bp fragment containing E1 showed expression (44% of the embryos) of the reporter mainly in the telencephalic region but also extending posteriorly to the hindbrain in agreement with the expression pattern of sox21 in zebrafish (Figure 2H). The 500-bp fragment containing the E2 element drives broad expression (67%) in the anterior neural tube, more specifically in whole forebrain and hindbrain regions. Additionally, ∼20% of the injected embryos showed enhanced reporter expression in the skeletal muscle, which was not observed with the enhancer control or E1 construct injected embryos, suggesting that in addition to neuronal enhancer activity, the E2-containing fragment possesses a skeletal muscle–enhancer activity in this transient transgenic reporter assay (Figure 2I). The observed expression pattern was similar to that of the pax7a gene, upstream of which E2 is located. The 1100-bp genomic fragment containing E3, on the other hand, showed weak activity similar to the enhancer control construct with no emerging tissue-specific pattern (Supplementary Figure S1 for a complete outcome of the zebrafish experiments), and thus this element was not considered to act as an enhancer, similarly to the case for the corresponding Ciona element (Figure 2L and Supplementary Figure S1). These results show that two out of three genomic fragments containing the selected oCNEs have the ability to work as enhancers both in vertebrates and in tunicates, and their patterns of expression is in agreement with that of neighboring genes. The third element was consistently found not to act as an enhancer in both organisms tested and in mouse via the enhancer browser, suggesting it might require testing in different developmental stages, or that it might have other functions which were not tested. Interestingly, according to the Broad HMM classification based on chromatin states in several cell lines, the corresponding human genomic region is classified in the UCSC genome browser as a poised promoter in ES cells, i.e. presents bivalent histone marks, H3K27me3 and H3K4me1/2 (data not shown).
oCNEs can act as enhancers in cross-transgenesis experiments
We also performed cross-transgenesis experiment to evaluate if the fragments were capable to work in a different background. Ciona fragments were injected in zebrafish, and similarly, zebrafish elements were tested in Ciona. All Ciona elements enhanced the activity of a minimal promoter when tested in zebrafish embryos (Figure 2P–R and Supplementary Figure S2). Conversely, all zebrafish elements showed activity in Ciona embryos (Figure 2M–O and Supplementary Figure S3). The E1 fragments showed the strongest activity in these cross-transgenesis experiments. Interestingly, both Ciona and zebrafish E1 fragment displayed highest activity in fish anterior neural tissue. Ciona and fish E1 also showed overlapping activity in Ciona mesenchyme. Likewise, the Ciona fragment containing E2 showed neuronal and muscle activity in zebrafish experiments, similar to what is observed with zebrafish E2. Thus it seems that the two strongest oCNE enhancer we tested, the E1 and E2 elements, have conserved at least some of their cis-regulatory specificity between the two species. This cis-regulatory activity is, however, differently interpreted in the two organisms, brain in zebrafish versus mesenchyme or muscle in Ciona, possibly as a result of changes in the expression profiles of trans regulators between these two species (18).
oCNEs overlap ultraconserved elements and are enriched in transcribed enhancers
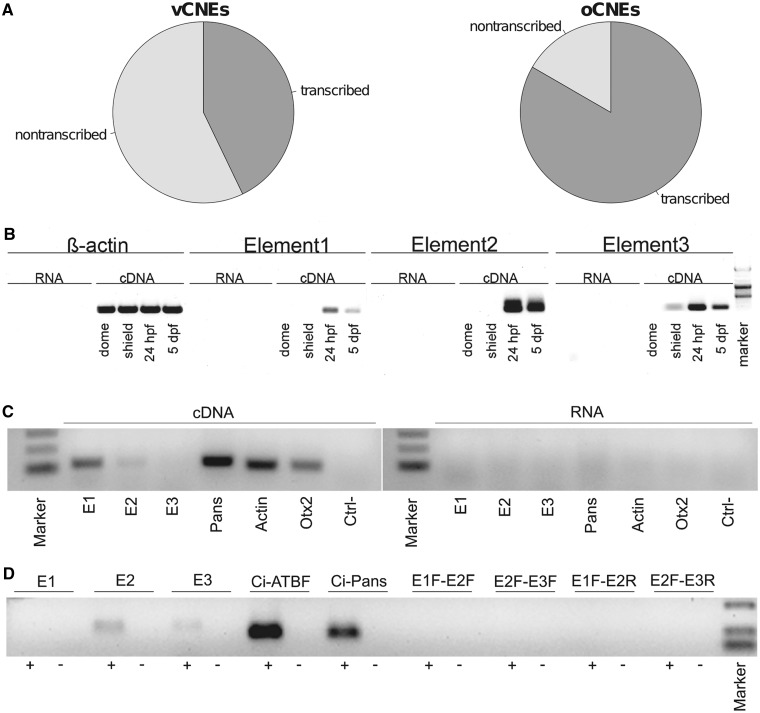
UCEs are extremely conserved non-coding sequences, whose function is not yet completely understood. They were shown to regulate transcription in mammal embryos as well as to be transcribed (48,70). UCEs are mostly associated with developmentally regulated genes and are enriched in gene deserts (2). The observation that two out of three validated genomic fragments containing oCNEs overlap UCEs in vertebrates led us to verify the overall proportion of oCNEs overlapping UCEs. Therefore, we calculated the overlap of the oCNEs with the extended set of 5404 vertebrate UCEs reported by Stephen et al. (47) discovering that the majority of oCNEs overlap known vertebrate UCEs (145 out of 183, ∼80%). The overlap is significantly higher than the overlap between vCNEs and UCEs (499 out of 900, 55%, P = 3.8e-09) pointing out that this is an oCNEs-specific enrichment. It is important to point out that such overlap could also be partially explained by our multi-steps procedure, which progressively selects for the highest conserved segments within each group. However, the usage of stringent parameters in the randomization steps rejects the hypothesis that these elements are conserved merely by chance. Extensive information about the functional validation of UCEs can be found in the enhancer browser (48). The database is composed of 1756 elements of which 50% (887) resulted to be positive as enhancers at the developmental stage tested. We calculated the overlap of these elements with the set of oCNEs. The results show that 85 out of 183 oCNEs overlap a conserved element tested in the enhancer browser. Of these, 66% (56) resulted to be functional enhancers in mouse. The difference between the two proportions is significant (P = 3.9E-03) indicating that oCNEs are enriched for elements found to be functional enhancers in mouse at developmental stage 11.5E. Several studies indicate that enhancers and UCEs can also be transcribed (49,70,71). To verify the possibility that oCNEs could act also at the transcript level, we tested the overlap between them and the published set of transcribed enhancers by Kim et al. (49). They showed that a subset of stimulus-dependent enhancers from mouse cortical neurons also show activity-regulated RNAPII binding, and therefore they are transcribed (49). The total set of intergenic functional enhancers contains 5117 genomic positions; of these, 2052 are also transcribed and are named eRNAs, while 3065 are not transcribed. The overlap between these two sets of enhancers and oCNEs shows that 15 oCNEs overlap eRNAs and only three overlap non-transcribed enhancers (Figure 3A). The differences between the overlaps in the two classes suggest that oCNEs are enriched for eRNAs (P = 0.0078). This is not a bias given by a priori enrichment in the vCNEs group, and indeed the same analysis in vCNEs group yield a similar proportion of conserved elements overlapping eRNAs and non-transcribed enhancers (P = 0.57). This result suggests that oCNEs could be enriched for a specific class of enhancers also able to be transcribed.
Figure 3.
Potential transcription of oCNEs: The oCNEs result to be enriched for eRNAs. Pie-charts in (A) show how vCNEs and oCNEs overlapping enhancers from Kim et al. (49) segregate between the classes of eRNAs and non-transcribed enhancers. Twenty-eight vCNEs overlap enhancers from Kim et al. and ∼40% of them overlap eRNAs. Conversely, 18 oCNEs overlap enhancers from Kim et al. and >80% of them overlap eRNAs. The three oCNEs used for the validation of the enhancer function were also validated for transcriptional activity in D. rerio (B), M. musculus (C) and C. intestinalis (D). Primers were designed to amplify a fragment of ∼100 bp around each element. As positive control, we used the following coding transcripts: bActin (D. rerio and M. musculus), Otx2 (M. musculus), Ci-atbf (C. intestinalis). Non-coding transcripts used were as follows: Ci-Pans (C. intestinalis) (51) and Pans (Mm.221244, the murine homolog of Ci-Pans) (52). All the used controls are known to be expressed at the time of the sampling. As negative control, we used DNAseI-digested RNA (indicated as RNA in B and C and as ‘-’ in D). In C. intestinalis, we also used different combinations of the forward/reverse primers. The absence of signal in the cDNA template PCR is indicative of the absence of genomic DNA contamination in the cDNA preparation demonstrating that the amplicons are real RNA products.
oCNEs can be transcribed
To validate if the identified elements are transcribed, we carried out RT-PCR from RNAs collected at four different stages of zebrafish development. Experiments were executed on the three elements for which we tested already the enhancer function. All the elements were found to be transcribed in a dynamic manner during development. While E1 and E2 appear to be expressed after shield stage, E3 starts to be transcribed at gastrula stage (Figure 3B). All three elements show a peak of transcription at 24 hpf, hinting at a potential role for transcription during the late gastrulation stages. Similar analyses were carried out in mouse as well as in sea squirt. In mouse, we tested expression at 8.5, 12.5 and adult stages. Expression was not detected at 12.5 and adult stages (data not shown), and conversely, expression of E1 and E2 but not E3 was detected at 8.5 (Figure 3C). Transcription of the three elements in C. intestinalis at the tail-bud stage showed expression of elements E2 and E3 (Figure 3D). These findings suggest that the identified oCNEs can be expressed during development in Olfactores and that the transcripts are produced at low levels as already shown for eRNA and more generally non-coding RNAs (49,72). Overlap analyses of zebrafish oCNEs against recently published dataset of RNA-seq data (73,74) did not give significant results, suggesting that their weak expression levels could need higher depth of sequencing to be detected.
To better understand if oCNEs transcripts could be associated to short or long RNAs, we analyzed the overlap of oCNEs with the UCSC human genome browser tracks collecting transcribed contigs from the ENCODE/CHSL RNA-seq data on Long (98 samples, 117 388 194 transcribed contigs) and Short (84 samples, 5 452 981 transcribed contigs) RNAs (53–55). The number of contigs overlapping each element indicates the number of samples in which an oCNEs overlap a transcribed region supporting the putative transcription of the element. Mapping of the oCNEs was compared with the mapping of a similar number of randomly selected elements. The results obtained indicate that 158 oCNEs overlap 4866 RNA contigs from the Long RNA-seq dataset, while 147 random elements overlap 3191 contigs. The difference between the two dataset is significant (P = 2.5E-77). The average number of samples in which each oCNEs result expressed in the Long RNA-seq dataset is ∼30, while it is ∼20 in the random set (Supplementary Figure S4). In the same analysis, using the Short RNA-seq data the oCNEs resulted to map on 18 contigs, while random regions overlap with seven; this difference is not significant (P = 0.08). We thus hypothesize, in the light of these results, that oCNEs are unlikely to be transcribed as short RNAs.
Functional enrichment analyses suggest oCNEs as hubs of homeobox gene regulatory networks
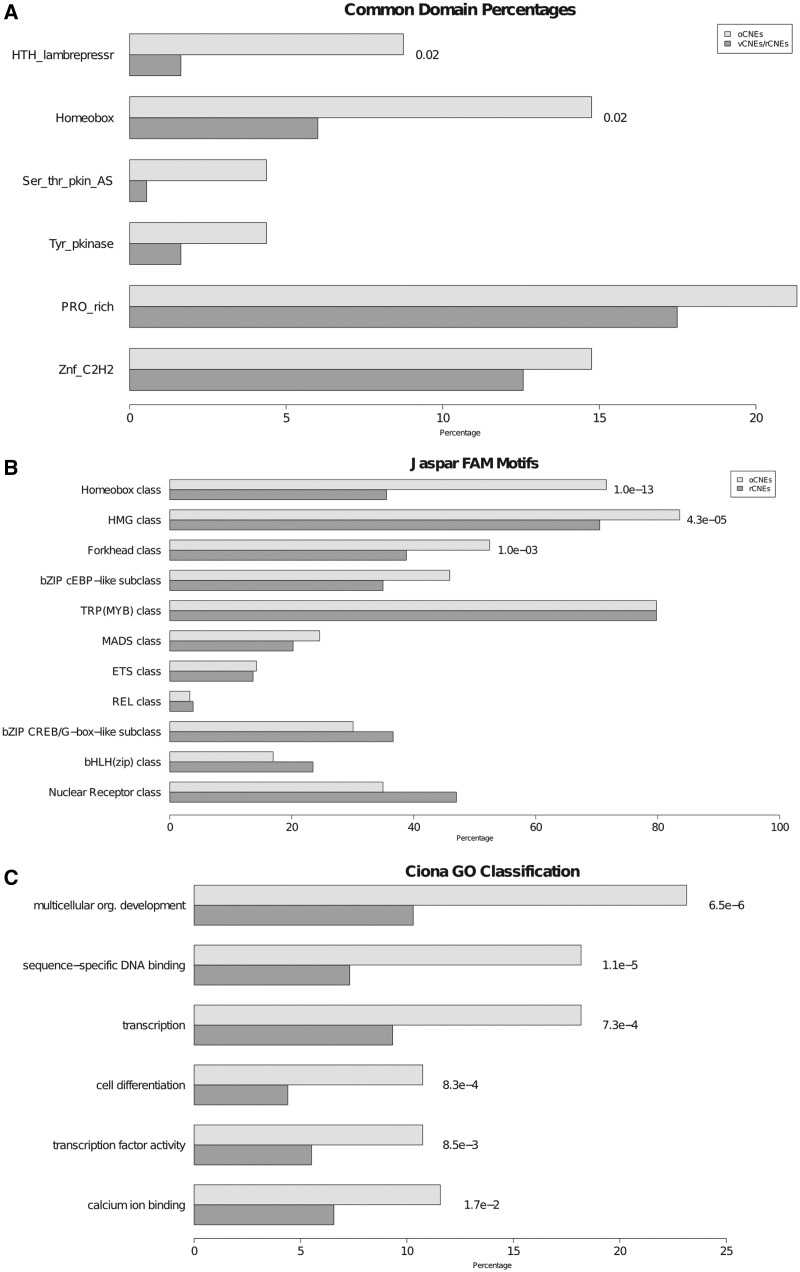
Genes and sequences associated with oCNEs were analyzed to define functional enrichments that could shed light on their specific cellular functions and origins. All the functional associations were analyzed considering as reference ‘universe’ the groups of genes (or sequences) containing at least one rCNE in sea squirt or one vCNE in mouse. Such a strategy is necessary to avoid false enrichments resulting from the fact that these elements are primarily conserved inside each group. First of all, we decided to verify whether genomic regions containing a specific oCNE were enriched for genes containing the same specific protein domains both in vertebrates and tunicates. Therefore, domain enrichment analyses were performed by (i) identifying if and which length interval in mouse and sea squirt showed significantly enriched frequency of common domains; and (ii) checking for the specific significantly enriched domains. The protein domains identified from genes transcribed in genomic intervals containing each oCNE were compared with those identified in randomly paired vCNE/rCNE regions. We performed the analysis over three length intervals around oCNEs, and the significance of the associations decreases proportionally with respect to the extension of the window, disappearing at ∼1 Mb (Supplementary Figure S5), in line with previous observations for the range of action of long-distance enhancers (62). Focusing on a window of 500 kb (adjusted P = 0.03), the common domains resulting significantly enriched in oCNE regions as opposed to random vCNEs/rCNEs pairs are the homeobox (adjusted P = 0.02) and the helix-turn-helix (HTH) lambdarepressor (adjusted P = 0.02), as shown in Figure 4A. The homeobox gene superfamily encodes transcription factors that act as master regulators of development through their ability to activate or repress a diverse range of downstream target genes (75). The HTH domain is a common denominator in basal and specific transcription factors from the three superkingdoms of life and is frequently present in homeobox genes (76).
Figure 4.
Functional enrichment analyses: A shows, for each domain, the percentage of oCNEs (light grey) and vCNEs/rCNEs random couples (dark grey) falling in intergenic regions associated to genes containing the same specific domain in all the species analyzed. Only domains for which the percentage is higher in oCNEs are reported. Adjusted P-values of the differences between the two groups are reported only if significant. Panel B shows, for each Jaspar fam motif, the percentage of C. intestinalis oCNEs (light grey) and C. intestinalis rCNEs (dark grey) containing at least one binding site for the specific motif. Adjusted P-values of the differences between the two groups are reported only if significant. C shows GO enrichments for each GO class associated to genes flanking tunicate oCNEs (light grey) and tunicate rCNEs (dark grey). Only oCNEs-associated significantly enriched classes are reported with the respective adjusted P-values.
Then, to check if oCNEs may indicate a common conserved regulatory mechanism, we performed a similar analysis focused on transcription factor binding site enrichments taking into account as significant only binding sites significantly enriched in all the groups of organisms. To this aim we used the transcription factor binding matrices from the Jaspar Family database (56), which provides generic matrixes for major families of transcription factors. We found common significant enrichments for binding sites recognized by the homeobox (Ciona adjusted P = 1.0E-13), the high mobility group (HMG; Ciona adjusted P = 4.3E-05) and the forkhead (Ciona adjusted P = 1.0E-03) transcription factors classes within oCNEs sequences (see Figure 4B for results in Ciona and Supplementary Table S8 for results in all the tested species). Interestingly, the HMG proteins are a superfamily of nuclear proteins that bind to DNA and nucleosomes and induce structural changes in the chromatin. They are important in chromatin domains dynamics and in regulating the expression of specific genes during development (77). Forkhead box (Fox) proteins are a superfamily of evolutionarily conserved transcriptional regulators, which control a wide spectrum of biological processes and are heavily used in developmental processes (78). Finally, GO enrichment analysis was performed on the set of genes associated with oCNEs and compared with the genes associated with rCNEs found in C. intestinalis. GO classifications for C. intestinalis were extracted from the Aniseed annotation database (see ‘Material and Methods’ section). Figure 4C shows the GO classes resulting specifically enriched in Ciona oCNEs: multicellular organismal development (adjusted P = 6.58E-06), sequence-specific DNA binding (adjusted P = 1.17E-05), transcription (adjusted P = 0.0007), cell differentiation (adjusted P = 0.0008), transcription factor activity (adjusted P = 0.008) and calcium ion binding (adjusted P = 0.017). Taken together, these results clearly indicate that the genes surrounding oCNEs as well as the transcription factors potentially binding oCNEs are significantly associated with genes involved in development and, more specifically, to morphogenesis and differentiation and these enrichments are significantly more specific than the ones related to rCNEs. GO enrichment analyses performed in mouse using either DAVID (58) or FATIGO (59) gave similar results when we compared oCNEs or vCNEs with rCNEs, but no enrichment was found comparing mouse oCNEs with vCNEs, suggesting that, in vertebrates, oCNEs and vCNEs belong to similar functional classes (data not shown).
Conservation in the Oikopleura and amphioxus genomes
To understand if oCNEs are retained in other sequenced model chordates, we searched for their presence in the Oikopleura dioica and amphioxus (Branchiostoma floridae) genomes. The pipeline we presented needs at least two sequenced and well annotated genomes belonging to the same class to analyze that specific class of organisms, and therefore we could not analyze them using our pipeline. Moreover, as we did not detect Ciona oCNEs sequences in the Oikopleura and amphioxus genomes by using Blastn, we decided to use information from all the organisms in oCNE blocks to build HMM (41) matrices from each oCNE multiple alignment based on the sequences conserved in all the analyzed species. We then scanned the Oikopleura and amphioxus genomes with the HMMs thus generated. This search yielded nine conserved elements in the Oikopleura and 13 in the amphioxus. Genes flanking and overlapping the elements thus discovered were annotated using Blast2GO (42) and considered as putative target genes. Annotations were manually checked against Ciona and mouse overlapping and flanking genes for each respective element (see Supplementary Table S6). Again, these elements resulted not to be located in the vicinity of evolutionarily related genes, although they appear to be associated to genes functionally related. Indeed, according to the Blast2GO classification, the top scoring biological processes represented in the associated genes are related to development and regulation (Supplementary Table S9). The number of conserved elements is small, and therefore, we cannot test for significance; however, the biological functions annotated by Blast2GO are remarkably similar to those enriched in the 183 original oCNE dataset. It is particularly interesting the presence of oCNEs in Oikopleura genomic loci containing putative orthologous for the Bmp and Lim homeobox genes. Indeed, these genes are also associated to oCNEs in vertebrates and ascidians. In the amphioxus, interesting genes associated to oCNEs are the putative homologs of Jumonji, Argonaute and Znf729. We conclude that only a small number of oCNEs is represented in the Oikopleura and amphioxus genomes, and these elements are not syntenic with ascidians or vertebrates but, again, their genomic loci result to be associated to functionally similar regions.
Finally, we checked if oCNE neighborhoods lacking synteny between Ciona and vertebrates could show evidences for common origin when taking into account the genome of amphioxus (i.e. are close together on the amphioxus genome). The results indicated that only three oCNEs could be associated to pairs of putative target Ciona/mouse genes localized on the same scaffold in the amphioxus genome (with a distance between them of 263 617, 2 448 029 and 898 799 bp). Randomizations showed that this result is not significant (1000 randomization produced an average of 6.4 associations with a standard deviation of 2.8). Interestingly, Hufton et al. reported the discovery of >1000 CNEs [defined as phylogenetically conserved non-coding elements (PCNEs)] among vertebrates or between vertebrates and amphioxus. Out of 183 oCNEs, 122 overlap the published set of vertebrate PCNEs, and 42 of them overlap the set conserved between vertebrates and amphioxus (data not shown). These PCNEs are conserved collinearly between vertebrates and amphioxus as a result of the methodology adopted. Our HMM approach could only map 4 out of these 42 oCNEs in Amphioxus, despite identifying some non-syntenic well-conserved oCNEs in this organism. This is probably because of the fact that the alignments by Hufton et al. were produced in a locus-specific way and with an estimated false-positive rate between 2 and 10% (based on two randomizations) as compared with our oCNE analysis, which was performed genome-wide at an FDR of 0.05%, and our HMM search, which was calibrated at high stringency, i.e. to yield only the original oCNE and close paralogs within the genome of origin, and thus only similar conserved elements in other genomes.
DISCUSSION
In this study, we developed a pipeline capable to identify, for the first time, CNEs spanning Olfactores genomes. Our analysis resulted in a set of 183 conserved non-coding blocks (oCNEs). We showed that oCNEs mainly overlap previously published UCEs and, although they are syntenic among vertebrates, they are found in non-syntenic loci in tunicates. Nevertheless, oCNEs are significantly associated with homeobox containing genes and genes involved in organismal development; also, they are significantly enriched for binding sites recognized by homeobox transcription factors. Such preponderance of homeobox genes associated to oCNEs, in the genomic context as well as in binding site predictions, could indicate a complex network of interactions which, during development, involve reciprocal regulatory relationship within this family of genes. The players of this network (usually defined as the ‘input’) appear to be the same genes in all the animal groups studied, but the regulatory interactions and the domains of expression encoded within these networks (often seen as the ‘output’), appears to be different in distant groups [see Cameron and Davidson (26) for a first proposal of the input/output theory]. Genomic fragments containing oCNEs act as domain-specific enhancers in developing embryos of sea squirt, mouse and zebrafish without retaining the same domain specificity between the groups. The cross-transgenesis experiments indicate that despite the long evolutionary distance separating the species under investigation, conserved oCNEs can retain enhancer effect in cross-species analysis and support the functional significance of these conserved sequences. While the specificity of enhancer effects is not fully retained, at least in the case of Ciona E1, anterior telencephalic activity is enriched in zebrafish, which is reminiscent to the zebrafish orthologous element resulting mostly specific to the anterior telencephalon. It is noteworthy that all elements tested appear to enhance the activity of a minimal promoter in fish as well as in Ciona. We chose to amplify larger fragments because the conservation between vertebrates and ascidians is limited to short sequences of ∼50 bps, which is unlikely to reflect the minimal functional unit. Consistent with this expectation oCNEs are anchored in longer regions conserved within each respective group. Thus oCNEs might represent a part of a specific regulatory element which, to work, would need support from sequence elements found in the flanking regions.
With constant refinements in the technologies capable to detect non-abundant transcripts, the observations that a large number of enhancers are also transcribed are tangibly increasing (49,54,70,71), suggesting that, at least in mammals, thousands of enhancers are transcribed. Interestingly, the oCNE dataset also shows significant overlap with the eRNA dataset. This enrichment is not a bias determined by the composition of vCNEs, indicating that oCNEs probably belong to a specific class of enhancers, which can also be transcribed. Furthermore, we indicate, by analyzing a large number of publicly available ENCODE datasets, that they are unlikely to transcribe short RNAs. It should be noted that for most eRNAs and UCEs analyzed, the full length and nature of the RNA molecules transcribed by these regions remains a largely unresolved question. Indeed, in this work, we demonstrated that oCNEs can effectively be transcribed even if we have not directly addressed the functional association between the transcription and the enhancer function. Further and more in-depth validations would need to be conducted to verify the extent, nature and specificity of oCNE expression.
It is important to specify that our results depend heavily on the methodology we used to identify oCNEs and that some homology relationships might be missing from current annotations. This raises the question whether oCNEs might be identified by mere chance. Our randomization-based filtering approach, which makes use of stringent FDR criteria indicating that <1 oCNE could be false, is pointing against this idea. On the contrary, given that other approaches were performed with more lenient statistical stringency, it is possible that we have missed some bona fide oCNEs, which might warrant future investigation. Similarly, our HMM search of oCNEs in other species such as amphioxus was performed stringently and might thus miss related and relevant CNEs, which could have diverged beyond the stringency of our approach. Manual curations of results and the significant overlaps with other relevant datasets such as eRNAs, UCEs, ENCODE data and the experimental evidence we produced are further proof of oCNEs’ biological relevance. A different and altogether more complex issue is to what extent oCNE-like elements could arise by convergent evolution. We do not have sufficient data to tackle appropriately this issue but we speculate that it could be unlikely if we consider a parsimonious scenario for the evolution of such elements. Finally, assembly errors could have generated some of the extensive non-orthologous shuffling we have observed. This is an important concern to address because many of these elements are found in gene deserts in which the lack of gene annotations can cause a higher proportion of assembly errors. However, in our pipeline this is unlikely because oCNEs originate from regionally conserved collinear regions in each group of organisms. Thus, to make an assembly error responsible for the generation of an oCNE, the same error should have occurred twice in the same collinear manner in at least two different organisms, which we believe to be highly improbable. It is possible, though, that assembly errors could cause some artificial duplication within the same genomic region of similar oCNEs, as seen in the duplication analysis within Ciona.
So, how can we explain the fact that such conserved regions are not conserved in a collinear fashion? The sequencing of new genomes could help us in shedding light on this point. Classically, CNEs are considered collinear regulatory regions conserved among lineages in terms of their position as well as in terms of their association to target genes whose sequences are conserved in their respective lineage but not among different lineages (6). oCNE elements do not appear to belong to this class, because they are well conserved among different lineages in terms of sequence while not being collinear. This is supported by the observation that, genes associated to oCNEs are significantly enriched for groups of genes in ascidians lacking clear vertebrate orthologs. Although they are not associated to the same potential target gene, they appear to maintain a clear preference for certain functional classes of genes. Despite a longer divergence time between amphioxus and vertebrates compared with Ciona and vertebrates, the conservation of synteny with vertebrates is greater for amphioxus than for Ciona (16). About 74% of amphioxus scaffolds show a significant presence of orthologs from the same human chromosome, while in Ciona, this proportion is ∼9%. The Oikopleura is the only known chordate genome to show no significant conservation of gene neighborhood with other chordates (79). Our sensitive pipeline has been able to find a single collinear element conserved between vertebrates and ascidians, and analysis in the amphioxus and Oikopleura genomes show the presence of a minority of non-collinear oCNEs. Such observations lead to speculation that these elements could have been present in a chordate ancestor and have been differentially lost or co-opted by different genes during the dramatic changes that brought to the differentiation of the chordate lineages. Particularly intriguing are the findings that early vertebrate whole genome duplications were predated by a period of intense genome rearrangement (80) and that, in addition to whole genome duplications, segmental and single-gene duplications shaped the genomes of extant vertebrates (81). A mechanism that can be taken into account for the generation of non-syntenic conserved elements in such a scenario can be accounted by partial rediploidization following local- or whole-genome duplications, which, in vertebrates, have been demonstrated to be at the basis of the retention of regulatory regions deriving by exons of lost duplicated genes (82). We screened oCNEs for specific overlap to cDNAs and single whole genomes to understand if they could result from rediploidization events but no such results were found. A different scenario to justify the unexpected variability observed in oCNEs, in terms of their location as well as of their expression domains, could be addressed to several peculiarities of the tunicate genomes. First, tunicate genomes are highly re-arranged and experienced extensive gene losses as compared with the non-duplicated early chordate karyotype. Putnam et al. (16) have identified 8437 gene families with members in amphioxus and other chordates that represent the descendants of genes found in the last common chordate ancestor. They also estimate that subsequent family expansions have generated ∼13 000 genes in amphioxus and vertebrates and ∼7000 in C. intestinalis. The lower number of tunicate genes is believed to be due to an extensive gene loss, which caused ∼2000 genes to be lost (83). The families of transcription factors that have lost the highest proportion of orthologs in tunicates are the homeobox, high-mobility group (HMG) and helix-loop-helix (HLH) [see (84) and its supplementary for a complete list of references and genes]. Intriguingly, these are the same gene families, which appear to be enriched in oCNEs. Hence, another mechanism that could justify the shuffling of oCNEs is that it could be associated with tunicate-specific gene losses and subsequent genomic rearrangements. If oCNEs were present in the chordate ancestor, they were probably co-opted by non-homologous but functionally similar genes, in tunicates, after the loss or the extreme derivation of the originally associated ones. A recent study shows that the roles of some Hox genes are not homologous to their vertebrate counterparts during Ciona larval development, further supporting the evidence that functional homology between tunicate and vertebrate genes is not always observed (85). In addition, gene expression dynamics of orthologous genes between developing C. intestinalis and D. rerio embryos were shown to be broadly divergent (18). Further support along this line is given by the fact that Hox and ParaHox genes in C. intestinalis are not organized in clusters, do not retain spatial and temporal developmental gene expression collinearity and contain transposable elements in their genomic loci (86,87). To us, this level of genomic and proteomic variability, unique to tunicates, could have occurred in concomitance with a peculiar rewiring of regulatory modules aimed at maintaining the chordate body plan. A final mechanism, which could be used to justify the shuffling of such elements, derives by the observation that they can be actively transcribed. Indeed, given that any type of RNA can serve as template for reverse transcription (88), the fact that oCNEs are transcribed suggests that they could have also been retrotransposed in new locations by the same mechanism involved, for example, in the creation of pseudogenes.
We thus propose that these conserved elements were shuffled either in an active (retroposition) or passive (rearrangements, rediploidization, derivation) fashion and co-opted by similar genes. The necessity for them to be shuffled is likely to have arisen during evolution of chordates to accommodate the coding variability, extensive gene gains and losses, genomic re-arrangements and the establishment of different developmental times to maintain a similar body plan for all the chordates.
Unfortunately, the impossibility to find genomic relics of shuffling events related to oCNEs makes it extremely difficult to demonstrate which mechanism took the leading part in their evolution. We searched for any such relics, but did not find any enrichment for specific k-mers, repeats, pseudogenes, chromatin interaction features in the genomic intervals overlapping or surrounding oCNEs, nor did oCNEs result to be derived by lost coding or non-coding exons (data not shown). When more chordate genomes and transcriptomes will be sequenced, it will be possible to answer more in-depth questions related to the evolutionary history of chordate regulatory elements. Nevertheless, the analysis herein presented is the first report of a sensitive and stringent pipeline that could be adopted to look for conservation of non-coding elements in distant and derivate groups of genomes as soon as new genomes are published. Moreover, the data provided constitute the first collection of non-coding elements conserved among Olfactores and represent an extremely valuable resource for future comparative, evolutionary and developmental studies. Finally we provide initial evidence that oCNEs can act as enhancers (also in cross-transgenesis) and are transcribed in different organisms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–9 and Supplementary Figures 1–5.
FUNDING
Seventh Framework Program of the European Commission (DOPAMINET and ZF Health projects) [223744 to E.S., F.M., P.L. and R.S., 242048 to F.M.]; the Italian ‘Progetto Bandiera RITMARE’ (to R.S.) R.S. has been supported by a Marie-Curie Research Early Stage Training Fellowship to develop part of the project at EBI. Funding for open access charge: RITMARE.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to acknowledge John Mattick for providing the extended set of UCEs data; Michael Greenberg, Tae-Kyung Kim and Jesse Gray for clarifications about the eRNAs dataset; Paolo Sordino and Salvatore D'Aniello for critical reading of the manuscript; Michael Brudno, Stefano Gustincich, Piero Carninci, Mariella Ferrante, Graziano Fiorito, Fiona McNish, Daniel Zerbino, Alison Meynert, Benedict Paten and Wolfgang Huber for helpful discussions and support. A special acknowledgement also goes to the late Parvesh Mahtani, who shared our enthusiasm for this project. We would like to thank two anonymous referees for their invaluable suggestions.
REFERENCES
- 1.Dermitzakis ET, Reymond A, Lyle R, Scamuffa N, Ucla C, Deutsch S, Stevenson BJ, Flegel V, Bucher P, Jongeneel CV, et al. Numerous potentially functional but non-genic conserved sequences on human chromosome 21. Nature. 2002;420:578–582. doi: 10.1038/nature01251. [DOI] [PubMed] [Google Scholar]
- 2.Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 3.Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plessy C, Dickmeis T, Chalmel F, Strähle U. Enhancer sequence conservation between vertebrates is favoured in developmental regulator genes. Trends Genet. 2005;21:207–210. doi: 10.1016/j.tig.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Vavouri T, Lehner B. Conserved noncoding elements and the evolution of animal body plans. Bioessays. 2009;31:727–735. doi: 10.1002/bies.200900014. [DOI] [PubMed] [Google Scholar]
- 6.Vavouri T, Walter K, Gilks W, Lehner B, Elgar G. Parallel evolution of conserved non-coding elements that target a common set of developmental regulatory genes from worms to humans. Genome Biol. 2007;8:R15. doi: 10.1186/gb-2007-8-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kermekchiev M, Pettersson M, Matthias P, Schaffner W. Every enhancer works with every promoter for all the combinations tested: could new regulatory pathways evolve by enhancer shuffling? Gene Expr. 1991;1:71–81. [PMC free article] [PubMed] [Google Scholar]
- 8.Kirchhamer CV, Yuh CH, Davidson EH. Modular cis-regulatory organization of developmentally expressed genes: two genes transcribed territorially in the sea urchin embryo, and additional examples. Proc. Natl Acad. Sci. USA. 1996;93:9322–9328. doi: 10.1073/pnas.93.18.9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Visel A, Akiyama JA, Shoukry M, Afzal V, Rubin EM, Pennacchio LA. Functional autonomy of distant-acting human enhancers. Genomics. 2009;93:509–513. doi: 10.1016/j.ygeno.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hufton AL, Mathia S, Braun H, Georgi U, Lehrach H, Vingron M, Poustka AJ, Panopoulou G. Deeply conserved chordate noncoding sequences preserve genome synteny but do not drive gene duplicate retention. Genome Res. 2009;19:2036–2051. doi: 10.1101/gr.093237.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aparicio S, Morrison A, Gould A, Gilthorpe J, Chaudhuri C, Rigby P, Krumlauf R, Brenner S. Detecting conserved regulatory elements with the model genome of the Japanese puffer fish, Fugu rubripes. Proc. Natl Acad. Sci. USA. 1995;92:1684–1688. doi: 10.1073/pnas.92.5.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Royo JL, Maeso I, Irimia M, Gao F, Peter IS, Lopes CS, D’Aniello S, Casares F, Davidson EH, Garcia-Fernández J, et al. Transphyletic conservation of developmental regulatory state in animal evolution. Proc. Natl Acad. Sci. USA. 2011;108:14186–14191. doi: 10.1073/pnas.1109037108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manzanares M, Wada H, Itasaki N, Trainor PA, Krumlauf R, Holland PW. Conservation and elaboration of Hox gene regulation during evolution of the vertebrate head. Nature. 2000;408:854–857. doi: 10.1038/35048570. [DOI] [PubMed] [Google Scholar]
- 14.Natale A, Sims C, Chiusano ML, Amoroso A, D’Aniello E, Fucci L, Krumlauf R, Branno M, Locascio A. Evolution of anterior Hox regulatory elements among chordates. BMC Evol. Biol. 2011;11:330. doi: 10.1186/1471-2148-11-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holland LZ, Albalat R, Azumi K, Benito-Gutiérrez E, Blow MJ, Bronner-Fraser M, Brunet F, Butts T, Candiani S, Dishaw LJ, et al. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 2008;18:1100–1111. doi: 10.1101/gr.073676.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Putnam NH, Butts T, Ferrier DEK, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu J-K, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- 17.Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- 18.Sobral D, Tassy O, Lemaire P. Highly divergent gene expression programs can lead to similar chordate larval body plans. Curr. Biol. 2009;19:2014–2019. doi: 10.1016/j.cub.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 19.Lemaire P, Smith WC, Nishida H. Ascidians and the plasticity of the chordate developmental program. Curr. Biol. 2008;18:R620–R631. doi: 10.1016/j.cub.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Britten RJ, Davidson EH. Gene regulation for higher cells: a theory. Science. 1969;165:349–357. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- 21.Zuckerkandl E. Molecular pathways to parallel evolution: I. Gene nexuses and their morphological correlates. J. Mol. Evol. 1994;39:661–678. doi: 10.1007/BF00160412. [DOI] [PubMed] [Google Scholar]
- 22.García-Bellido A. Symmetries throughout organic evolution. Proc. Natl Acad. Sci. USA. 1996;93:14229–14232. doi: 10.1073/pnas.93.25.14229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsong AE, Miller MG, Raisner RM, Johnson AD. Evolution of a combinatorial transcriptional circuit: a case study in yeasts. Cell. 2003;115:389–399. doi: 10.1016/s0092-8674(03)00885-7. [DOI] [PubMed] [Google Scholar]
- 24.Ihmels J, Bergmann S, Gerami-Nejad M, Yanai I, McClellan M, Berman J, Barkai N. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science. 2005;309:938–940. doi: 10.1126/science.1113833. [DOI] [PubMed] [Google Scholar]
- 25.Prud’homme B, Gompel N, Rokas A, Kassner VA, Williams TM, Yeh S-D, True JR, Carroll SB. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature. 2006;440:1050–1053. doi: 10.1038/nature04597. [DOI] [PubMed] [Google Scholar]
- 26.Cameron RA, Davidson EH. Flexibility of transcription factor target site position in conserved cis-regulatory modules. Dev. Biol. 2009;336:122–135. doi: 10.1016/j.ydbio.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 27.Oda-Ishii I, Bertrand V, Matsuo I, Lemaire P, Saiga H. Making very similar embryos with divergent genomes: conservation of regulatory mechanisms of Otx between the ascidians Halocynthia roretzi and Ciona intestinalis. Development. 2005;132:1663–1674. doi: 10.1242/dev.01707. [DOI] [PubMed] [Google Scholar]
- 28.Lowe CB, Bejerano G, Haussler D. Thousands of human mobile element fragments undergo strong purifying selection near developmental genes. Proc. Natl Acad. Sci. USA. 2007;104:8005–8010. doi: 10.1073/pnas.0611223104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanges R, Kalmar E, Claudiani P, D’Amato M, Muller F, Stupka E. Shuffling of cis-regulatory elements is a pervasive feature of the vertebrate lineage. Genome Biol. 2006;7:R56. doi: 10.1186/gb-2006-7-7-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chuzhanova NA, Krawczak M, Nemytikova LA, Gusev VD, Cooper DN. Promoter shuffling has occurred during the evolution of the vertebrate growth hormone gene. Gene. 2000;254:9–18. doi: 10.1016/s0378-1119(00)00308-5. [DOI] [PubMed] [Google Scholar]
- 31.Ueda M, Arimura S, Yamamoto MP, Takaiwa F, Tsutsumi N, Kadowaki K. Promoter shuffling at a nuclear gene for mitochondrial RPL27. Involvement of interchromosome and subsequent intrachromosome recombinations. Plant Physiol. 2006;141:702–710. doi: 10.1104/pp.105.075564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D. Evolution’s cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl Acad. Sci. USA. 2003;100:11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Margulies EH, Cooper GM, Asimenos G, Thomas DJ, Dewey CN, Siepel A, Birney E, Keefe D, Schwartz AS, Hou M, et al. Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome. Genome Res. 2007;17:760–774. doi: 10.1101/gr.6034307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hubbard TJP, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JGR, Korf I, Lapp H, et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, Green ED, Sidow A, Batzoglou S. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 2003;13:721–731. doi: 10.1101/gr.926603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- 39.Brudno M, Chapman M, Göttgens B, Batzoglou S, Morgenstern B. Fast and sensitive multiple alignment of large genomic sequences. BMC Bioinformatics. 2003;4:66. doi: 10.1186/1471-2105-4-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tassy O, Dauga D, Daian F, Sobral D, Robin F, Khoueiry P, Salgado D, Fox V, Caillol D, Schiappa R, et al. The ANISEED database: digital representation, formalization, and elucidation of a chordate developmental program. Genome Res. 2010;20:1459–1468. doi: 10.1101/gr.108175.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 42.Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roure A, Rothbächer U, Robin F, Kalmar E, Ferone G, Lamy C, Missero C, Mueller F, Lemaire P. A multicassette Gateway vector set for high throughput and comparative analyses in ciona and vertebrate embryos. PLoS One. 2007;2:e916. doi: 10.1371/journal.pone.0000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gehrig J, Reischl M, Kalmár E, Ferg M, Hadzhiev Y, Zaucker A, Song C, Schindler S, Liebel U, Müller F. Automated high-throughput mapping of promoter-enhancer interactions in zebrafish embryos. Nat. Methods. 2009;6:911–916. doi: 10.1038/nmeth.1396. [DOI] [PubMed] [Google Scholar]
- 46.Kawakami K. Transgenesis and gene trap methods in zebrafish by using the Tol2 transposable element. Methods Cell Biol. 2004;77:201–222. doi: 10.1016/s0091-679x(04)77011-9. [DOI] [PubMed] [Google Scholar]
- 47.Stephen S, Pheasant M, Makunin IV, Mattick JS. Large-scale appearance of ultraconserved elements in tetrapod genomes and slowdown of the molecular clock. Mol. Biol. Evol. 2008;25:402–408. doi: 10.1093/molbev/msm268. [DOI] [PubMed] [Google Scholar]
- 48.Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 49.Kim T-K, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 51.Alfano C, Teresa Russo M, Spagnuolo A. Developmental expression and transcriptional regulation of Ci-Pans, a novel neural marker gene of the ascidian, Ciona intestinalis. Gene. 2007;406:36–41. doi: 10.1016/j.gene.2007.05.026. [DOI] [PubMed] [Google Scholar]
- 52.Karali M, Peluso I, Marigo V, Banfi S. Identification and characterization of microRNAs expressed in the mouse eye. Invest. Ophthalmol. Vis. Sci. 2007;48:509–515. doi: 10.1167/iovs.06-0866. [DOI] [PubMed] [Google Scholar]
- 53.Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M, Sloan CA, Rosenbloom KR, Roe G, Rhead B, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2012;41:D64–D69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bryne JC, Valen E, Tang M-HE, Marstrand T, Winther O, Da Piedade I, Krogh A, Lenhard B, Sandelin A. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lenhard B, Wasserman WW. TFBS: computational framework for transcription factor binding site analysis. Bioinformatics. 2002;18:1135–1136. doi: 10.1093/bioinformatics/18.8.1135. [DOI] [PubMed] [Google Scholar]
- 58.Dennis G, Sherman B, Hosack D, Yang J, Gao W, Lane H, Lempicki R. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:R60. [PubMed] [Google Scholar]
- 59.Al-Shahrour F, Diaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 60.Brudno M, Malde S, Poliakov A, Do CB, Couronne O, Dubchak I, Batzoglou S. Glocal alignment: finding rearrangements during alignment. Bioinformatics. 2003;19:i54–i62. doi: 10.1093/bioinformatics/btg1005. [DOI] [PubMed] [Google Scholar]
- 61.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- 62.Vavouri T, McEwen GK, Woolfe A, Gilks WR, Elgar G. Defining a genomic radius for long-range enhancer action: duplicated conserved non-coding elements hold the key. Trends Genet. 2006;22:5–10. doi: 10.1016/j.tig.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 63.Kikuta H, Laplante M, Navratilova P, Komisarczuk AZ, Engström PG, Fredman D, Akalin A, Caccamo M, Sealy I, Howe K, et al. Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res. 2007;17:545–555. doi: 10.1101/gr.6086307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miura Y, Tam T, Ido A, Morinaga T, Miki T, Hashimoto T, Tamaoki T. Cloning and characterization of an ATBF1 isoform that expresses in a neuronal differentiation-dependent manner. J. Biol. Chem. 1995;270:26840–26848. doi: 10.1074/jbc.270.45.26840. [DOI] [PubMed] [Google Scholar]
- 65.Jung C-G, Kim H-J, Kawaguchi M, Khanna KK, Hida H, Asai K, Nishino H, Miura Y. Homeotic factor ATBF1 induces the cell cycle arrest associated with neuronal differentiation. Development. 2005;132:5137–5145. doi: 10.1242/dev.02098. [DOI] [PubMed] [Google Scholar]
- 66.Miwata K, Chiba T, Horii R, Yamada L, Kubo A, Miyamura D, Satoh N, Satou Y. Systematic analysis of embryonic expression profiles of zinc finger genes in Ciona intestinalis. Dev. Biol. 2006;292:546–554. doi: 10.1016/j.ydbio.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 67.Sandberg M, Källström M, Muhr J. Sox21 promotes the progression of vertebrate neurogenesis. Nat. Neurosci. 2005;8:995–1001. doi: 10.1038/nn1493. [DOI] [PubMed] [Google Scholar]
- 68.Basch ML, Bronner-Fraser M, García-Castro MI. Specification of the neural crest occurs during gastrulation and requires Pax7. Nature. 2006;441:218–222. doi: 10.1038/nature04684. [DOI] [PubMed] [Google Scholar]
- 69.Corbo JC, Levine M, Zeller RW. Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development. 1997;124:589–602. doi: 10.1242/dev.124.3.589. [DOI] [PubMed] [Google Scholar]
- 70.Licastro D, Gennarino VA, Petrera F, Sanges R, Banfi S, Stupka E. Promiscuity of enhancer, coding and non-coding transcription functions in ultraconserved elements. BMC Genomics. 2010;11:151. doi: 10.1186/1471-2164-11-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei C-L, Natoli G. A large fraction of extragenic RNA Pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, Fan L, Sandelin A, Rinn JL, Regev A, et al. Systematic identification of long non-coding RNAs expressed during zebrafish embryogenesis. Genome Res. 2011;22:577–591. doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Christensen KL, Patrick AN, McCoy EL, Ford HL. The six family of homeobox genes in development and cancer. Adv. Cancer Res. 2008;101:93–126. doi: 10.1016/S0065-230X(08)00405-3. [DOI] [PubMed] [Google Scholar]
- 76.Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 77.Bianchi ME, Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Curr. Opin. Genet. Dev. 2005;15:496–506. doi: 10.1016/j.gde.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 78.Hannenhalli S, Kaestner KH. The evolution of Fox genes and their role in development and disease. Nat. Rev. Genet. 2009;10:233–240. doi: 10.1038/nrg2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Denoeud F, Henriet S, Mungpakdee S, Aury J-M, Da Silva C, Brinkmann H, Mikhaleva J, Olsen LC, Jubin C, Canestro C, et al. Plasticity of animal genome architecture unmasked by rapid evolution of a pelagic tunicate. Science. 2010;330:1381–1385. doi: 10.1126/science.1194167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hufton AL, Groth D, Vingron M, Lehrach H, Poustka AJ, Panopoulou G. Early vertebrate whole genome duplications were predated by a period of intense genome rearrangement. Genome Res. 2008;18:1582–1591. doi: 10.1101/gr.080119.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Olinski RP, Lundin L-G, Hallböök F. Conserved synteny between the Ciona genome and human paralogons identifies large duplication events in the molecular evolution of the insulin-relaxin gene family. Mol. Biol. Evol. 2006;23:10–22. doi: 10.1093/molbev/msj002. [DOI] [PubMed] [Google Scholar]
- 82.Dong X, Navratilova P, Fredman D, Drivenes Ø, Becker TS, Lenhard B. Exonic remnants of whole-genome duplication reveal cis-regulatory function of coding exons. Nucleic Acids Res. 2010;38:1071–1085. doi: 10.1093/nar/gkp1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hughes AL, Friedman R. Loss of ancestral genes in the genomic evolution of Ciona intestinalis. Evol. Dev. 2005;7:196–200. doi: 10.1111/j.1525-142X.2005.05022.x. [DOI] [PubMed] [Google Scholar]
- 84.Imai KS, Hino K, Yagi K, Satoh N, Satou Y. Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: towards a comprehensive understanding of gene networks. Development. 2004;131:4047–4058. doi: 10.1242/dev.01270. [DOI] [PubMed] [Google Scholar]
- 85.Ikuta T, Satoh N, Saiga H. Limited functions of Hox genes in the larval development of the ascidian Ciona intestinalis. Development. 2010;137:1505–1513. doi: 10.1242/dev.046938. [DOI] [PubMed] [Google Scholar]
- 86.Ferrier DEK, Holland PWH. Ciona intestinalis ParaHox genes: evolution of Hox/ParaHox cluster integrity, developmental mode, and temporal colinearity. Mol. Phylogenet. Evol. 2002;24:412–417. doi: 10.1016/s1055-7903(02)00204-x. [DOI] [PubMed] [Google Scholar]
- 87.Ikuta T, Yoshida N, Satoh N, Saiga H. Ciona intestinalis Hox gene cluster: its dispersed structure and residual colinear expression in development. Proc. Natl Acad. Sci. USA. 2004;101:15118–15123. doi: 10.1073/pnas.0401389101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brosius J. RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene. 1999;238:115–134. doi: 10.1016/s0378-1119(99)00227-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.